Introduction
Human gliomas are a heterogeneous group of primary
malignant brain tumors, which most commonly occur in the central
nervous system of children and adults (1). Glioblastoma multiforme (GBM), the most
aggressive form of glioma, exhibits advanced features of
malignancy, including rapid tumor cell proliferation, apoptosis
resistance, florid necrosis and angiogenesis (2). These tumor properties are associated
with poor clinical outcome by conferring resistance to chemotherapy
and radiotherapy, and by promoting neurological debilitation
leading to individuals succumbing to mortality within 12–18 months
of diagnosis (3).
Gliomas can be categorized based on the type of
glial cell, which they are most histologically similar to, the
location of the tumor and the aggressiveness of the cancer cells.
Tumors, which are most similar to astrocytes are specifically
termed astrocytomas and can be further classified into grades I–IV
based on the criteria set by the World Health Organization (WHO)
(4), with higher grades corresponding
to more aggressive tumors. Grade I and II astrocytomas correspond
to low-grade tumors, which are predominantly non-malignant. Grade
III and IV astrocytomas are high-grade, malignant tumors. Grade III
astrocytomas are also known as anaplastic astrocytomas, whereas
grade IV astrocytomas, commonly referred to as glioblastoma, are
the most aggressive of all gliomas. GBMs are also the most common
type of glioma with an annual incident rate of 3.19/100,000 in the
USA (5,6).
Matrix metalloproteinases (MMPs) are zinc-dependent
endopeptidases, and their expression is regulated by proteolytic
activation and by selective inhibitory proteins. The majority of
the extracellular matrix (ECM) components are substrates of MMPs
(7). MMPs have also been reported to
process several bioactive factors, apoptotic chemokines and cell
signaling factors, which affect immune responses (8). The overexpression of certain members of
the MMP family has been correlated with invasion, metastasis and
poor prognosis. MMP9 has been implicated in the invasion and
metastasis of head and neck squamous cell carcinoma (9,10). The
knockout of MMP9 has been reported to lead to reduced skin and
pancreatic carcinogenesis and metastasis with delayed tumor
vascularization (10,11). In addition, the knockout of MMP 23, 28
and 29 lead to enhanced tumorigenesis and metastasis in certain
animal models of cancer (11). The
present study hypothesized that MMP9 may be an important factor in
the progression and prognosis of glioma through proliferation and
angiogenesis.
The aim of the present study was to investigate the
role of MMP9 in glioma. The results demonstrated that MMP9 was
upregulated in glioma tissues and its expression was correlated
with tumor grade. It was also observed that the overexpression of
MMP9 in a glioma cell line accelerated tumor growth and induced a
significant increase in clonogenic potential. It was shown that an
increase in the number of copies of MMP was significantly
associated with the increased expression of MMP in gliomas.
Materials and methods
Tissue specimens and patients
Tumor tissues were collected from 62 patients with
gliomas who underwent successful tumor resection or biopsy at the
Department of Neurosurgery of Xijing Hospital, Fourth Military
Medical University (Xi'an, China) between March 2012 and March
2015. The patients included 45 men and 17 women, with a median age
of 62 years (range, 32–88 years). In addition, the results of
survival analyses were collected from 80 patients with grade III
gliomas who underwent successful tumor resection or biopsy at the
Department of Neurosurgery of Xijing Hospital, Fourth Military
Medical University between July 2001 and July 2005. The present
study was performed in accordance with The Code of Ethics of the
World Medical Association (Declaration of Helsinki) and approved by
the Institutional Review Board of Xijing Hospital, Fourth Military
Medical University. Written informed consent was obtained from each
patient or their legally authorized representative. All patients
underwent preoperative computed tomography and magnetic resonance
imaging. All tissue sections were reviewed by two pathologists
without knowledge of clinical outcomes. Following collection during
surgery, half of the tissue sample from the bulk of the tumor was
immediately frozen in liquid nitrogen and stored at −80°C. The
remaining half was processed for primary tumor cultures.
Cell culture and transfection
The human U87 glioma cells (American Type Culture
Collection, Manassas, VA, USA) were maintained in RPMI 1640 medium
supplemented with 10% fetal bovine serum (FBS; Sijiqing, Hangzhou,
China), 2 mM L-glutamine, 50 IU/ml penicillin and 50 µg/ml
streptomycin sulfate, and cultured in 5% CO2 at 37°C.
The U87 cells were transfected with 2 µg of pIRES2-enhanced green
fluorescent protein (EGFP)-MMP9 (Clontech Laboratories, Inc.,
Mountainview, CA, USA) or pIRES2-EGFP (Clontech Laboratories, Inc.)
using 5 µl Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. At 5 h
post-transfection, the medium was replaced with complete medium,
and the cells were incubated for a further 48 h. The cells were
then selected by G418 (1 mg/ml concentration). After 2 weeks, the
G418-resistant colonies were isolated and pooled, and were
confirmed using fluorescence-activated cell sorting, showing that
>90% cells were EGFP positive.
MTT and colony-forming assays
The U87 cells were seeded in triplicate in 96-well
plates (1×103 cells/well) and were cultured in 200 µl
medium for 48 h prior to the addition of MTT. For the MTT assay,
half of the medium (100 µl) was removed and an equal volume of
fresh medium containing 20% MTT (5 mg/ml) was added. The cells were
incubated for a further 4 h at 37°C. The medium was then removed,
and 150 µl of dimethyl sulfoxide (Sigma-Aldrich; Merck Millipore,
Darmstadt, Germany) was added to each well, and mixed by shaking at
room temperature for 10 min. The absorbance was measured at 490 nm.
To perform a colony-forming assay, the U87 cells
(2×103/well) were embedded in medium containing 0.33%
agar gel and 10% FBS in 12-well plates, pre-coated with 0.5% agar
solution, in triplicate. The gel was covered with normal medium and
was cultured routinely. The number of colonies, defined as
containing >50 cells, were counted following 10 days of
incubation under an inverted microscope (IX71; Olympus Corporation,
Tokyo, Japan).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from the cultured cells
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Complementary DNA (cDNA)
was prepared using a reverse transcription system (Toboyo Co.,
Ltd., Osaka, Japan) according to the manufacturer's protocol. qPCR
was performed in triplicate using a SYBR Premix EX Taq kit (Takara
Bio, Inc., Otsu, Japan) and the ABI PRISM 7300 real-time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.) with GAPDH as
an internal control. The total reaction system consisted of 30 µl
with 15 µl SYBR Premix, 1 µl cDNA, 1 µl upstream primer, 1 µl
downstream primer and 12 µl double-distilled H2O. The
thermocycling conditions were as follows: 95°C for 1 min, followed
by 95°C 30 sec and 60°C for 34 sec for 35 cycles. The following
primers for were used for qPCR: GAPDH, forward
5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse 5′-TGGTGAAGACGCCAGTGGA-3′;
and MMP9, forward 5′-GTGCTGGGCTGCTGCTTTGCTG-3′ and reverse
5′-GTCGCCCTCAAAGGTTTGGAAT-3′. RT-qPCR quantification was performed
with the 2−ΔΔCq method (12).
Analysis of copy number variation
Genomic DNA was extracted from the fresh and frozen
glioma tissues using an automated DNA extractor (EZ1; Qiagen GmbH,
Hilden, Germany) and quantified using a Nanodrop spectrophotometer
(Thermo Fisher Scientific, Inc.). Sequencing of the MMP9 coding
region was performed in 62 patients. Primers were designed using
Primer 5.0 software to amplify the five coding exons of the MMP9
(RefSeq, https://www.ncbi.nlm.nih.gov/gene/4318; NM_004994.2)
gene, including the intronic flanking sequences. The primer
sequences were as follows: MMP9 forward, 5′-TACTCTGCCTGCACCACCGA-3′
and reverse 5′-TCTCTCATCATTTCTCAGAT-3′. The amplified products were
subsequently purified and sequenced. For the analysis of variations
in MMP9 gene copy number, four normalization assays mapping to
HSA21 and four normalization DNAs were systematically included in
each run, as described previously (13). Gene dosage segments were classified as
chromosomal ‘gain’ or ‘loss’ if the absolute value of the predicted
dosage was >0.75 times the interquartile range of the difference
between the observed and predicted values for each region. The
sequencing reactions were performed as reported previously
(14).
Immunohistochemistry
The antibodies used for immunohistochemistical
analysis included rabbit polyclonal anti-MMP9 antibody (cat. no.
ab38898; Abcam, Cambridge, MA, USA) and peroxidase goat anti-rabbit
IgG antibody (cat. no. PI-1000; Vector Laboratories, Inc.,
Burlington, CA, USA). Immunohistochemical staining was performed
using peroxidase complex methods. Tissue sections (4 µm thick) were
mounted on slides, and then deparaffinized and rehydrated through
xylene baths and graded concentrations of alcohol. Antigen
retrieval was performed via pressure cooker treatment at 100°C for
100 sec in 0.01 mmol/l citrate buffer (pH 6). The sections were
immersed in 0.3% hydrogen peroxide for 12 min to inactivate the
endogenous peroxidase, and then incubated with 10% blocking serum
for 30 min to reduce nonspecific binding. The primary antibody
against MMP9 was diluted at 1:400, and the sections were incubated
with the diluted primary antibody overnight at 4°C. Subsequently,
the slides were incubated with the secondary antibody (peroxidase
goat anti-rabbit IgG antibody, diluted 1:2,000) at room temperature
for 2 h. Diaminobenzidine was used as a chromogen, and commercial
hematoxylin was used for counterstaining. The staining was
visualized with a BX51 microscope (Olympus Corporation).
Western blot analysis
The glioma tissues were minced in RIPA lysis buffer
(Beyotime Institute of Biotechnology, Inc., Haimen, China)
containing 0.1 mM PMSF. The tissue extracts were collected by
centrifugation at 14,000 × g for 5 min at 4°C. The protein
concentration in the extracts was determined using BCA Protein
Assay reagents (Pierce; Thermo Fisher Scientific, Inc.), according
to the manufacturer's protocol. The samples (20 µg/lane) were
analyzed using 12% SDS polyacrylamide gel electrophoresis, followed
by blotting onto a nitrocellulose membrane. The membranes were
probed using primary antibodies, followed by horseradish
peroxidase-conjugated goat anti-mouse or rabbit antibodies. The
primary antibodies included rabbit polyclonal IgG anti-MMP9 (cat.
no. ab38898; Abcam) and monoclonal anti-β-actin (cat. no. ABT264;
Sigma-Aldrich; Merck Millipore). The primary antibodies were
diluted 1:1,000 and incubated overnight at 4°C. The secondary
antibody (peroxidase goat anti-rabbit IgG antibody; cat. no.
PI-1000; Vector Laboratories, Inc.) was diluted 1:4,000 and
incubated for 2 h at room temperature. The membrane was developed
using chemoluminescent reagents (SuperSingnal West Femto Maximum
Sensitivity Substrate; Pierce; Thermo Fisher Scientific, Inc.) and
quantification was performed using Tanon 5200 Multi software (Tanon
Science & Technology Co., Ltd. Shanghai, China).
Statistical analysis
SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA)
was used for statistical analysis. The results are expressed as the
mean ± standard deviation. Differences in the expression of MMP9 in
glioma tissues between tumor grades (WHO grades II, III and IV)
were assessed using analysis of variance. The difference in
survival rates between the two groups, shown by Kaplan-Meier
curves, was examined using a log-rank test. Student's t-test was
used to determine the difference in absorbance and number of
colonies between two groups. Histograms of MMP9 copy number
frequency distributions with respect to tumor grades were produced.
The linear trend of MMP9 copy numbers across tumor grades was
determined using the Mantel-Haenszel χ2-test. P<0.05
was considered to indicate a statistically significant
difference.
Results
MMP9 is upregulated in higher-grade
gliomas
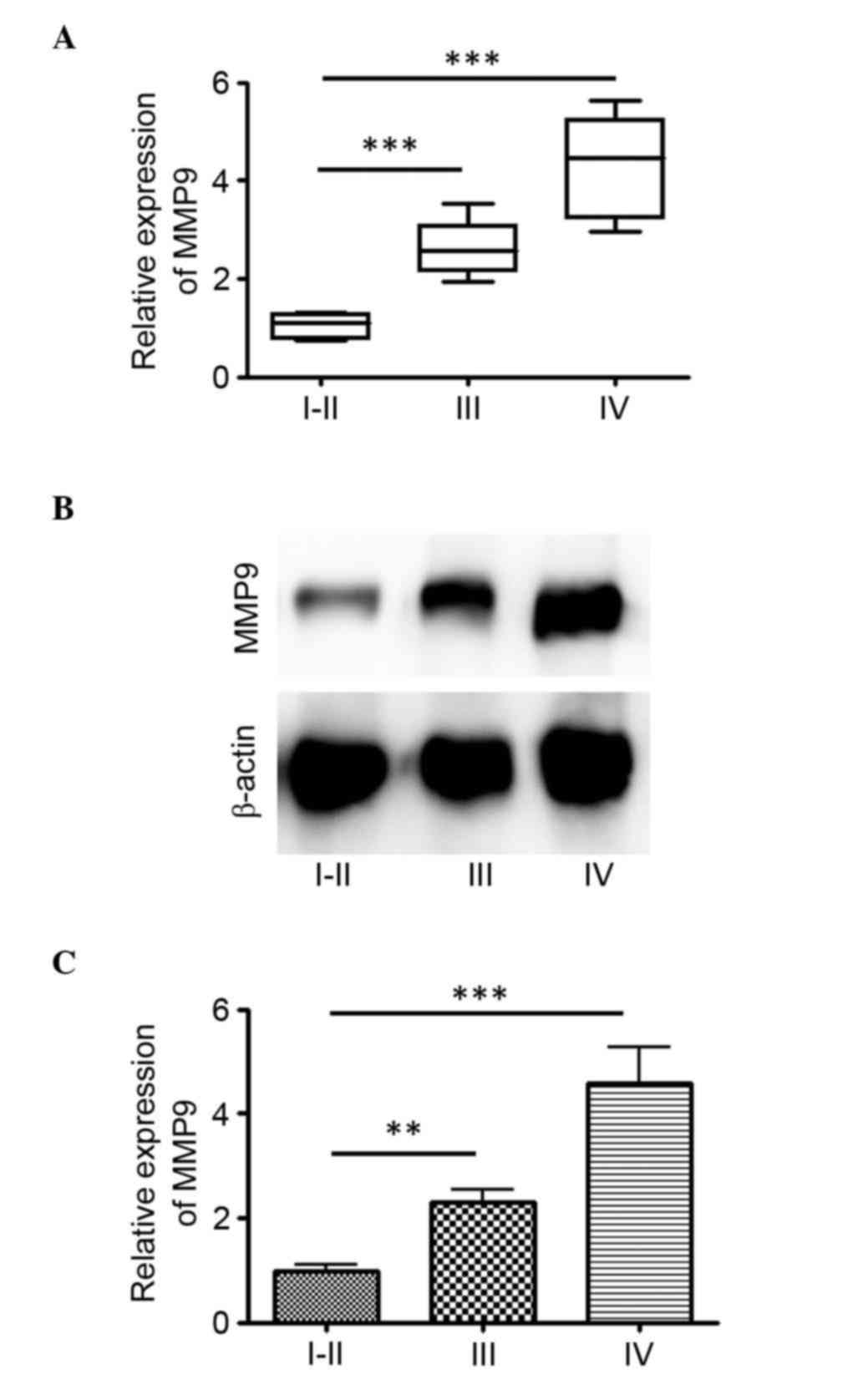
The RNA expression level of MMP9 was determined from
62 glioma tissues, comprising 11 WHO grades I and II, 18 WHO grade
III and 33 WHO grade IV. The relative expression levels of MMP9
ranged between 0.75 and 5.62 in the tumor tissues. A statistically
significant higher relative expression of MMP9 was found in grade
IV tumor tissues, compared with grades I–II and III (P<0.001)
tumor tissues (Fig. 1A). In the
glioma tumor tissues, the median relative expression of MMP9 was
4.28 in GBM with an interquartile range (IQR) of 2.96–5.62, 2.61 in
WHO grade III (IQR, 1.95–3.52) and 0.85 in WHO grade I–II (IQR,
0.55–1.61) glioma (Fig. 1A). The
protein expression levels of MMP9 in these glioma tissues were then
examined. There was also a statistically significant higher
relative expression in grade IV tumor tissues, compared with grades
I–II and III tumor tissues (P<0.001; Fig. 1B and C). These results indicated that
MMP9 was upregulated in gliomas of higher grade.
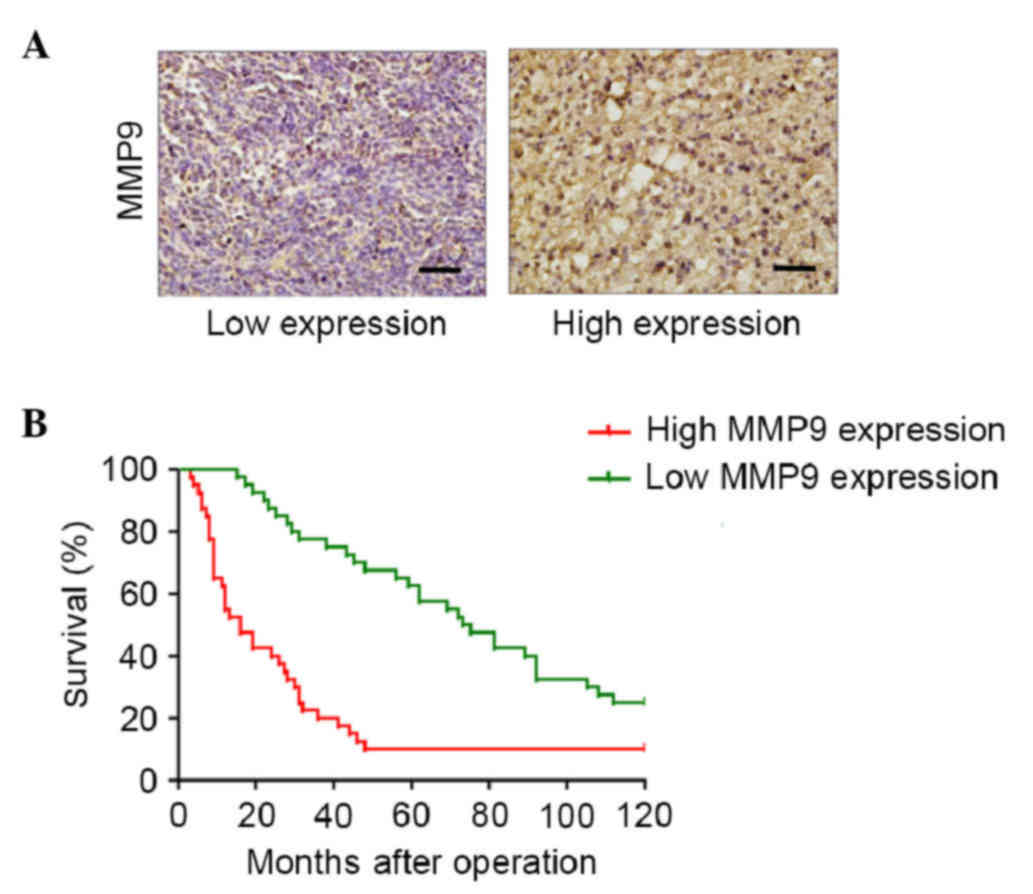
High tissues expression levels of MMP9
are associated with unfavorable clinical outcomes in paitnets with
WHO grade III gliomas
The present study detected the expression of MMP9 in
glioma tumor samples. The median expression level of MMP9 was 50%
(mean, 50%; range, 10–80%; Fig. 2A).
The association between the expression of MMP9 and overall survival
rate was examined in 80 patients with grade III gliomas, who
underwent successful tumor resection or biopsy at the Department of
Neurosurgery of Xijing Hospital, Fourth Military Medical University
between March 2001 and March 2005. Univariate analysis was
performed using a log-rank test, which indicated that patients with
a high expression of MMP9 had a significantly lower overall
survival rate, compared with those with a low expression of MMP9
(Fig. 2B). These data suggested that
the expression levels of MMP9 may represent an independent
predictor of survival rates in patients with WHO grade III
gliomas.
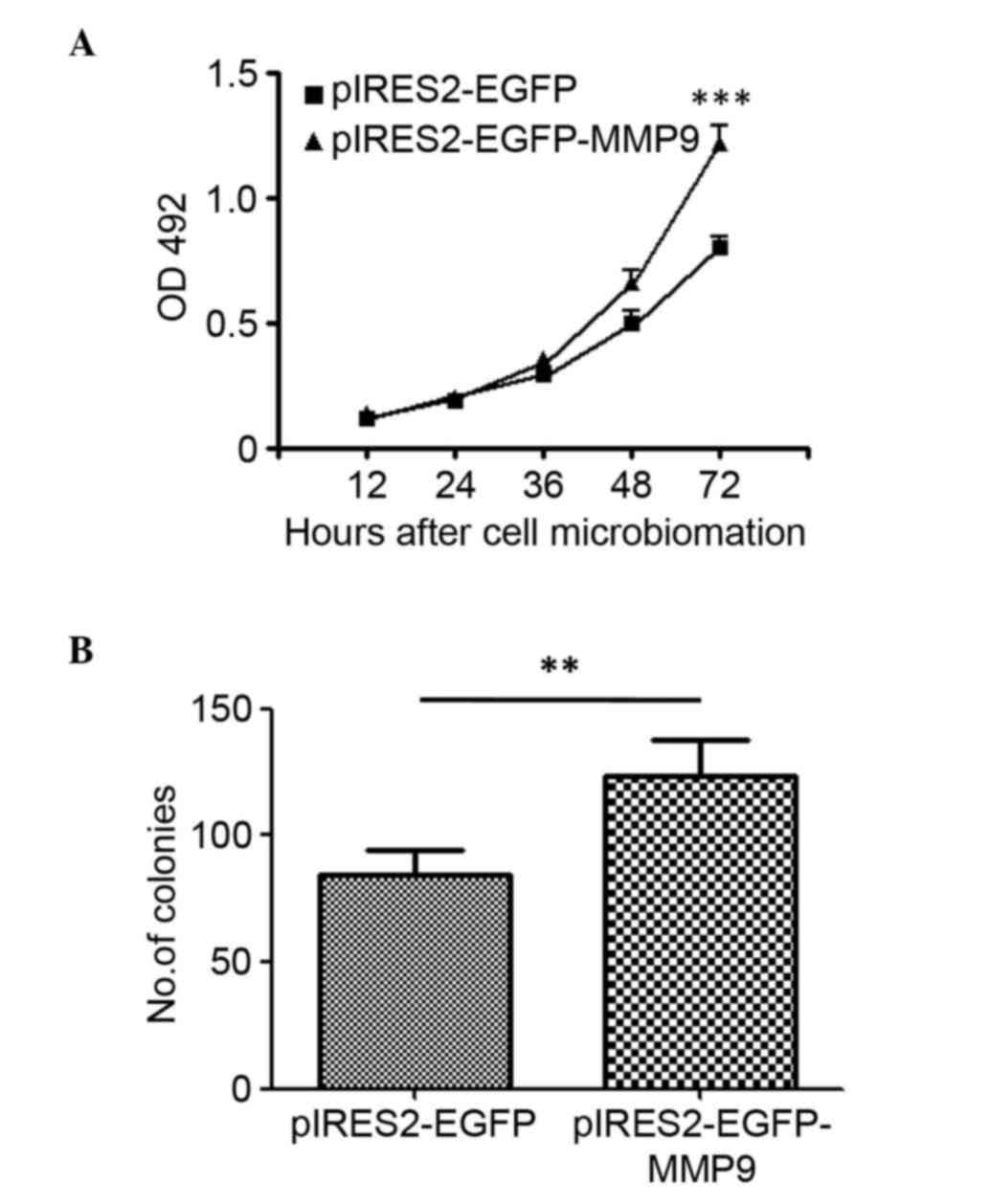
MMP9 induces cell proliferation in
glioblastoma
To evaluate the effect of MMP9 in glioma cell
proliferation, U87 glioma cells were transfected with
pIRES2-EGFP-MMP9 to construct a cell line stably expressing MMP9.
The cell viability was measured using an MTT assay at 12, 24, 36,
48 and 72 h post-microbiomation, respectively. The results showed
that the overexpression of MMP9 significantly increased cell
proliferation (P<0.001; Fig. 3A).
Subsequently, the present investigated whether the overexpression
of MMP9 affected the clonogenic potential of glioma cells as an
indirect index of their tumorigenic potential. The U87 glioma
cells, transfected to express MMP9 or with a control vector, were
plated at limiting dilution and the formation of large colonies
(>50 cells) was assessed 10 days later. The enforced expression
of MMP9 induced a significant increase in the clonogenic potential
(P<0.01), with an average of 123.3 colonies in the
MMP9-transfected cells, compared with 84.7 colonies in the
control-transfected cultures (Fig.
3B).
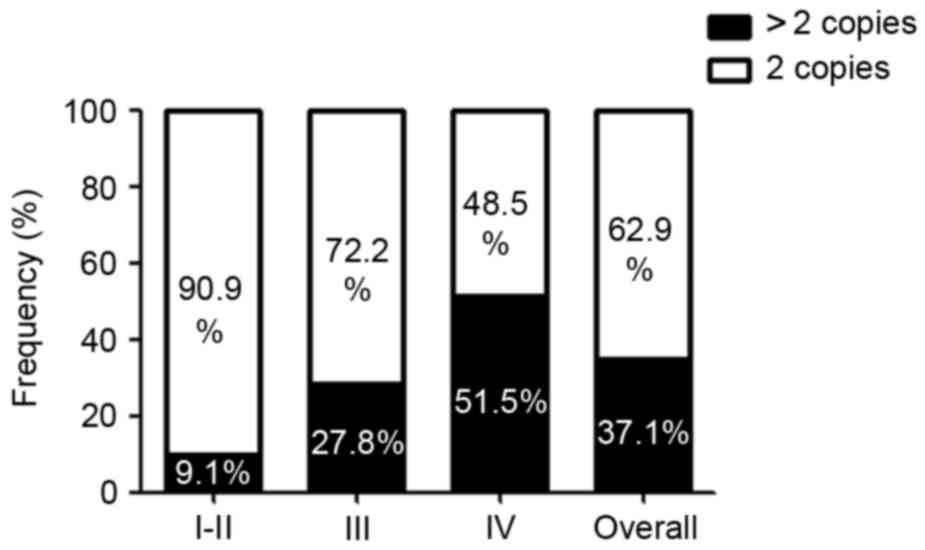
Increase in MMP9 copy numbers in
glioma
To investigate the mechanisms, which may cause the
increase in the expression of MMP9 in glioma, the coding and
untranslated regions of MMP9 were sequenced in 62 patients,
detecting any likely pathogenic variant (data not shown).
Subsequently, to determine MMP9 copy numbers, RT-qPCR analysis was
performed. The MMP9 gene copy number increased in 23/62 (37.1%) of
the analyzed glioma tissue samples. A linear trend of MMP9 copy
number across tumor grades was found (Fig. 4). Specifically, an increase in MMP9
copy number was observed in ~9.1% of the grade I and II gliomas,
27.8% of the grade III gliomas of and 51.5% of the glioblastomas
(Fig. 4).
Discussion
MMPs are critical in tumor cell growth, migration,
invasion, metastasis and angiogenesis (15). MMP9 functions predominantly as a
collagenase by degrading type IV collagen, a major component of the
ECM and basement membrane (16,17). In
view of its broad functions in tumor invasion and metastasis, MMP9
may be a valuable prognostic biomarker in glioma. Previous
meta-analysis has shown a significant correlation between high
expression levels of MMP9 and poor prognosis in gastric cancer
(18), breast cancer (19), non-small cell lung cancer (20) and colorectal cancer (21). MMP9 has also been implicated in the
invasion and metastasis of head and neck squamous cell carcinoma
(9). Knockout of MMP9 leads to
reduced skin and pancreatic carcinogenesis and metastasis, showing
delayed tumor vascularization (9–11). In the
present study, the prognostic value of MMP9 in glioma was
investigated. It was hypothesized that MMP9 may be a valuable
factor in determining the progression and prognosis of gliomas.
Human gliomas are the most common and
life-threatening type of neurological malignancy in adults
(22). The present study was the
first, to the best of our knowledge, to investigate the role of
MMP9 in glioma. The experimental evidence showed that the
expression levels of MMP9 were significantly increased in glioma
and were associated with glioma WHO grades in the tissues
collected. A higher expression levels of MMP9 in tissues was an
independent predictor of survival rates in WHO grade III tumors. In
addition, the overexpression of MMP9 promoted cell growth and
induced a significant increase in the clonogenic potential of U87
glioblastoma cell lines. The present study also investigated the
molecular mechanism underlying the observed increase in the
expression of MMP9. The experimental data suggested that the
overexpression of MMP9 in glioblastoma cells may have occurred
primarily through the increase in gene copy number. In the present
study, the analysis of survival rates revealed a significant
increase in disease progression and decrease in survival rates of
patients with WHO grade III tumors expressing higher levels of
MMP9, compared with those with lower expression levels. These
results suggested that the overexpression of MMP9 may be necessary
for the transition to the aggressive phenotype typical of WHO grade
III gliomas, suggesting the likely involvement of the MMP9 gene in
gliomagenesis and disease progression.
Acknowledgements
This study was supported by the Social Development
Research project in Shaanxi province, China (grant no.
2010K14-02-05). The authors would like to thank the members of the
laboratories at the Department of Neurosurgery, Xijing Hospital of
the Fourth Military Medical University.
Glossary
Abbreviations
Abbreviations:
|
MMP9
|
matrix metalloproteinase 9
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
WHO
|
World Health Organization
|
|
GBM
|
glioblastoma multiforme
|
|
ECM
|
extracellular matrix
|
|
FBS
|
fetal bovine serum
|
|
RNA
|
ribonucleic acid
|
|
cDNA
|
complementary DNA
|
References
|
1
|
Marumoto T and Saya H: Molecular biology
of glioma. Adv Exp Med Biol. 746:2–11. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ohgaki H, Dessen P, Jourde B, Horstmann S,
Nishikawa T, Di Patre PL, Burkhard C, Schüler D, Probst-Hensch NM,
Maiorka PC, et al: Genetic pathways to glioblastoma: A
population-based study. Cancer Res. 64:6892–6899. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Furnari FB, Fenton T, Bachoo RM, Mukasa A,
Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al:
Malignant astrocytic glioma: Genetics, biology, and paths to
treatment. Genes Dev. 21:2683–2710. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hervey-Jumper SL and Berger MS: Maximizing
safe resection of low- and high-grade glioma. J Neurooncol.
130:269–282. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ostrom QT, Gittleman H, Liao P, Rouse C,
Chen Y, Dowling J, Wolinsky Y, Kruchko C and Barnholtz-Sloan J:
CBTRUS statistical report: Primary brain and central nervous system
tumors diagnosed in the United States in 2007–2011. Neuro Oncol.
16:(Suppl 4). iv1–iv63. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Thomas AA, Brennan CW, DeAngelis LM and
Omuro AM: Emerging therapies for glioblastoma. JAMA Neurol.
71:1437–1444. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Husmann K, Arlt MJ, Muff R, Langsam B,
Bertz J, Born W and Fuchs B: Matrix Metalloproteinase 1 promotes
tumor formation and lung metastasis in an intratibial injection
osteosarcoma mouse model. Biochim Biophys Acta. 1832:347–354. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Korpi JT, Hagström J, Lehtonen N,
Parkkinen J, Sorsa T, Salo T and Laitinen M: Expression of matrix
metalloproteinases-2, −8, −13, −26, and tissue inhibitors of
metalloproteinase-1 in human osteosarcoma. Surg Oncol. 20:e18–e22.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Deraz EM, Kudo Y, Yoshida M, Obayashi M,
Tsunematsu T, Tani H, Siriwardena SB, Keikhaee MR, Qi G, Iizuka S,
et al: MMP-10/stromelysin-2 promotes invasion of head and neck
cancer. PLoS One. 6:e254382011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Folgueras AR, Pendás AM, Sánchez LM and
López-Otin C: Matrix metalloproteinases in cancer: From new
functions to improved inhibition strategies. Int J Dev Biol.
48:411–424. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Overall CM and Kleifeld O: Tumour
microenvironment-opinion: Validating matrix metalloproteinases as
drug targets and anti-targets for cancer therapy. Nat Rev Cancer.
6:227–239. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Howald C, Merla G, Digilio MC, Amenta S,
Lyle R, Deutsch S, Choudhury U, Bottani A, Antonarakis SE, Fryssira
H, et al: Two high throughput technologies to detect segmental
aneuploidies identify new Williams-Beuren syndrome patients with
atypical deletions. J Med Genet. 43:266–273. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Micale L, Augello B, Fusco C, Selicorni A,
Loviglio MN, Silengo MC, Reymond A, Gumiero B, Zucchetti F,
D'Addetta EV, et al: Mutation spectrum of MLL2 in a cohort of
Kabuki syndrome patients. Orphanet J Rare Dis. 6:382011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Egeblad M and Werb Z: New functions for
the matrix metalloproteinases in cancer progression. Nat Rev
Cancer. 2:161–174. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liotta LA, Tryggvason K, Garbisa S, Hart
I, Foltz CM and Shafie S: Metastatic potential correlates with
enzymatic degradation of basement membrane collagen. Nature.
284:67–68. 1980. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Stetler-Stevenson WG: Type IV collagenases
in tumor invasion and metastasis. Cancer Metastasis Rev. 9:289–303.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang QW, Liu L, Chen R, Wei YQ, Li P, Shi
HS and Zhao YW: Matrix metalloproteinase-9 as a prognostic factor
in gastric cancer: A meta-analysis. Asian Pac J Cancer Prev.
13:2903–2908. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Song J, Su H, Zhou YY and Guo LL:
Prognostic value of matrix metalloproteinase 9 expression in breast
cancer patients: A meta-analysis. Asian Pac J Cancer Prev.
14:1615–1621. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Peng WJ, Zhang JQ, Wang BX, Pan HF, Lu MM
and Wang J: Prognostic value of matrix metalloproteinase 9
expression in patients with non-small cell lung cancer. Clin Chim
Acta. 413:1121–1126. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li CY, Yuan P, Lin SS, Song CF, Guan WY,
Yuan L, Lai RB, Gao Y and Wang Y: Matrix metalloproteinase 9
expression and prognosis in colorectal cancer: A meta-analysis.
Tumour Biol. 34:735–741. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Micale L, Fusco C, Fontana A, Barbano R,
Augello B, De Nittis P, Copetti M, Pellico MT, Mandriani B,
Cocciadiferro D, et al: TRIM8 downregulation in glioma affects cell
proliferation and it is associated with patients survival. BMC
Cancer. 15:4702015. View Article : Google Scholar : PubMed/NCBI
|