Introduction
Breast cancer is the fifth most common cause of
cancer-related mortalities and the second most common form of
non-skin-associated cancer worldwide (1). At present, the most important prognostic
factors used to guide decisions regarding adjuvant systemic
treatment are tumor size, nodal status, tumor clinical stage,
hormone receptor [progesterone receptor (PGR) and estrogen receptor
(ER)], human epidermal growth factor receptor 2 (HER2) status
(2), and histological grade. Various
other clinicopathological factors, including proliferation index
and novel molecular markers, have been investigated to improve the
prediction of clinical outcome (3,4). Despite
improvements in risk stratification, the current prognostic factors
exhibit moderate accuracy in classifying breast tumors according to
their clinical behaviors. In breast cancer, axillary lymph nodes
are typically the initial site of metastasis (5). The presence of lymph node metastasis
predicts the development of distant metastases and is considered
one of the most informative prognostic factors when evaluating
patients with breast cancer (6–8).
Several studies have identified correlations between
clinicopathological parameters of patients with breast cancer and a
high risk of developing lymph node metastases. Among the parameters
most significantly correlated with lymph node involvement are
histological grade, tumor size and age (9,10).
Furthermore, number and proportion of evaluated sentinel lymph node
biopsies (SLNBs) have been correlated with metastases in the
axillary lymph nodes (11–13). An SLN is defined as one of the first
nodes to collect lymphatic fluid from a malignant tumor and
malignant cells (1). The result of
the SLNB indicates whether complete axillary lymph node dissection
(ALND) should be performed (14),
which allows complete evaluation of lymph nodes. However, ALND is
associated with significant morbidity and is not associated with a
significant increase in patient survival (15). Prognostic and predictive tools are
required to more accurately select patients for lymph node
dissection and spare large numbers of patients undergoing this
procedure when it is not necessary.
The advent of polymerase chain reaction (PCR)-based
diagnostic methods, in particular reverse transcription-
quantitative PCR (RT-qPCR), made the detection of SLN metastasis
(16) and axillary lymph node
involvement in breast cancer possible, which resulted in the
identification of several molecular markers with significant
potential prognosis in relation to risk of axillary and systemic
metastases (17–20). The purpose of the present study was to
determine whether genes that had been previously described as
prognostic markers in breast cancer (21) are also predictors of lymph node
involvement in breast carcinoma. The expression of 10 genes
[protein phosphatase Mg2+/Mn2+ dependent 1D (PPM1D),
β-1,3-N-acetylglucosaminyltransferase (B3GNT7), neural precursor
cell expressed developmentally down-regulated 9 (NEDD9), prohibitin
(PHB), phosphoinositide-3-kinase regulatory subunit 5 (PIK3R5),
phosphatidylinositol-5-phosphate 4-kinase type IIα (PIP4K2A),
TRF1-interacting ankyrin-related ADP-ribose polymerase 2 (TNKS2),
BCL2 associated agonist of cell death (BAD), G2 and S-phase
expressed 1 (GTSE1) and PAX interacting protein 1 (PAXIP1)] was
analyzed by RT-qPCR in the primary tumor tissues of patients with
and without lymph node involvement, in addition to primary tumors
and lymph node metastases of the same patients in order to
determine whether these genes have predictive power in relation to
risk of axillary metastases.
Materials and methods
Samples and patients
A total of 50 primary tumor samples were collected
from 49 patients that underwent segmental resection or mastectomy
(1 patient presented with bilateral tumors); 41 samples were
obtained from Hospital São Francisco de Assis (Jacareí, Brazil) and
9 were from other hospitals, including Hospital Antoninho da Rocha
Marmo, Santos Dumont Hospital and Hospital Pio XII (São José dos
Campos, Brazil). Frozen primary breast tumor tissues were collected
from women with negative (n=27) and positive (n=23) lymph node
status, and, of the positive lymph node cases, primary breast
tumors and paired lymph node metastases tissues were acquired from
11 patients. Written informed consent was obtained from all
patients during the collection period, and the study was reviewed
and approved by the Research Ethics Committee of the University of
Taubaté (Taubaté, Brazil) (CEP 554/11). Table I provides a summary of the
clinicopathological data from the 50 tumor samples from 49 patients
with breast carcinoma related to the status of axillary lymph
nodes.
 | Table I.Clinicopathological data of 50 tumor
samplesa from 49
patients with primary breast carcinoma associated with the status
of axillary lymph nodes. |
Table I.
Clinicopathological data of 50 tumor
samplesa from 49
patients with primary breast carcinoma associated with the status
of axillary lymph nodes.
| Features | n | Node-positive n
(%) | Node-negative n
(%) |
|---|
| Age, years | 49 |
|
|
|
≤50 |
| 9 (18) | 5 (10) |
|
>50 |
| 14 (29) | 21 (43) |
| Histology | 50 |
|
|
|
Invasive ductal |
| 21 (42) | 21 (42) |
| Others
(in situ, lobular) |
| 2 (4) | 6 (8) |
| Grade | 46b |
|
|
|
1+2 |
| 13 (28) | 14 (30) |
| 3 |
| 10 (22) | 9 (20) |
| ER status | 35b |
|
|
|
Negative |
| 2 (6) | 5 (14) |
|
Positive |
| 11 (31) | 17 (49) |
| PGR status | 45b |
|
|
|
Negative |
| 4 (9) | 6 (13) |
|
Positive |
| 9 (20) | 26 (58) |
| HER2 status | 32b |
|
|
|
HER2- |
| 4 (13) | 12 (38) |
|
HER+ |
| 8 (25) | 8 (25) |
| Ki67 status, % | 27b |
|
|
|
>25 |
| 4 (15) | 5 (19) |
|
≤25 |
| 5 (19) | 13 (48) |
| T stage | 48b |
|
|
| T1 +
T2 |
| 19 (40) | 24 (50) |
| T3 +
T4 |
| 3 (6) | 2 (4) |
| Tumor size, cm | 47b |
|
|
| ≤2 |
| 10 (21) | 11 (23) |
|
>2 |
| 13 (28) | 13 (28) |
Immediately after surgery, tumor tissue samples were
frozen and stored at −80°C. To ensure consistency, diagnosis of
every specimen was made by a single breast pathologist (Center for
Diagnostic Medicine, Pathology and Cytology, São José dos Campos,
Brazil). Whenever necessary, tissue samples were macrodissected
with a scalpel to guarantee that only sections comprised of ≥90%
tumor cells were used for RNA isolation and subsequent gene
expression analysis. A total of 5 non-tumor breast tissue samples
from patients undergoing mammary reduction, which had been
histopathologically confirmed as healthy, were used as controls.
Histopathological classification was performed according to the
International Classification of Disease for Oncology from the World
Health Organization (22), and the
clinical stage was determined according to the Union for
International Cancer Control Tumor-Node-Metastasis (23) classification. The malignancy of
carcinoma infiltration was scored according to the Bloom and
Richardson grading system (24).
Total RNA isolation, quantification
and synthesis of cDNA
Total RNA was extracted from 50 macrodissected
primary tumor samples, 11 lymph nodes and 5 healthy breast tissues
using the RNeasy® Lipid Tissue Mini kit (Qiagen, São
Paulo, Brazil) according to the manufacturer's protocol. RNA
samples were purified with 0.1% acetate-ethanol, resuspended in
RNase-free ultra-pure water and stored at −80°C until use. RNA
quality was analyzed by 1% agarose gel electrophoresis, and the
concentration and quality was measured using a NanoDrop-1000
spectrophotometer v.3.7 (Labtrade, Sao Paulo, Brazil). To avoid DNA
contamination, RNA samples were treated with the RNase-Free DNase
Set (Qiagen) according to the manufacturer's protocol.
cDNA synthesis reactions were carried out in a
Peltier Thermal Cycler (MJ-96G; Biocycle Co., Ltd, Hangzhou,
China). Total RNA (1 ug) from each sample was reverse transcribed
in a 20 µl final volume containing 10 µl 2x first-strand buffer (10
mm mgcl2, and 1 mmde per dntp), 0.5 µl Oligo
(dT)20, 0.5 µl random primers, 1 µl annealing buffer and
2 µl SuperScript™ III Reverse Transcriptase enzyme (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA). RT was carried
out for 50 min at 50°C, 10 min at 25°C, followed by 50 min at 50°C.
The reaction mixture was subsequently inactivated for 5 min at
85°C.
Evaluation of transcript expression by
RT-qPCR
RT-qPCR reactions for the 10 study genes and the
reference gene (MRLP19) were carried out in duplicate on an ABI
Prism 7000 Sequence Detection system (Thermo Fisher Scientific,
Inc.) using Platinum®SYBR® Green qPCR
SuperMix-UDG (Applied Biosystems; Thermo Fisher Scientific, Inc.)
in a total volume of 10 µl according to manufacturer's protocol.
Primers were designed using Primer Express software (v3.0; Applied
Biosystems; Thermo Fisher Scientific, Inc.) and the sequences are
presented in Table II. In order to
avoid amplification of contaminating genomic DNA, the primers were
placed at the junction between the two exons or in a different
exon. RT was carried out for 1 cycle of 95°C for 10 min, followed
by 40 cycles of 15 sec at 95°C and 1 min at 60°C. A dissociation
curve was included in all experiments. For all primers,
amplification curves were constructed with serial dilutions of
healthy breast and breast carcinoma cDNA (100, 20, 4, 0.8 and 0.16
ng/µl). Standard curves of the targets and reference genes
demonstrated similar amplification efficiencies (>90%).
Quantitative data was analyzed using the Sequence Detection system
software (v1.0; Applied Biosystems; Thermo Fisher Scientific,
Inc.). The details of the gene-specific RT-qPCR assays are
presented in Table II.
 | Table II.Details of the gene-specific primers
used in reverse transcription-quantitative polymerase chain
reaction assays. |
Table II.
Details of the gene-specific primers
used in reverse transcription-quantitative polymerase chain
reaction assays.
| Gene | Gene name | Function | Size of PCR
product, bp | Primer
sequences |
|---|
| PPM1D | Protein
phosphatase, Mg2+/Mn2+ dependent 1D | Regulators of cell
stress response pathways | 81 | Forward:
5′-TTGGAATATGATTCCACCACAAGA-3′ |
|
|
|
|
| Reverse:
5′-CCATGCTCACCCATCAGGTATTT-3′ |
| B3GNT7 |
β-1,3-N-acetylglucosaminyltransferase | Cell migration,
cell cycle and survival | 81 | Forward:
5′-CCACGTCCCCTTCATTTTCA-3′ |
|
|
|
|
| Reverse:
5′-TGCCGGTCAGCCAGAAATT-3′ |
| NEDD9 | Neural precursor
cell expressed developmentally down-regulated protein 9 | Regulation of
invasion, apoptosis and cell cycle | 81 | Forward:
5′-AGGCCCCTGACTGTAGCAGC-3′ |
|
|
|
|
| Reverse:
5′-CCTTACCCTGTAGGTGGACGTAATC-3′ |
| PHB | Prohibitin | Negative regulator
of proliferation and tumor suppressor | 81 | Forward:
5′-CCAGCATCGGAGAGGACTATGAT-3′ |
|
|
|
|
| Reverse:
5′-CAAAGCGAGCCACCACTGAC-3′ |
| PIK3R5 |
Phosphoinositide-3-kinase regulatory
subunit 5 | Cell growth,
proliferation and differentiation | 79 | Forward:
5′-ACGCTACGTGTTGTGGTCTTTG-3′ |
|
|
|
|
| Reverse:
5′-CCAGCCGCCGAAGGTT-3′ |
| PIP4K2A | Phosphatidylinosito
l-5-phosphate 4-kinase type IIα | Regulation of
secretion, proliferation and differentiation | 65 | Forward:
5′-GGCCGAAATGCACAACATC-3′ |
|
|
|
|
| Reverse:
5′-GGGTGATCCCATGACATTCC-3′ |
| TNKS2 | TRF1-interacting
ankyrin-related ADP-ribose polymerase 2 | Promotion of
increased telomere length | 81 | Forward:
5′-AAGATACACTCACCGGAGAAAAGAAG-3′ |
|
|
|
|
| Reverse:
5′-GGAGACCCATGAAATAGCATTCG-3′ |
| BAD | BCL2-antagonist of
cell death protein | Regulators of
programmed cell death | 81 | Forward:
5′-CTTTAAGAAGGGACTTCCTCGCC-3′ |
|
|
|
|
| Reverse:
5′-AAGACTCGCGTCCAGCTGG-3′ |
| GTSE1 | G2 and S
phase-expressed protein 1 | Regulator of the
DNA damage | 63 | Forward:
5′-CGGAGAAGCCCAAGAAAGAGAT-3′ |
|
|
|
|
| Reverse:
5′-CCTTCTCAGCTGGGATTTTTGT-3′ |
| PAX1P1 | PAX transcription
activation domain interacting protein 1 | Genome stability
and progression mitosis | 81 | Forward:
5′-AATGGCTTATTTGGCAGGTGC-3′ |
|
|
|
|
| Reverse:
5′-CCAGTTGGTTCTTTACAGATGAGGACT-3′ |
| MRPL19 | Mitochondrial
ribosomal protein L19 | Mammalian
mitochondrial ribosomal | 70 | Forward:
5′-CAGAGATCAGGAAGAGGACTTGGA-3′ |
|
|
|
|
| Reverse:
5′-TCTCGACACCTTGTCCTTCGA-3′ |
Measurements of gene expression were calculated
using the relative ΔΔCq method, in which the mean Cq value of each
target gene in each sample is subtracted from the mean Cq value of
the reference gene (25). The
transcripts of housekeeping genes MRLP19, PPIA, and GAPDH, were
quantified as previously described (26), and the MRLP19 gene was selected as an
endogenous control, which provided increased accuracy and
resolution in the quantitation of gene expression data,
facilitating the detection of smaller changes in gene expression
than otherwise possible. Normalized expression levels of target
gene tumor samples were expressed in fold-change relative to their
abundance in a pool of non-tumor breast tissue control samples,
which was calculated as follows: 2−(∆Cq test sample −
∆Cq control sample).
Statistical analysis
Statistical analyses comparing clinicopathological
characteristics (including age, histological type, histological
grade, ER, PGR, HER2 and Ki-67 status, and tumor stage and size)
with the presence or absence of lymph node involvement were
performed using IBM SPSS v20.0 and the χ2 test. P≤0.05
was considered to indicate a statistically significant
difference.
For the analysis of RT-qPCR, the median
2−∆∆Cq values for each analyzed group were compared
using Fisher's exact test [performed using GraphPad InStat software
v5.0 (GraphPad Software, Inc., La Jolla, CA, USA)] to determine
whether the relative fold change was significantly different
between the two groups. P≤0.05 was considered to indicate a
statistically significant difference.
Results
Clinicopathological
characteristics
A total of 50 frozen primary breast tissues were
obtained from 49 patients (1 patient presented with bilateral
tumors). The average age of the patients at diagnosis was 57 years
(range, 53–60 years). According to clinicopathological data, 53% of
patients were lymph node-negative and 47% were lymph node-positive,
with the average number of metastatic lymph nodes being 6.04
(range, 1–21). The average age at diagnosis did not differ
significantly between those with (53 years) and without (60 years)
lymph node metastasis.
Patients were chosen based on sample availability
opposed to clinical parameters; therefore, the analyzed tumors
represent a variety of pathological characteristics and tumor
types. The majority of specimens were from patients with invasive
ductal metastasis (84%), predominantly size T1 and T2 (90%). The
majority of patients had a positive ER (80%), PgR (78%) and HER2
(50%) status. A total of 2 patients were diagnosed with stage-IV
breast cancer. A few months after diagnosis, 4 patients developed
distant metastasis and 1 succumbed to the disease. None of the
evaluated pathological features differed significantly between
groups [primary tumors in women with negative and positive lymph
node status: Age, P=0.2047; histological type, P=0.2609;
histological grade, P=1.0000; ER status, P=0.6889; PGR status,
P=0.4411; HER2 status, P=0.2734; Ki-67 status, P=0.4221; tumor
stage, P=0.6492; and tumor size, P=1.0000 (Fisher's exact test, P
≤0.05)]. These results indicate that the clinicopathological
characteristics in the analyzed samples are not associated with the
presence or absence of lymph node metastasis (Table I).
Gene expression
In the present study, the expression of 10 genes
(PPM1D, B3GNT7, NEDD9, PHB, PIK3R5, PIP4K2A, TNKS2, BAD, GTSE1 and
PAXIP1), which were selected from 58 genes in a previous microarray
study (21), was assessed by RT-qPCR
in 50 samples from 49 patients with breast cancer with lymph node
metastasis in two comparisons: i) Primary tumors without lymph node
involvement (n=27) compared with primary tumors with lymph node
involvement (n=23) according to the clinicopathological data
(analysis 1); ii) and primary tumor (n=11) samples compared with
corresponding lymph node metastases (n=11) (analysis 2).
When comparing patients with primary tumors without
lymph node involvement (n=27 samples) with patients with lymph node
involvement (n=23 samples) (analysis 1), no statistically
significant difference was detected for the majority of the genes
evaluated. Only the PIK3R5 gene exhibited increased
expression in primary tumor samples with lymph node involvement
compared with primary tumors without involvement (P=0.0347). The
PIP4K2A gene demonstrated a tendency of increased expression
in primary tumors with lymph node involvement compared with those
without impairment. For all assessed genes, expression in primary
tumors (n=11 samples) compared with paired lymph node metastases
(n=11 samples) (analysis 2) did not demonstrate any significant
differences (P≤0.05). Data from analyses 1 and 2 are presented in
Figs. 1 and 2, respectively.
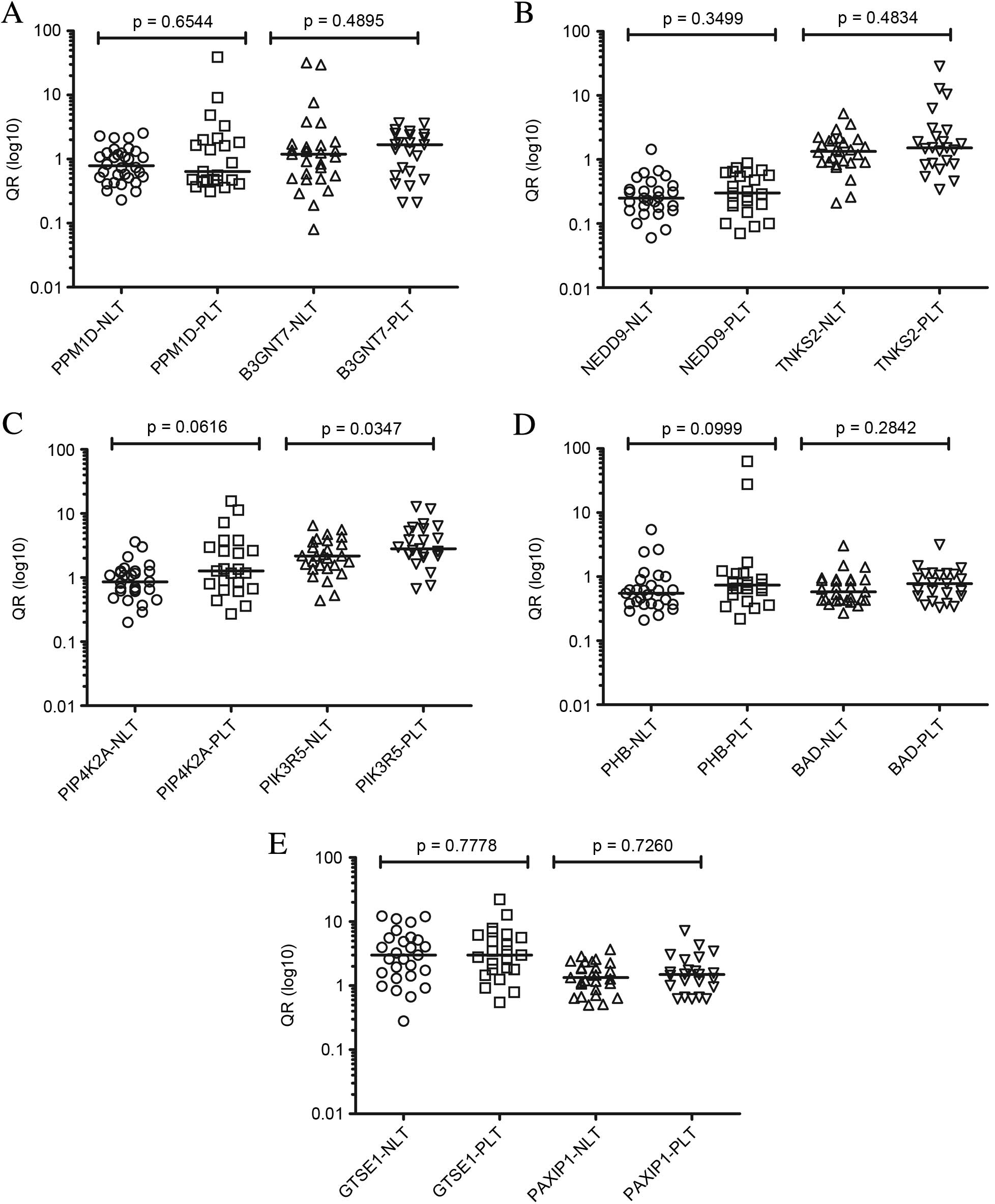 | Figure 1.Comparison between levels of
expression by reverse transcription-quantitative polymerase chain
reaction of the transcripts (A) PPM1D and B3GNT7, (B) NEDD9 and
TNKS2, (C) PIP4K2A and PIK3R5, (D) PHB and BAD and (E) GTSE and
PAXIP1 of primary tumors with and without lymph node involvement
(analysis 1) with respective P-values. QR, relative quantification;
NLT, negative lymph node tumor; PLT, positive lymph node tumor;
PPM1D, protein phosphatase Mg2+/Mn2+ dependent 1D; B3GNT7,
β-1,3-N-acetylglucosaminyltransferase; NEDD9, neural precursor cell
expressed developmentally down-regulated 9; TNKS2, TRF1-interacting
ankyrin-related ADP-ribose polymerase 2; PIP4K2A,
phosphatidylinositol-5-phosphate 4-kinase type IIα; PIK3R5,
phosphoinositide-3-kinase regulatory subunit 5; PHB, prohibitin;
BAD, BCL2 associated agonist of cell death; GTSE, G2 and S-phase
expressed 1; PAXIP1, PAX interacting protein 1. |
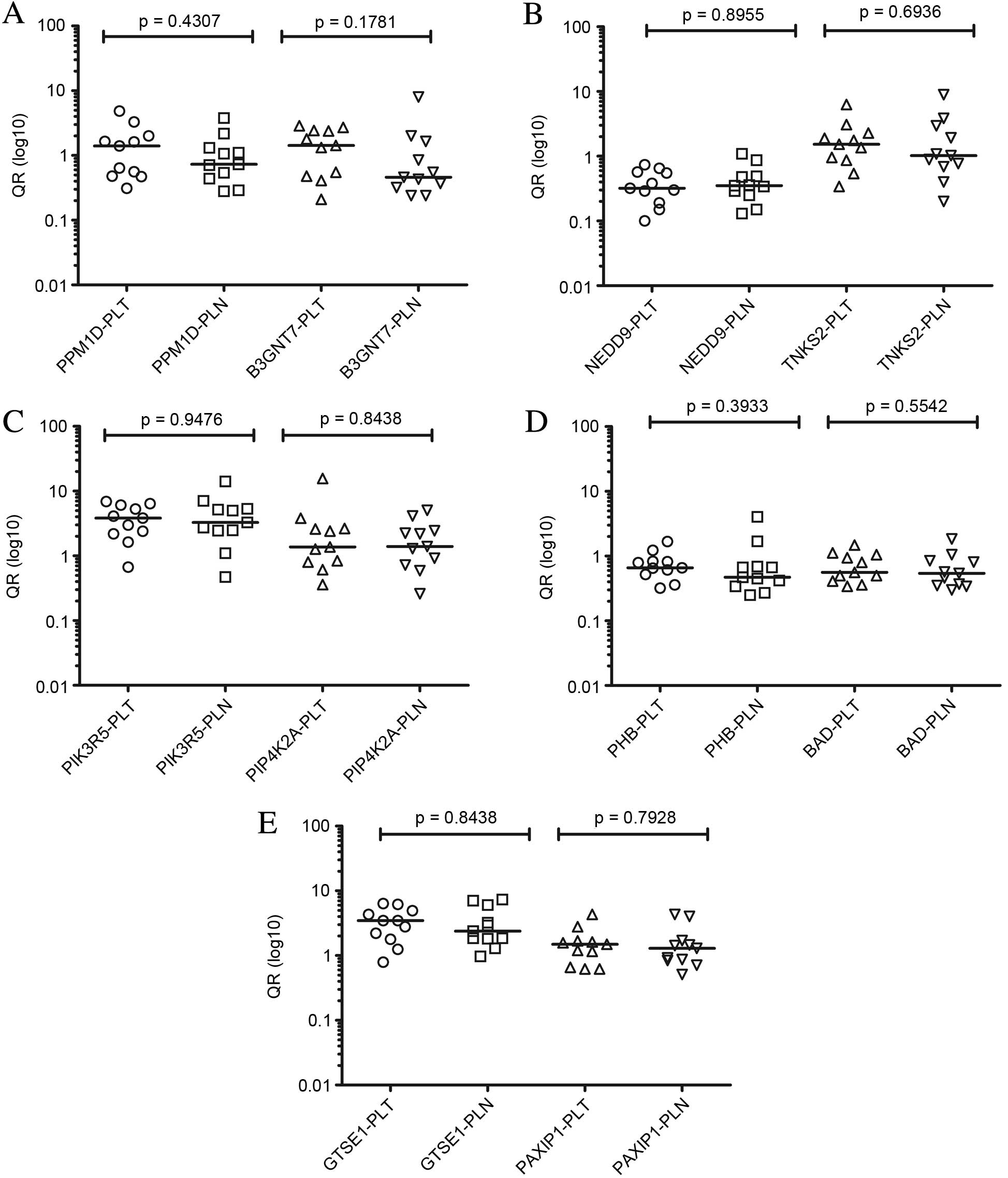 | Figure 2.Comparison between levels of
expression by reverse transcription-quantitative polymerase chain
reaction of the transcripts (A) PPM1D and B3GNT7, (B) NEDD9 and
TNKS2, (C) PIP4K2A and PIK3R5, (D) PHB and BAD and (E) GTSE and
PAXIP1 of primary tumors and paired lymph node metastases (analysis
2) with respective P-values. QR, relative quantification; PLT,
positive lymph node tumor; PLN, positive lymph node; PPM1D, protein
phosphatase Mg2+/Mn2+ dependent 1D; B3GNT7,
β-1,3-N-acetylglucosaminyltransferase; NEDD9, neural precursor cell
expressed developmentally down-regulated 9; TNKS2, TRF1-interacting
ankyrin-related ADP-ribose polymerase 2; PIP4K2A,
phosphatidylinositol-5-phosphate 4-kinase type IIα; PIK3R5,
phosphoinositide-3-kinase regulatory subunit 5; PHB, prohibitin;
BAD, BCL2 associated agonist of cell death; GTSE, G2 and S-phase
expressed 1; PAXIP1, PAX interacting protein 1. |
Discussion
Attempts have been made to characterize factors that
may predict an increasing risk of nodal involvement, which is the
most significant independent prognostic factor in breast cancer and
remains the most important feature for defining risk category.
Identification of genes involved in the stabilization of metastasis
in the lymph nodes may increase the understanding of the metastatic
process (27).
Differentially-expressed genes may represent those involved in the
initiation of metastasis, which alter angiogenesis, cell motility
and invasion, therefore allowing primary tumor cells with
metastatic potential to disseminate (28). Furthermore, determining the status of
these genes may provide key information to establish the potential
of these markers as predictors of lymph node involvement. Hence,
these markers would serve as molecular targets against which novel
therapeutics could be developed to prevent the early stages of
metastasis. The identification of molecular markers may spare women
at low risk of lymph node metastasis from unnecessary surgical
procedures, including ALND, and the ensuing complications of lymph
node disruption. In addition, this may allow identification of the
8–10% of node-positive women diagnosed by SLNB as node-negative
(29), thus leading to a more
accurate prognosis and delineation of a specific treatment for each
patient (30). However, based on
literature, the data are preliminary and controversial. To date, in
spite of assessment of hundreds of markers, few have been used in
clinical practice for treatment or prognosis in breast cancer.
To this end, efforts have been made to develop a
molecular signature of breast tumors that differs between patients
with and without lymph node metastasis. Certain studies have
detected a considerable number of genes differentially-expressed in
the two groups (31–33). By contrast, a number of research
groups have been unable to develop molecular signatures predictive
of lymph node metastasis (33–36).
Similarly, a previous study failed to detect an effective power in
the molecular signature of primary breast tumors associated with
lymph node metastasis (27). This
study, through evaluation of 41 samples of primary tumors without
lymph node involvement and 35 samples with lymph node involvement
by microarray analysis, identified only 13 differentially-expressed
genes that correctly classified 90% of negative lymph nodes and
only 66% of lymph node positive tumors (27). The authors suggested that a single
molecular classifier for lymph node metastasis may not exist for
several factors, including the paucity of cells within the primary
tumor with metastatic potential, tumor heterogeneity, effect of the
microenvironment or inherited host susceptibility to metastasis
(27).
In the present study, the expression of only 1 gene
(PIK3R5) of 10 analyzed was significantly different in analysis 1,
with increased expression in samples of primary tumors with lymph
node involvement compared with primary tumors without lymph node
involvement. This gene serves a role in cell growth,
differentiation, proliferation, motility, survival and
intracellular transport, and its route is associated with the
progression of melanoma (37,38). The PIK3R5 gene is associated
with inhibition of autophagy (promoting tumor growth) (39) and certain authors have suggested that
autophagy also works as a cytoprotective mechanism (40). In the current study, the
PIP4K2A gene exhibited a tendency for increased expression
in primary tumors with lymph node involvement compared with those
without impairment. This gene is involved in various processes,
including cell proliferation, differentiation and motility
(41). Myhre et al (41) suggested that PIP4K2A affects
the metastatic process in breast cancer after observing that it was
highly expressed in tumors in patients who developed distant
metastases compared with patients without metastasis. Increasing
the number of samples within a similar study may potentially
confirm the predictive value of this gene. Based on these results,
the present study observed that of the 10 genes analyzed, only
PIK3R5 may be considered a predictor of lymph node
involvement in breast carcinoma, and that although other genes have
been characterized to be involved with the development of distant
metastasis (21), they did not have
predictive potential. The results obtained in the current study are
consistent with several studies in the literature that failed to
obtain a molecular signature with predictive power (27,35,36,42).
Based on these results, it may be concluded that tumor prognosis is
independent of the presence or absence of lymph node
involvement.
The absence of a signature for lymph node metastasis
may be assigned to biological properties of primary tumors,
including the nature and number of cells within the primary tumor
with metastatic potential. Several studies have reported the
presence of small subpopulations of cells with full metastatic
potential in localized regions of the primary tumor and that the
genetic signatures from these rare cells could be masked by the
majority of tumor cells that do not have full metastatic capacity
(43,44). These studies do not exclude the
ability of the gene expression signatures derived from primary
tumors to predict which tumors may metastasize (27,45,46).
In addition, evaluation of gene expression
differences between primary breast tumors and matched metastatic
lymph nodes should allow genes involved in the metastatic process
to be identified. However, in the majority of studies, the status
of assessed genes is determined only at the primary tumor, and to
the best of our acknowledge, few studies have been published in the
literature regarding the evaluation of gene expression, both by
microarray and RT-qPCR, which compare primary breast tumors and
paired lymph node metastases in breast cancer (47–50).
Contrary to those results (27,43–46), Feng
et al (47) hypothesized that
metastases in the lymph nodes must originate from a fraction of
metastatic cells from primary tumors, and genes
differentially-expressed between the primary tumor and
corresponding axillary metastasis must serve a key role in
metastasis in breast cancer. This study performed a microarray
analysis of 21,000 well-characterized genes, and 79 genes with
differential expression in 14/26 cases analyzed distinguished
primary tumors and corresponding lymph node metastasis samples,
establishing a pattern of changes in gene expression associated
with the metastatic process (47).
Despite identifying similarities between primary breast and paired
lymph node metastases, Ellsworth et al (50) detected 51 genes that were
differentially-expressed between these two groups; 13 of these
genes with higher expression in lymph node metastasis are largely
involved in signal transduction, transcription and immune response.
This study detected similar classes of genes involved in the
comparison of primary tumor with matched lymph node metastases to
those obtained by Feng et al (47). However, additional studies observed
contradictory results that support a model in which genes involved
in changes in extracellular matrix stability are critical to the
early metastatic process, while those involved in immune response,
signal transduction and proliferation are important for
colonization at the secondary site (48,49).
In the present study, the expression analysis of 10
genes from primary tumors and corresponding lymph node metastases
(analysis 2) was conducted. The results did not detect significant
differences in the expression of any of the evaluated genes in each
group. These results are consistent with the hypothesis that only a
fraction of cells, which are phenotypically and biologically
heterogeneously localized in certain regions of the primary tumor,
have higher metastatic potential (27,51). This
may explain why changes in expression of specific genes with
predictive potential, including PIK3R5, cannot be detected
in the primary tumor; molecular alterations of these rare cells are
able to be masked by cells of the primary tumor that lack a high
metastatic capacity. An additional factor that may explain the
inability to detect the differential expression of this gene
between primary tumors and corresponding lymph nodes is that it
could only be detected if the tumors of the majority of these
patients were already involved in the metastatic process. In the
current study, only 2/11 patients with positive lymph nodes
presented with distant metastases. Presently, for therapeutic
purposes, it is assumed that the molecular phenotype of the primary
tumor is the same as that for lymph node involvement; however, it
should be noted that there is great heterogeneity in tumors, and
that, over time, affected lymph nodes may acquire novel biological
characteristics and different forms of invasion, blood or lymph,
thus leading to failures in treatment (52).
In conclusion, the present study identified that
PIK3R5 exhibited differential expression between
node-positive and node-negative primary tumors. This gene serves a
role in cell growth, differentiation, proliferation, motility,
survival and intracellular transport, and the results of the
current study demonstrate that it should be considered as a
predictor of lymph node involvement in breast carcinoma. Although
the majority of the evaluated genes have been characterized in
previous studies as prognostic markers involved in the development
of distant metastases, they did not have predictive potential in
the present study. Further studies with a larger number of samples
are required to confirm these results, and novel molecular markers
are necessary to effectively discriminate patients with and without
the propensity to develop lymph node metastasis, therefore sparing
low-risk women from the morbidities associated with surgical
evaluation and reducing the false-negative rate associated with
SLNB.
Acknowledgements
The authors would like to thank Miss Jéssica Camila
da Silva for her technical assistance regarding RNA extraction, Dr
Edgard Franco de Moraes Coutinho (Esteticlin Clinic, São José dos
Campos, Brazil) for providing healthy control tissues, Dr Jose
Spartaco Vial (Clinica Pro Onco, São José dos Campos, Brazil) for
providing tumor tissues and Miss Alene Alder-Rangel (Alder's
English Services, University of Paraíba Valley, São José dos
Campos, Brazil) for reviewing the English in the original
manuscript. The present study was partially supported by a research
grant from the Foundation for Research Support of the State of São
Paulo (FAPESP), Brazil.
References
|
1
|
Tang Y, Xu F, Tao K, Qian N and Toi M:
Clinical applications of sentinel lymph node biopsy in ductal
carcinoma in situ of the breast: A dilemma. Tohoku J Exp Med.
224:1–5. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Calhoun BC and Collins LC: Predictive
markers in breast cancer: An update on ER and HER2 testing and
reporting. Semin Diagn Pathol. 32:362–369. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Howat JM, Barnes DM, Harris M and Swindell
R: The association of cytosol oestrogen and progesterone receptors
with histological features of breast cancer and early recurrence of
disease. Br J Cancer. 47:629–640. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fisher ER, Costantino J, Fisher B and
Redmond C: Pathologic findings from the National Surgical Adjuvant
Breast Project (Protocol 4). Discriminants for 15-year survival.
National Surgical Adjuvant Breast and Bowel Project Investigators.
Cancer. 71:(Suppl 6). 2141–2150. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pantel K and Brakenhoff RH: Dissecting the
metastatic cascade. Nat Rev Cancer. 4:448–456. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ponzone R, Maggiorotto F, Mariani L,
Jacomuzzi ME, Magistris A, Mininanni P, Biglia N and Sismondi P:
Comparison of two models for the prediction of nonsentinel node
metastases in breast cancer. Am J Surg. 193:686–692. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gurleyik G, Aker F, Aktekin A and Saglam
A: Tumor characteristics influencing non-sentinel lymph node
involvement in clinically node negative patients with breast
cancer. J Breast Cancer. 14:124–128. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wallwiener CW, Wallwiener M, Kurth RR,
Röhm C, Neubauer H, Banys MJ, Staebler A, Schönfisch B, Meuer SC,
Giese T and Fehm TN: Molecular detection of breast cancer
metastasis in sentinel lymph nodes by reverse transcriptase
polymerase chain reaction (RT-PCR): Identifying, evaluating and
establishing multi-marker panels. Breast Cancer Res Treat.
130:833–844. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cavalli LR: Molecular markers of breast
axillary lymph node metastasis. Expert Rev Mol Diagn. 9:441–454.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yenidunya S, Bayrak R and Haltas H:
Predictive value of pathological and immunohistochemical parameters
for axillary lymph node metastasis in breast carcinoma. Diagn
Pathol. 6:182011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cserni G, Burzykowski T, Vinh-Hung V,
Kocsis L, Boross G, Sinkó M, Tarján M, Bori R, Rajtár M, Tekle E,
et al: Axillary sentinel node and tumour-related factors associated
with non-sentinel node involvement in breast cancer. Jpn J Clin
Oncol. 34:519–524. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hung WK, Chan MC, Mak KL, Chong SF, Lau Y,
Ho CM and Yip AW: Non-sentinel lymph node metastases in breast
cancer patients with metastatic sentinel nodes. ANZ J Surg.
75:27–31. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Marrazzo A, Boscaino G, Marrazzo E,
Taormina P and Toesca A: Breast cancer subtypes can be determinant
in the decision making process to avoid surgical axillary staging:
A retrospective cohort study. Int J Surg. 21:156–161. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Veronesi U, Galimberti V, Zurrida S,
Pigatto F, Veronesi P, Robertson C, Paganelli G, Sciascia V and
Viale G: Sentinel lymph node biopsy as an indicator for axillary
dissection in early breast cancer. Eur J Cancer. 37:454–458. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Newman EA and Newman LA: Lymphatic mapping
techniques and sentinel lymph node biopsy in breast cancer. Surg
Clin North Am. 87353–364. (viii)2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Blumencranz PW, Pieretti M, Allen KG and
Blumencranz LE: Molecular analysis of breast sentinel lymph nodes.
Surg Oncol Clin N Am. 20467–485. (viii)2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Backus J, Laughlin T, Wang Y, Belly R,
White R, Baden J, Min C Justus, Mannie A, Tafra L, Atkins D and
Verbanac KM: Identification and characterization of optimal gene
expression markers for detection of breast cancer metastasis. J Mol
Diagn. 7:327–336. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Abdul-Rasool S, Kidson SH, Panieri E, Dent
D, Pillay K and Hanekom GS: An evaluation of molecular markers for
improved detection of breast cancer metastases in sentinel nodes. J
Clin Pathol. 59:289–297. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Patani NR, Dwek MV and Douek M: Predictors
of axillary lymph node metastasis in breast cancer: A systematic
review. Eur J Surg Oncol. 33:409–419. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kurosumi M and Takei H: Significance and
problems of histopathological examination and utility of real-time
reverse transcriptase-polymerase chain reaction method for the
detection of sentinel lymph node metastasis in breast cancer.
Breast Cancer. 14:342–349. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Canevari RA, Marchi FA, Domingues MA, de
Andrade VP, Caldeira JR, Verjovski-Almeida S, Rogatto SR and Reis
EM: Identification of novel biomarkers associated with poor patient
outcomes in invasive breast carcinoma. Tumor Biol. Aug 2–2016.(Epub
ahead of print). View Article : Google Scholar
|
|
22
|
World Health Organization, . International
Classification of Diseases for Oncology. Third. World Health
Organization; Geneva: 2000
|
|
23
|
UICCTMN, . Classificação dos Tumores
MalignosMinistério da Saúde. 7th. Rio de; Janeiro: 2007
|
|
24
|
Bloom HJ and Richardson WW: Histological
grading and prognosis in breast cancer; a study of 1409 cases of
which 359 have been followed for 15 years. Br J Cancer. 11:359–377.
1957. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
McNeill RE, Miller N and Kerin MJ:
Evaluation and validation of candidate endogenous control genes for
real-time quantitative PCR studies of breast cancer. BMC Mol Biol.
8:1072007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ellsworth RE, Field LA, Love B, Kane JL,
Hooke JA and Shriver CD: Differential gene expression in primary
breast tumors associated with lymph node metastasis. Int J Breast
Cancer. 2011:1427632011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nguyen DX and Massagué J: Genetic
determinants of cancer metastasis. Nat Rev Genet. 8:341–352. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Michaelson JS, Silverstein M, Sgroi D,
Cheongsiatmoy JA, Taghian A, Powell S, Hughes K, Comegno A, Tanabe
KK and Smith B: The effect of tumor size and lymph node status on
breast carcinoma lethality. Cancer. 98:2133–2143. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mansel RE, Goyal A and Newcombe RG:
ALMANAC Trialists Group: Internal mammary node drainage and its
role in sentinel lymph node biopsy: The initial ALMANAC experience.
Clin Breast Cancer. 5:279–286. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bertucci F, Houlgatte R, Benziane A,
Granjeaud S, Adélaïde J, Tagett R, Loriod B, Jacquemier J, Viens P,
Jordan B, et al: Gene expression profiling of primary breast
carcinomas using arrays of candidate genes. Hum Mol Genet.
9:2981–2991. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang E, Cheng SH, Dressman H, Pittman J,
Tsou MH, Horng CF, Bild A, Iversen ES, Liao M, Chen CM, et al: Gene
expression predictors of breast cancer outcomes. Lancet.
361:1590–1596. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Abba MC, Sun H, Hawkins KA, Drake JA, Hu
Y, Nunez MI, Gaddis S, Shi T, Horvath S, Sahin A, et al: Breast
cancer molecular signatures as determined by SAGE: Correlation with
lymph node status. Mol Cancer Res. 5:881–890. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
van't Veer LJ, Dai H, van de Vijver MJ, He
YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ,
Witteveen AT, et al: Gene expression profiling predicts clinical
outcome of breast cancer. Nature. 415:530–536. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Weigelt B, Hu Z, He X, Livasy C, Carey LA,
Ewend MG, Glas AM, Perou CM and Van't Veer LJ: Molecular portraits
and 70-gene prognosis signature are preserved throughout the
metastatic process of breast cancer. Cancer Res. 65:9155–9158.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lu X, Lu X, Wang ZC, Iglehart JD, Zhang X
and Richardson AL: Predicting features of breast cancer with gene
expression patterns. Breast Cancer Res Treat. 108:191–201. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gray-Schopfer V, Wellbrock C and Marais R:
Melanoma biology and new targeted therapy. Nature. 445:851–857.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shull AY, Latham-Schwark A, Ramasamy P,
Leskoske K, Oroian D, Birtwistle MR and Buckhaults PJ: Novel
somatic mutations to PI3K pathway genes in metastatic melanoma.
PLoS One. 7:e433692012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ávalos Y, Canales J, Bravo-Sagua R,
Criollo A, Lavandero S and Quest AF: Tumor suppression and
promotion by autophagy. BioMed Res Int. 2014:6039802014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Orzechowski A, Bettuzzi S, Pawlikowska P
and Pająk B: Control of autophagy in cancer. Biomed Res Int.
2015:6987402015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Myhre S, Mohammed H, Tramm T, Alsner J,
Finak G, Park M, Overgaard J, Børresen-Dale AL, Frigessi A and
Sørlie T: In silico ascription of gene expression differences to
tumor and stromal cells in a model to study impact on breast cancer
outcome. PLoS One. 5:e140022010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
van de Vijver MJ, He YD, van't Veer LJ,
Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C,
Marton MJ, et al: A gene-expression signature as a predictor of
survival in breast cancer. N Engl J Med. 347:1999–2009. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kang Y, Siegel PM, Shu W, Drobnjak M,
Kakonen SM, Cordón-Cardo C, Guise TA and Massagué J: A multigenic
program mediating breast cancer metastasis to bone. Cancer Cell.
3:537–549. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ding L, Ellis MJ, Li S, Larson DE, Chen K,
Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, et al:
Genome remodelling in a basal-like breast cancer metastasis and
xenograft. Nature. 464:999–1005. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fidler IJ and Kripke ML: Genomic analysis
of primary tumors does not address the prevalence of metastatic
cells in the population. Nat Genet. 34:232003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Welch DR: Microarrays bring new insights
into understanding of breast cancer metastasis to bone. Breast
Cancer Res. 6:61–64. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
47
|
Feng Y, Sun B, Li X, Zhang L, Niu Y, Xiao
C, Ning L, Fang Z, Wang Y, Zhang L, et al: Differentially expressed
genes between primary cancer and paired lymph node metastases
predict clinical outcome of node-positive breast cancer patients.
Breast Cancer Res Treat. 103:319–329. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Suzuki M and Tarin D: Gene expression
profiling of human lymph node metastases and matched primary breast
carcinomas: Clinical implications. Mol Oncol. 1:172–180. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Vecchi M, Confalonieri S, Nuciforo P,
Viganò MA, Capra M, Bianchi M, Nicosia D, Bianchi F, Galimberti V,
Viale G, et al: Breast cancer metastases are molecularly distinct
from their primary tumors. Oncogene. 27:2148–2158. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ellsworth RE, Seebach J, Field LA, Heckman
C, Kane J, Hooke JA, Love B and Shriver CD: A gene expression
signature that defines breast cancer metastases. Clin Exp
Metastasis. 26:205–213. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yokota J: Tumor progression and
metastasis. Carcinogenesis. 21:497–503. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chambers AF, Naumov GN, Vantyghem SA and
Tuck AB: Molecular biology of breast cancer metastasis. Clinical
implications of experimental studies on metastatic inefficiency.
Breast Cancer Res. 2:400–407. 2000. View
Article : Google Scholar : PubMed/NCBI
|
















