Introduction
Colon cancer is one of the most common types of
gastroenteric tumors for males and females, accounting for a large
proportion of cancer-associated mortalities worldwide (1,2). Due to
changes in human environment, nutritional habits and life style,
the incidence rate of colon cancer has increased worldwide over the
last 20 years (3,4). Colon cancer occurs in a multi-step
process, and metastasis is the major cause of morbidity and
mortality, with ~1/3 of patients with colon cancer ultimately
developing metastatic disease (5).
The use of chemotherapy and surgical resection for the treatment of
malignant colon cancer is increasing, but the results of these
treatments are typically poor (6).
Therefore, investigation into the molecular mechanism underlying
the pathogenesis and progression of colon cancer, and the search
for specific, sensitive biomarkers for the early diagnosis and
prognosis prediction of colon cancer is required.
There are 11 galectin family members identified in
humans at present (7), including
galectin-9, which is a type 1 tandem repeat containing a C-terminal
coding region determinant (C-CRD) of 149 amino acids and an
N-terminal CRD of 148 amino acids (8). The protein was first identified as an
eosinophil chemoattractant and activation factor (9,10), and
later confirmed as a physiological ligand of T-cell immunoglobulin
and mucin domain 3 (11). Galectin-9
is known to exhibit a variety of biological functions, including
cell aggregation, adhesion, proliferation, apoptosis and modulation
of inflammation (12,13). Attention has previously been focused
on the molecular mechanism of galectin-9 in malignant tumors. For
example, Nobumoto et al (14)
confirmed that galectin-9 suppressed tumor metastasis by blocking
adhesion to the endothelium and extracellular matrices. A study by
Zhang et al (15) demonstrated
that galectin-9 acted as a prognostic factor with antimetastatic
potential in hepatocellular carcinoma. However, the expression and
role of galectin-9 in human colon cancer have not been fully
verified.
MicroRNAs (miRNAs or miRs) are a class of non-coding
single-stranded RNA molecules with ~22–24 nucleotides (16). miRNAs serve a pivotal role in the
regulation of target gene expression by binding to the
3′-untranslated regions (3′-UTR) of their target messenger RNA
(mRNA), leading to mRNA degradation or inhibition of translation
into protein (17,18). Currently, >2,042 mature miRNAs have
been identified in humans, which constitute a large network that
regulates the expression of ≤30% of all cellular proteins (19,20). The
expression of miRNAs is regulated developmentally and spatially,
and increasing evidence has demonstrated that miRNAs modulate a
variety of cellular functions, including cell differentiation,
proliferation and death (21).
Numerous studies have indicated the involvement of miRNAs in the
progression and metastasis of numerous types of cancer, suggesting
that miRNAs may be used in future therapeutic and diagnostic
applications (22–24).
At present, the upstream regulatory miRNA of
galectin-9 is undefined. The purpose of the present study was to
investigate the upstream regulatory miRNA of galectin-9 in colon
cancer. The present study demonstrated that elevated expression
levels of galectin-9 and miR-455-5p in colon cancer were associated
with HT29 cell proliferation and apoptosis, and confirmed that
miR-455-5p directly targets galectin-9 3′-UTR and negatively
regulates galectin-9 expression in colon cancer cells.
Materials and methods
Tissue collection
Paired resected surgical specimens from primary
tumors and corresponding adjacent non-tumor sites were obtained
from 10 patients that underwent primary surgical resection of colon
cancer between June and October 2013 at the Department of
Gastrointestinal Surgery of Wuhan Union Hospital (Wuhan, China).
Tissue specimens were confirmed separately by two experienced
pathologists under double-blinded conditions. None of the patients
received any therapy prior to operation. The demographic features
and clinicopathological data were reviewed in the patients' medical
records. The present study was approved by the Ethics Committee of
Wuhan Union Hospital and performed with informed consent obtained
from all patients. All samples were frozen in liquid nitrogen and
stored at −80°C for future molecular analyses.
Cell culture
The human colon cancer HT29 cell line was purchased
from the Shanghai Institute of Biochemistry and Cell Biology
(Shanghai, China), and cultured in Dulbecco's modified Eagle's
medium (DMEM; HyClone; GE Healthcare Life Sciences, Logan, UT, USA)
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), 1 U/ml penicillin (Gibco;
Thermo Fisher Scientific, Inc.) and 1 µg/ml streptomycin (Gibco;
Thermo Fisher Scientific, Inc.). All cultures were maintained at
37°C in a humidified atmosphere with 5% CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA and miRNA were extracted from the cells
and tissues using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Wilmington, DE, USA) according to the
manufacturer's protocol. RNA quantity and quality were determined
using 1% agarose gel electrophoresis and an optical density 260/280
absorption ratio of >1.8 using the NanoDrop 2000 (Thermo Fisher
Scientific, Waltham, MA, USA). Complementary DNA was synthesized
using a PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd.,
Dalian, China) and a MyCycler™ thermal cycler (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) according to the
manufacturer's protocol. The cycling conditions were as follows:
37°C for 15 min, followed by 85°C for 15 sec and then 4°C. RT-qPCR
was performed using SYBR® Premix Ex Taq™II (Takara
Biotechnology Co., Ltd.) and Mx3000P qPCR system (Agilent
Technologies, Inc., Santa Clara, CA, USA). Each reaction was
performed in a total volume of 20 µl, containing 10 µl
SYBR® Premix Ex Taq™II, 2 µl primers, 2 µl template
complementary DNA, 0.4 µl ROX Reference Dye (50X; Takara
Biotechnology Co., Ltd.) and 5.6 µl distilled H2O.
Cycling conditions were as follows: 95°C for 30 sec, followed by 40
cycles of amplification (95°C for 5 sec, 60°C for 30 sec and 72°C
for 30 sec). RT-qPCR was performed in triplicate for each sample.
The expression of each type of RNA and miRNA was defined from the
quantification cycle (Cq), and relative expression levels were
calculated using the 2−∆∆Cq method (25). Human GAPDH and U6 were used as the
housekeeping genes for the amplifications. The PCR primers used in
the present study were described previously (26).
Western blot analysis
Proteins were extracted from the cells and tissues
using radioimmunoprecipitation assay lysis buffer (Beyotime
Institute of Biotechnology, Haimen, China), and the lysates were
cleared by centrifugation at 13,523 × g at 4°C for 15 min.
Subsequent to their concentration being measured with the Pierce
BCA Protein Assay kit (Thermo Fisher Scientific, Inc.), the
proteins were mixed with SDS loading buffer, separated by 10%
SDS-PAGE and transferred onto a polyvinylidene difluoride membrane
(EMD Millipore, Billerica, MA, USA). Subsequent to the blockage of
nonspecific binding sites for 1 h with 5% nonfat milk, the blots
were incubated with rabbit anti-human galectin-9 (dilution,
1:1,000; catalog no. YT1841; ImmunoWay Biotechnology Company,
Plano, TX, USA) or anti-β-actin (dilution, 1:2,000; catalog no.
12,620; Cell Signaling Technology, Inc., Danvers, MA, USA) primary
antibodies at 4°C overnight, followed by incubation with goat
anti-rabbit horseradish peroxidase-conjugated secondary antibody
(dilution, 1:2,000; catalog no. 7074; Cell Signaling Technology,
Inc.) at room temperature for 1 h. Proteins were visualized with
Pierce™ ECL Western Blotting Substrate (Thermo Fisher Scientific,
Inc.). The immunoblots were visualized using ImageJ software
version 1.49 (National Institutes of Health, Bethesda, MD,
USA).
Flow cytometric analysis for
apoptosis
HT29 cells transfected with Galectin-9/pcDNA3.1
vector were seeded in 6-well plates (1×106 cells/well)
and cultured with DMEM in a humidified chamber at 37°C in 5%
CO2 for 24 h. Cell apoptosis was evaluated using an
Annexin-V-FLUOS Staining kit (catalog no. 11858777001; Roche
Applied Science, Mannheim, Germany). Briefly, 1×106
cells were washed with PBS and centrifuged at 200 × g at
room temperature for 5 min. Then, the cell pellets were resuspended
in 100 µl Annexin-V-FLUOS labeling solution (containing 2 µl
Annexin-V-FLUOS labeling reagent, 2 µl propidium iodide solution
and 96 µl incubation buffer) and incubated for 10–15 min at
15–25°C. Apoptosis was assessed by flow cytometry using FACSCalibur
(BD Biosciences, Franklin Lakes, NJ, USA). Non-transfected HT29
cells were used as a negative control. Each group was independently
evaluated three times.
Cell proliferation assay
At 24 h post-transfection, the cells were digested
using trypsin (Wuhan Amyjet Scientific Co., Ltd., Wuhan, China) and
washed twice with PBS (Sangon Biotech Co., Ltd., Shanghai, China),
and then seeded into 96-well plates at a concentration of
2×103 cells/well. Cell Counting kit-8 (CCK-8; Dojindo
Molecular Technologies, Inc., Kumamoto, Japan) was used to assess
the cell proliferation activity at 0, 24, 48 and 72 h. A total of
10 µl CCK-8 was added to each well, and following 2 h of incubation
at 37°C, the optical density value at 450 nm was determined with a
scan reader (MTX Lab Systems, Inc., Vienna, VA, USA).
Target prediction
miRNAs that target galectin-9 were identified by
examining the galectin-9 3′-UTR with bioinformatics algorithms that
predict miRNA target sites. Specifically, miRanda (www.microrna.org) and TargetScan (www.targetscan.org) were used for the analysis of the
alignment between miRNAs and the 3′-UTR of galectin-9.
Plasmid construction, miRNA synthesis
and transfection
The plasmid (p) cytomegalovirus (CMV)
-galectin-9-3′-UTR wild-type (WT), pCMV-galectin-9-3′-UTR mutant
(MU) and galectin-9 overexpression vector (Galectin-9/pcDNA3.1)
were constructed as described previously (26). The specific primers for galectin-9
3′UTR-WT were: Forward, 5′-ATAGAATTCGCGGCTTCCTGGCCCTG-3′ and
reverse, 5′-CGCAAGCTTTGAATGTGCCAACAAGCA-3′. The specific primers
for galectin-9 3′UTR-MU were: Forward,
5′-AATGAAAATGCTTGTTGGAATTCTCAAAGCTTATCGAT-3′ and reverse,
5′-TTTCCAGGAGGGGTGAAGAATTCGTGCACGGTGCAAGG-3′. miR-455-5p and
miR-control (used as a negative control) mimics were synthesized by
Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The cells were
transiently transfected using Lipofectamine 2000 transfection
reagent (Invitrogen; Thermo Fisher Scientific, Inc., USA) for 24 h,
according to the protocol of the manufacturer.
Luciferase reporter gene assay
The HT29 cells were plated in a 96-well plate and
co-transfected with miR-455-5p or control mimics and
pCMV-galectin-9 3′-UTR-WT or pCMV-galectin-9 3′-UTR-MU, in addition
to the pRL-TK vector (Promega Corporation, Madison, WI, USA), using
Lipofectamine 2000. The cells were collected 24 h after
transfection, and the luciferase activity was analyzed using the
Dual-Luciferase Reporter Assay System (Promega Corporation) in a
Modulus single-tube multimode reader (Turner BioSystems, Inc.;
Promega Corporation). The pRL-TK vector (Promega Corporation,
Madison, WI, USA) that provided the constitutive expression of
Renilla luciferase was co-transfected as an internal control
to correct for differences between transfection and harvest
efficiencies. The transfections were performed at least twice in
independent experiments.
Statistical analysis
The Student's t-test was used to evaluate
statistical significance. P<0.05 was considered to indicate a
statistically significant difference. All experiments were repeated
>3 times, and the results from a representative experiment were
selected to draw diagrams and data analysis. All data were
statistically analyzed using GraphPad Prism 5.0 (GraphPad Software,
Inc., La Jolla, CA, USA).
Results
Expression and role of galectin-9 in
colon cancer tissue and in colon cancer cell proliferation and
apoptosis
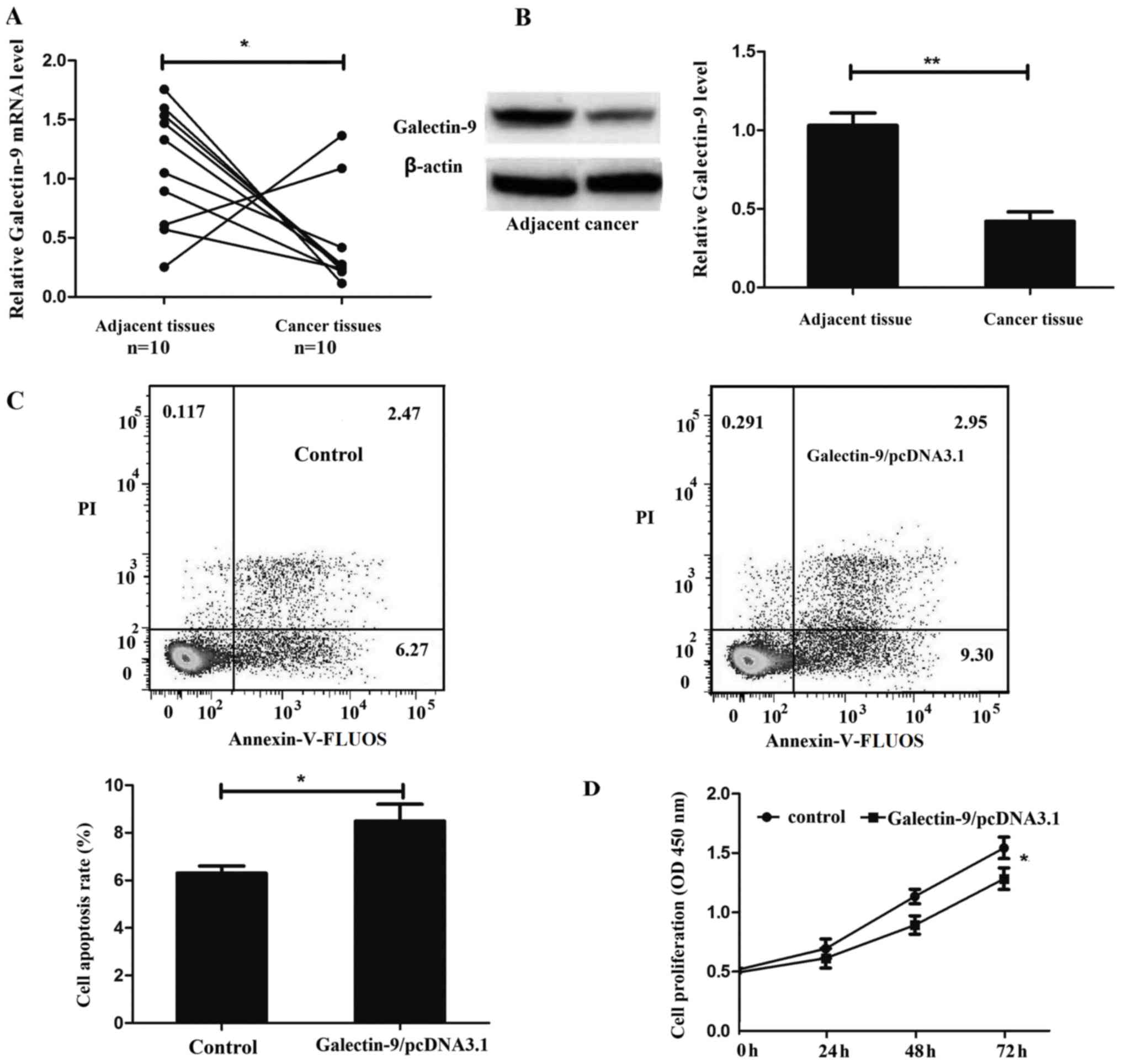
The expression levels of galectin-9 in colon cancer
tissues and the corresponding adjacent tissues from 10 patients
were determined using RT-qPCR and western blot analysis. Galectin-9
was revealed to be significantly downregulated in colon cancer
tissue at the mRNA and protein level compared with the
corresponding adjacent tissue (P=0.0405 and P=0.0037; Fig. 1A and B, respectively).
To investigate the role of galectin-9 in colon
cancer cell apoptosis, flow cytometric analysis was evaluated using
an Annexin-V-FLUOS Staining kit. The present study revealed that
overexpression of galectin-9 promoted HT29 cell apoptosis (Fig. 1C).
In order to additionally examine the role of
galectin-9 in colon cancer cell proliferation, the present study
constructed the galectin-9 overexpression vector
Galectin-9/pcDNA3.1 and transfected it into HT29 cells. CCK-8 assay
revealed that overexpression of galectin-9 inhibited HT29 cell
proliferation (Fig. 1D).
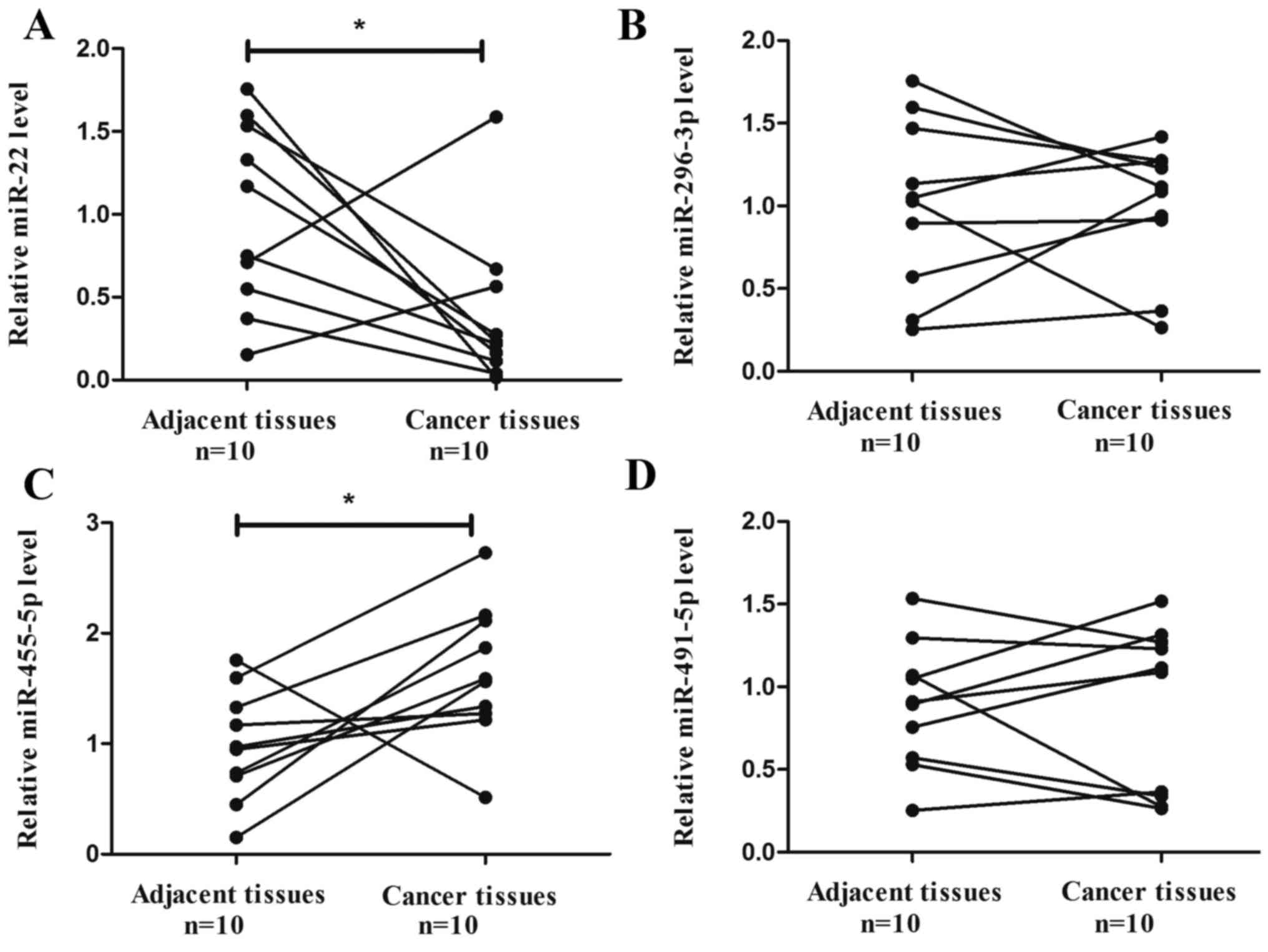
Expression of candidate miRNAs and
role of miR-455-5p in colon cancer
The target prediction programs miRanda and
TargetScan were used to predict and identify miRNAs that may target
galectin-9. The present study identified four miRNAs (miR-22,
296-3p, 455-5p and 491-5p) that were potential regulators of
galectin-9 (26).
The expression of the aforementioned four miRNAs in
colon cancer tissues and corresponding adjacent tissues was
determined. The results indicated that the expression of miR-22 was
significantly downregulated in colon cancer tissue compared with
the corresponding adjacent tissue (P=0.0397; Fig. 2A), while miR-455-5p was significantly
upregulated in colon cancer tissue compared with the corresponding
adjacent tissue (P=0.0346; Fig. 2C).
miR-296-3p and miR-491-5p did not exhibit a significant difference
in expression between colon cancer tissue and the corresponding
adjacent tissue (P=0.9063 and P=0.9477; Fig. 2B and D, respectively). miR-455-5p was
selected in the present study for further experiments, as its
expression level exhibited an inverse correlation with galectin-9
expression.
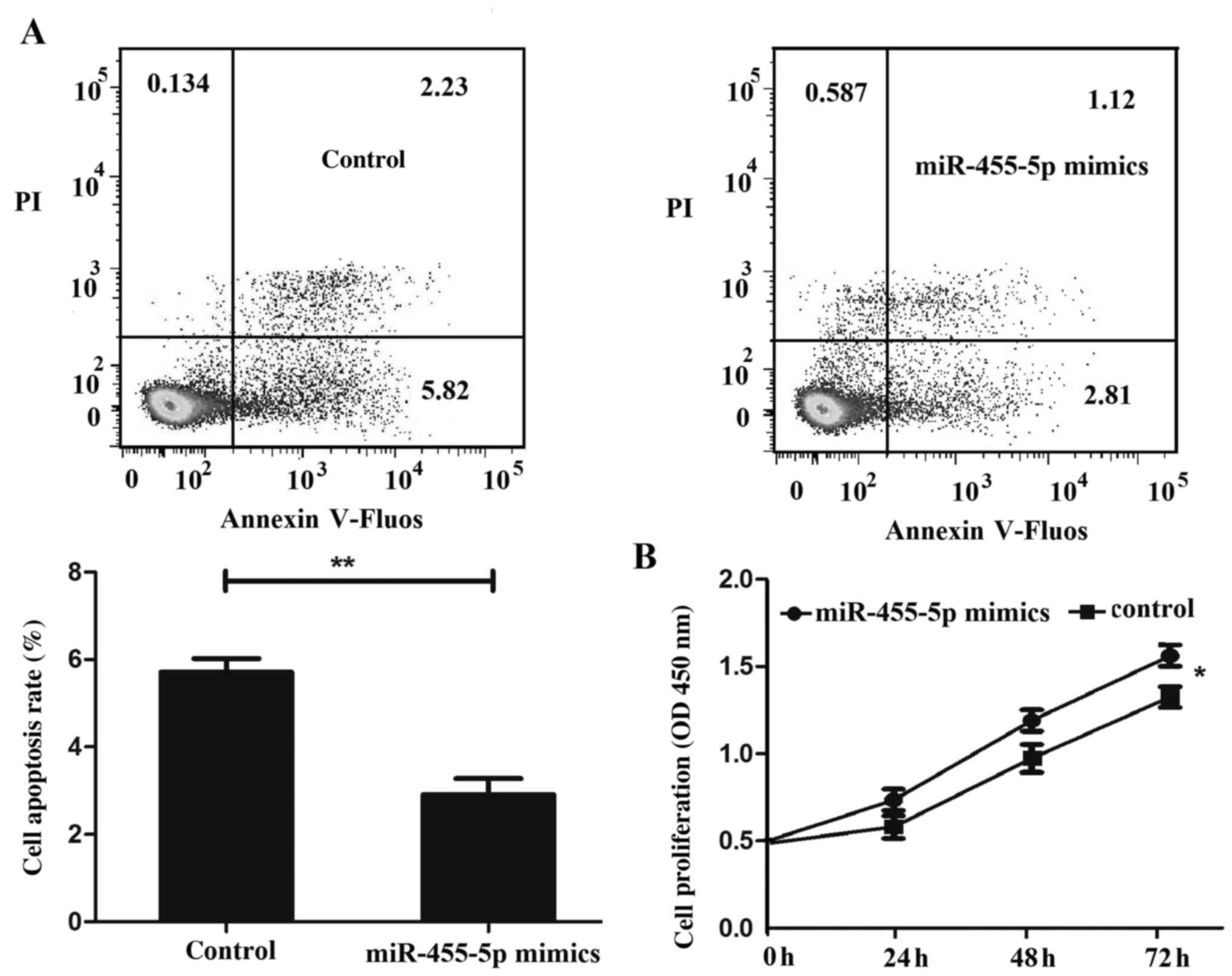
The present study subsequently examined the role of
miR-455-5p in colon cancer cell apoptosis and proliferation,
revealing that miR-455-5p inhibited HT29 cell apoptosis (Fig. 3A) and promoted HT29 cell proliferation
(Fig. 3B).
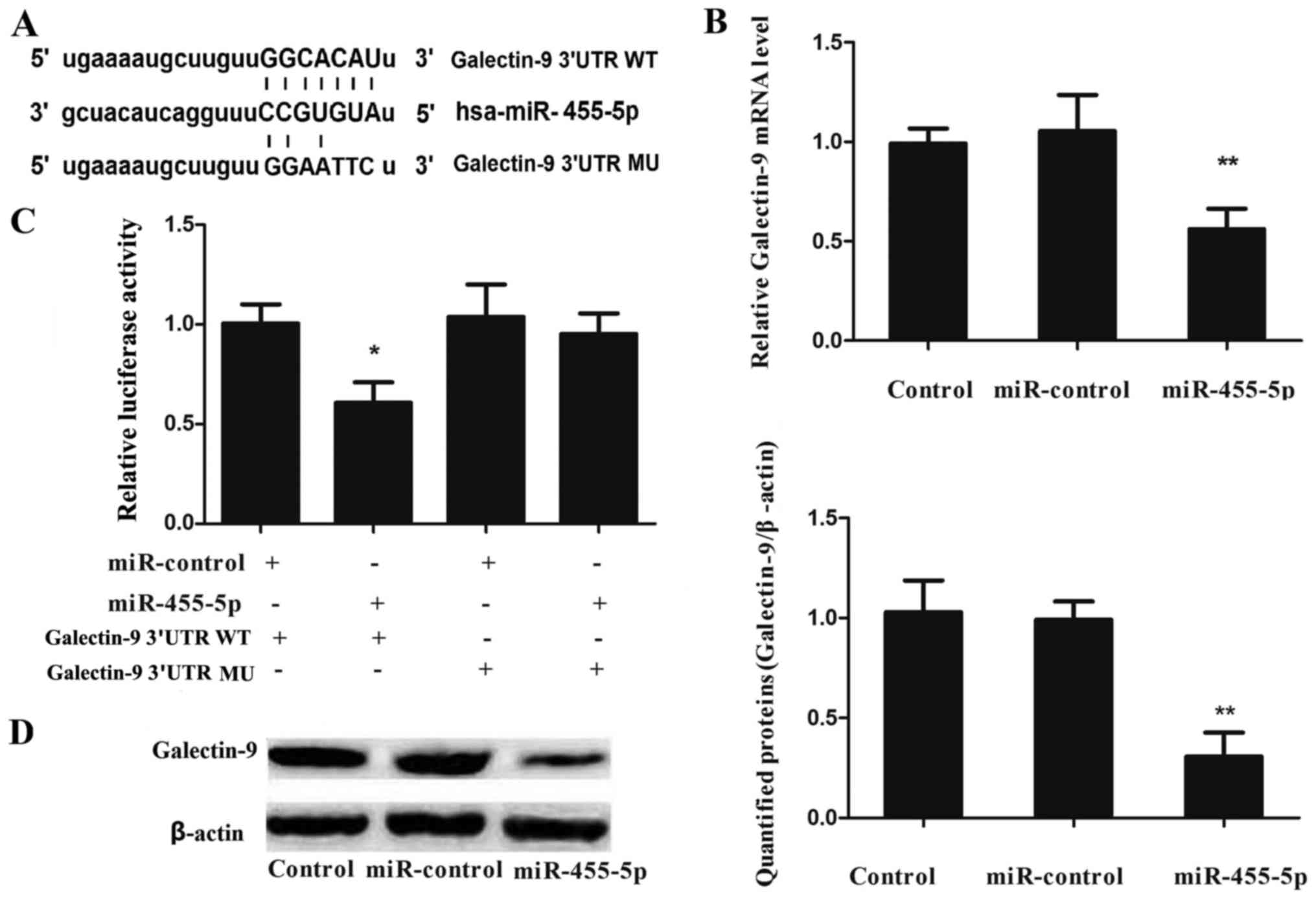
miR-455-5p directly targets and
downregulates galectin-9 in HT29 cells
To validate whether galectin-9 was a direct target
of miR-455-5p, the present study constructed galectin-9 3′-UTR-WT
and galectin-9 3′-UTR-MU plasmids (Fig.
4A), and transfected them into HT29 cells with miR-control or
miR-455-5p mimic, respectively. Luciferase activity was measured 24
h after transfection. The luciferase activity in the cells
co-transfected with miR-455-5p mimic and galectin-9 3′-UTR-WT
plasmid significantly decreased compared with that in cells
co-transfected with miR-control mimic or galectin-9 3′-UTR-MU
plasmid (Fig. 4B).
To investigate the potential correlation between
miR-455-5p and galectin-9 mRNA/protein expression in colon cancer
cells, miR-control or miR-455-5p mimic were transfected into HT29
cells, and the mRNA/protein expression levels of galectin-9 were
examined by RT-qPCR and western blot analysis, respectively. The
expression of galectin-9 mRNA (P=0.0043; Fig. 4C) and protein (P=0.0027; Fig. 4D) were observed to be significantly
downregulated subsequent to transfection with miR-22 mimics,
compared with the results obtained for the miR-control. These
findings indicate that galectin-9 is a direct downstream target of
miR-455-5p in HT29 cells.
Discussion
Galectin-9 has been detected extracellularly and
intracellularly, and the expression of the protein is widely
distributed in tissues (27,28). Indeed, the available results suggest
that galectin-9 expression is frequently altered when comparing
tumor tissue with normal tissue (29). In addition, one study supports the
hypothesis that galectin-9 is involved in several aspects of tumor
progression (30). Galectin-9 also
induces the apoptosis of various cell types, including human
melanoma, T cell and leukemia cell lines (13,31,32). The
present study explored the expression and potential role of
galectin-9 in colon cancer, and noticed that galectin-9 was
significantly downregulated in colon cancer tissue compared with
corresponding adjacent tissue. Furthermore, overexpression of
galectin-9 inhibited HT29 cell proliferation and promoted HT29 cell
apoptosis. These results suggest that galectin-9 is important in
colon cancer.
Numerous studies highlight the impact of miRNA on
the tumorigenesis of human carcinoma (33–35). To
define the upstream regulatory miRNA of galectin-9, the target
prediction programs miRanda and TargetScan were used to predict
miRNAs that possibly target galectin-9. miR-22, 296-3p, 455-5p and
491-5p were identified as potential miRNAs, and their expression
levels were measured in colon cancer and corresponding adjacent
tissues. Of these four miRNAs, miR-455-5p was significantly
upregulated, while miR-22 was downregulated, in colon cancer tissue
compared with the corresponding adjacent tissue. By contrast,
miR-296-3p and 491-5p did not exhibit a significant difference.
miR-455-5p has recently been shown to be important in the
progression of numerous types of malignancy. Liu et al
(36) identified that miR-455-5p was
associated with anaplastic lymphoma kinase expression. Shoshan
et al (37) confirmed that
miR-455-5p contributes to melanoma growth and metastasis through
the downregulation of the tumor-suppressor gene cytoplasmic
polyadenylation element binding protein 1. However, the exact role
with respect to influencing cell proliferation and apoptosis, and
the regulatory mechanism of miR-455-5p in colon cancer remains
unclear. The present study demonstrated that miR-455-5p promoted
HT29 cell proliferation and inhibited HT29 cell apoptosis. These
results suggested that miR-455-5p serves an oncogenic role in colon
cancer. In addition, miR-455-5p and galectin-9 expression exhibited
an inverse correlation and role in influencing cell proliferation
and apoptosis, which provides a foundation for additional
investigation into their association.
To explore whether miR-455-5p is involved in the
regulation of galectin-9 expression, a Dual-Luciferase Reporter
Assay System was employed. The luciferase reporter assay indicated
that the luciferase activity of the reporter containing the
wide-type 3′-UTR of the galectin-9 gene decreased following
treatment with miR-455-5p mimic, indicating that miR-455-5p
suppresses gene expression through miR-455-5p-binding sequences at
the 3′-UTR of galectin-9. In addition, RT-qPCR and western blot
analysis revealed that the mRNA and protein expression of
galectin-9 was inhibited by treatment with the miR-455-5p mimic in
HT29 cells. These data suggest that miR-455-5p reduces galectin-9
expression by inhibiting translation and/or causing mRNA
instability.
In summary, the present study provides evidence that
miR-455-5p mediates the downregulation of galectin-9 in colon
cancer. The present study demonstrated that miR-455-5p promoted
HT29 cell proliferation and inhibited HT29 cell apoptosis by
suppressing galectin-9 expression. miRNA-455-5p functions as a
potential oncogene in colon cancer, and the miRNA-455-5p/galectin-9
axis provides a novel insight into the pathogenesis of colon
cancer.
References
|
1
|
Neri F, Dettori D, Incarnato D, Krepelova
A, Rapelli S, Maldotti M, Parlato C, Paliogiannis P and Oliviero S:
TET1 is a tumour suppressor that inhibits colon cancer growth by
derepressing inhibitors of the WNT pathway. Oncogene. 34:4168–4176.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Slattery ML, Pellatt DF, Mullany LE, Wolff
RK and Herrick JS: Gene expression in colon cancer: A focus on
tumor site and molecular phenotype. Genes Chromosomes Cancer.
54:527–541. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
van Duijnhoven FJ, Bueno-De-Mesquita HB,
Ferrari P, Jenab M, Boshuizen HC, Ros MM, Casagrande C, Tjønneland
A, Olsen A, Overvad K, et al: Fruit, vegetables, and colorectal
cancer risk: The European Prospective Investigation into Cancer and
Nutrition. Am J Clin Nutr. 89:1441–1452. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
McCormack VA and Boffetta P: Today's
lifestyles, tomorrow's cancers: Trends in lifestyle risk factors
for cancer in low- and middle-income countries. Ann Oncol.
22:2349–2357. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kan JY, Hsu YL, Chen YH, Chen TC, Wang JY
and Kuo PL: Gemifloxacin, a fluoroquinolone antimicrobial drug,
inhibits migration and invasion of human colon cancer cells. Biomed
Res Int. 2013:1597862013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Abbadessa B, Agnew J, Bonomo G, et al:
Overall survival is negatively impacted by postoperative
complications following curative resection of rectal cancer but not
colon cancer. Dis Colon Rectum. 56:E261–E262. 2013.
|
|
7
|
Leffler H, Carlsson S, Hedlund M, Qian Y
and Poirier F: Introduction to galectins. Glycoconj J. 19:433–440.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wiersma VR, de Bruyn M, Helfrich W and
Bremer E: Therapeutic potential of Galectin-9 in human disease. Med
Res Rev. 33:(Suppl 1). E102–E126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Matsumoto R, Hirashima M, Kita H and
Gleich GJ: Biological activities of ecalectin: A novel
eosinophil-activating factor. J Immunol. 168:1961–1967. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Matsumoto R, Matsumoto H, Seki M, Hata M,
Asano Y, Kanegasaki S, Stevens RL and Hirashima M: Human ecalectin,
a variant of human galectin-9, is a novel eosinophil
chemoattractant produced by T lymphocytes. J Biol Chem.
273:16976–16984. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Niwa H, Satoh T, Matsushima Y, Hosoya K,
Saeki K, Niki T, Hirashima M and Yokozeki H: Stable form of
galectin-9, a Tim-3 ligand, inhibits contact hypersensitivity and
psoriatic reactions: A potent therapeutic tool for Th1- and/or
Th17-mediated skin inflammation. Clin Immunol. 132:184–194. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abedin MJ, Kashio Y, Seki M, Nakamura K
and Hirashima M: Potential roles of galectins in myeloid
differentiation into three different lineages. J Leukoc Biol.
73:650–656. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kashio Y, Nakamura K, Abedin MJ, Seki M,
Nishi N, Yoshida N, Nakamura T and Hirashima M: Galectin-9 induces
apoptosis through the calcium-calpain-caspase-1 pathway. J Immunol.
170:3631–3636. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nobumoto A, Nagahara K, Oomizu S, Katoh S,
Nishi N, Takeshita K, Niki T, Tominaga A, Yamauchi A and Hirashima
M: Galectin-9 suppresses tumor metastasis by blocking adhesion to
endothelium and extracellular matrices. Glycobiology. 18:735–744.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang ZY, Dong JH, Chen YW, Wang XQ, Li
CH, Wang J, Wang GQ, Li HL and Wang XD: Galectin-9 acts as a
prognostic factor with antimetastatic potential in hepatocellular
carcinoma. Asian Pac J Cancer Prev. 13:2503–2509. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kozak M: Faulty old ideas about
translational regulation paved the way for current confusion about
how microRNAs function. Gene. 423:108–115. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qu J, Zhao L, Zhang P, Wang J, Xu N, Mi W,
Jiang X, Zhang C and Qu J: MicroRNA-195 chemosensitizes colon
cancer cells to the chemotherapeutic drug doxorubicin by targeting
the first binding site of BCL2L2 mRNA. J Cell Physiol. 230:535–545.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ren XS, Yin MH, Zhang X, Wang Z, Feng SP,
Wang GX, Luo YJ, Liang PZ, Yang XQ, He JX and Zhang BL:
Tumor-suppressive microRNA-449a induces growth arrest and
senescence by targeting E2F3 in human lung cancer cells. Cancer
Lett. 344:195–203. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kasashima K, Nakamura Y and Kozu T:
Altered expression profiles of microRNAs during TPA-induced
differentiation of HL-60 cells. Biochem Biophys Res Commun.
322:403–410. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McManus MT: MicroRNAs and cancer. Semin
Cancer Biol. 13:253–258. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Miao J, Wu S, Peng Z, Tania M and Zhang C:
MicroRNAs in osteosarcoma: Diagnostic and therapeutic aspects.
Tumour Biol. 34:2093–2098. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang Q, Jiang W, Zhuang C, Geng Z, Hou C,
Huang D, Hu L and Wang X: microRNA-22 downregulation of galectin-9
influences lymphocyte apoptosis and tumor cell proliferation in
liver cancer. Oncol Rep. 34:1771–1778. 2015.PubMed/NCBI
|
|
27
|
Thijssen VL, Hulsmans S and Griffioen AW:
The galectin profile of the endothelium: Altered expression and
localization in activated and tumor endothelial cells. Am J Pathol.
172:545–553. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Türeci O, Schmitt H, Fadle N, Pfreundschuh
M and Sahin U: Molecular definition of a novel human galectin which
is immunogenic in patients with Hodgkin's disease. J Biol Chem.
272:6416–6422. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu Z, Han H, He X, Li S, Wu C, Yu C and
Wang S: Expression of the galectin-9-Tim-3 pathway in glioma
tissues is associated with the clinical manifestations of glioma.
Oncol Lett. 11:1829–1834. 2016.PubMed/NCBI
|
|
30
|
Heusschen R, Griffioen AW and Thijssen VL:
Galectin-9 in tumor biology: A jack of multiple trades. Biochim
Biophys Acta. 1836:177–185. 2013.PubMed/NCBI
|
|
31
|
Saita N, Goto E, Yamamoto T, Cho I,
Tsumori K, Kohrogi H, Maruo K, Ono T, Takeya M, Kashio Y, et al:
Association of galectin-9 with eosinophil apoptosis. Int Arch
Allergy Immunol. 128:42–50. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kobayashi T, Kuroda J, Ashihara E, Oomizu
S, Terui Y, Taniyama A, Adachi S, Takagi T, Yamamoto M, Sasaki N,
et al: Galectin-9 exhibits anti-myeloma activity through JNK and
p38 MAP kinase pathways. Leukemia. 24:843–850. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang YW, Shi DB, Chen X, Gao C and Gao P:
Clinicopathological significance of microRNA-214 in gastric cancer
and its effect on cell biological behaviour. PLoS One.
9:e913072014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu CL, Iqbal J, Teruya-Feldstein J, Shen
Y, Dabrowska MJ, Dybkaer K, Lim MS, Piva R, Barreca A, Pellegrino
E, et al: microRNA expression profiling identifies molecular
signatures associated with anaplastic large cell lymphoma. Blood.
122:2083–2092. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shoshan E, Mobley AK, Braeuer RR, Kamiya
T, Huang L, Vasquez ME, Salameh A, Lee HJ, Kim SJ, Ivan C, et al:
Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma
growth and metastasis. Nat Cell Biol. 17:311–321. 2015. View Article : Google Scholar : PubMed/NCBI
|