Introduction
Breast cancer is one of the most common types of
cancer diagnosed in women worldwide (1), and nipple discharge is one of the most
common symptoms in women presenting with breast complaints
(2). Intraductal papilloma, a benign
tumor, is the most common cause of abnormal nipple discharge
(3); however, ~5–15% of cases are
associated with a malignant lesion (4,5). There are
no clear guidelines on what differentiates benign with malignant
etiology based on clinical and radiographic assessments (6), and >20% of patient with cancer have
no obvious clinical or radiological evidence of breast carcinoma
(7). The use of nipple discharge
cytology for diagnosis is currently limited due to its low
sensitivity of 7% (8); therefore, the
identification of novel biomarkers in nipple discharge to detect
breast cancer at an early stage is essential for improving the
treatment and survival of patients.
MicroRNA (miRNA/miR) is a class of non-coding small
RNA that can induce mRNA degradation and/or translational
repression, through binding to the 3′ untranslated region (UTR) of
mRNAs and thereby negatively regulating the transcription of target
genes (9). It has been demonstrated
that miRNAs are associated with numerous essential biological
processes, including proliferation, apoptosis, development and
differentiation, in addition to human cancer initiation and
progression (10,11). Previous studies have demonstrated that
the deregulation of miRNAs in human cancer is tissue- and/or
time-specific, highlighting their potential roles as biomarkers
(12,13). For example, let-7 was identified as a
diagnostic biomarker for certain lung cancer subtypes, in addition
to a prognostic marker for disease progression and response to
treatment (14–16). Cui et al (17) reported that miR-21 and miR-106a in
gastric secretions are potential diagnostic biomarkers for gastric
cancer. Improved understanding of the molecular mechanisms by which
miRNAs function in breast cancer will be beneficial for the
development of novel approaches for the early detection and
monitoring of breast cancer. A previous study identified a panel of
miRNAs, including miR-3646, −4484 and −4732-5p, in nipple discharge
as potential diagnostic biomarkers for breast cancer (18). In the present study, the potential
functions of miR-3646, −4484 and −4732-5p, and the miRNA-associated
pathways in breast cancer, were analyzed.
Materials and methods
Target gene prediction
The target gene prediction databases Targetscan
(version 6.2; http://www.targetscan.org) and MicroRNA Target
Prediction and Functional Study Database (miRDB; http://mirdb.org/miRDB) (19) were used to identify potential targets
of miR-3646, −4484 and −4732-5p.
Enrichment analysis
A gene ontology (GO) term enrichment analysis
(http://www.geneontology.org) (20) was performed to identify biological
processes, molecular functions and cellular components associated
with the target genes of miR-3646, miR-4484 and miR-4732-5p
predicted by Targetscan. Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway enrichment analysis (http://www.genome.jp/kegg) (21) was used to identify the miRNA targets
associated with signaling pathways in breast cancer. P<0.05 was
considered to indicate a statistically significant difference. For
better visualization, the top ten GO terms and pathways (if any)
were presented. A protein-protein interaction analysis using the
Search Tool for the Retrieval of Interacting Genes (version 9.1;
http://string91.embl.de/) database (22) was performed to test whether the
targets were associated with each other.
Transcription factor (TF)-mRNA network
construction
The potential TF-mRNA pairs were obtained from the
tfbsConsSites table, which contains transcription factor binding
sites conserved in the human/mouse/rat alignment (downloaded from
the University of California Santa Cruz genome browser; http://genome.ucsc.edu/cgi-bin/hgTables). Binding
sites located in the promoter region (from-2,000 bp to +500 bp of
the transcription start site) of specified coding genes and with Z
scores >2.33 were selected for further processing. Subsequently,
Cytoscape version 3.2.0 software (http://www.cytoscape.org/) was used to visualize the
TF-mRNA regulatory network.
Results
Potential target genes of
miR-3646,-4484 and −4732-5p
The miRDB and Targetscan databases were used to
predict the target genes of miR-3646, −4484 and −4732-5p. The
numbers of target genes predicted by the databases for miR-3646,
−4484 and −4732-5p were 574, 12 and 10, respectively (Fig. 1A-C). Based on the prediction databases
scores and the literature, a number of the target genes were
selected for further investigation (Fig.
1D). The selected target genes included adiponectin, C1Q and
collagen domain containing adiponectin (ADIPOQ), A-kinase anchoring
protein 12, coiled-coil domain containing (CCDC) 50, CCDC6,
cytoplasmic polyadenylation element-binding protein 1 (CPEB1), DnaJ
heat shock protein family (Hsp40) member B4 (DNAJB4) and eukaryotic
translation initiation factor 4E (EIF4E) for miR-3646; amyloid
precursor protein (APP), EIF4E, methionine-R-sulfoxide reductase
B3, patched 2 and TATA-box binding protein associated factor 4 for
miR-4484; and ALKBH5, ankyrin repeat domain 17, BCL2-associated
transcription factor 1 (BCLAF1) and protein phosphatase 1E for
miR-4732-5p. Notably, EIF4E was identified as a mutual target of
miR-3646 and miR-4484.
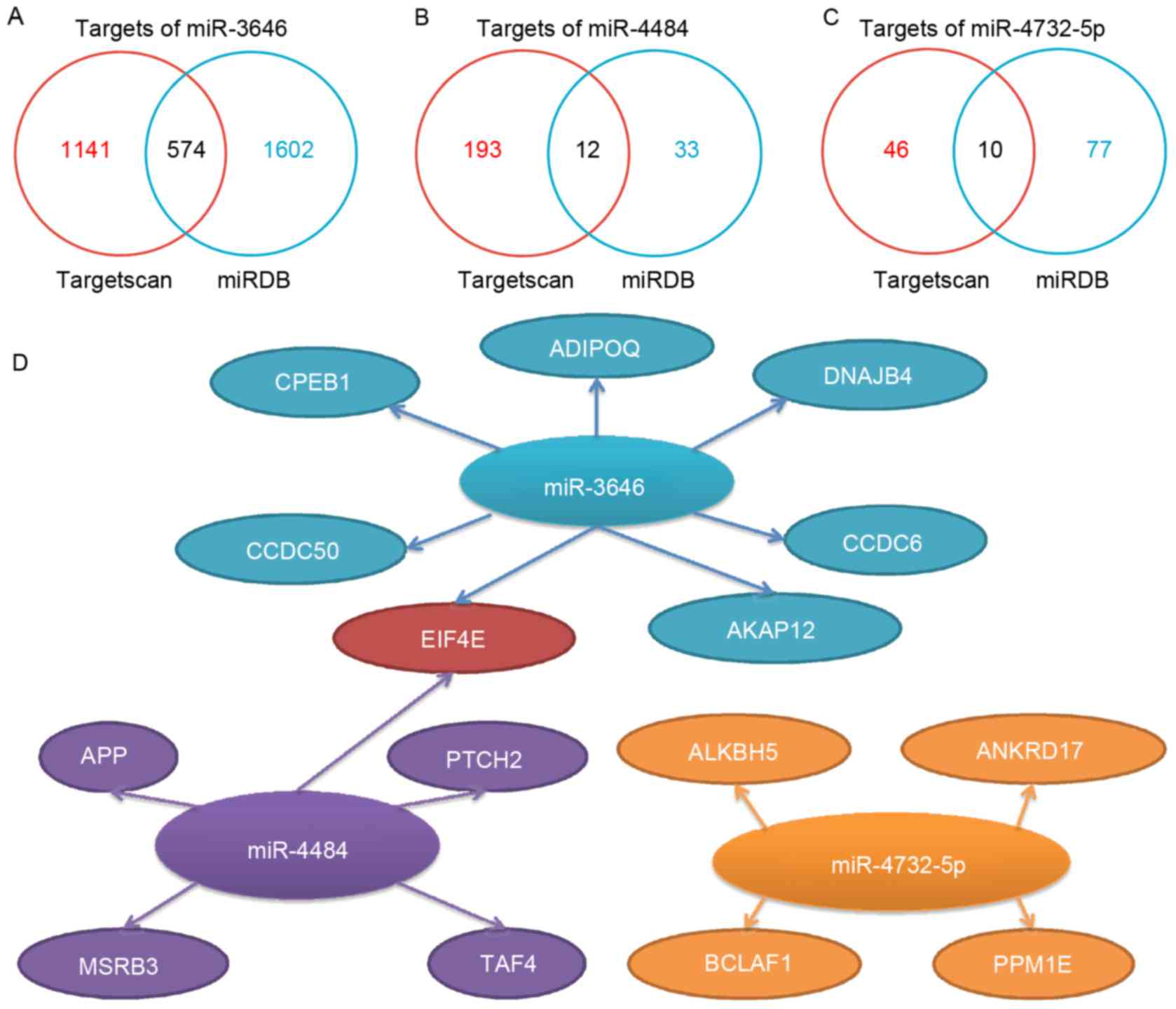 | Figure 1.Predicted target genes for miR-3646,
−4484 and −4732-5p. The number of the potential targets predicted
by Targetscan and miRDB are illustrated for miR (A) −3646, (B)
−4484 and (C) −4732-5p. (D) Representative predicted targets of
miR-3646, −4484, −4732-5p. The main criteria for selecting
representative genes are based on their functions from previous
cancer-associated studies, in combination with the targets scores
provided by Targetscan (cumulative weighted context and score) and
miRDB (Target Score). miR, microRNA; miRDB, MicroRNA Target
Prediction and Functional Study Database; CPEB1, cytoplasmic
polyadenylation element-binding protein 1; APP, amyloid precursor
protein; ADIPOQ, C1Q and collagen domain containing adiponectin;
DNAJB4, DnaJ heat shock protein family (Hsp40) member B4; CCDC,
coiled-coil domain containing; AKAP12, A-kinase anchoring protein
12; EIF4E, eukaryotic translation initiation factor 4E; PTCH2,
patched 2; TAF4, TATA-box binding protein associated factor 4;
MSRB3, methionine-R-sulfoxide reductase B3; ANKRD17, ankyrin repeat
domain 17; PPM1E, protein phosphatase 1E; BCLAF1, BCL2-associated
transcription factor 1. |
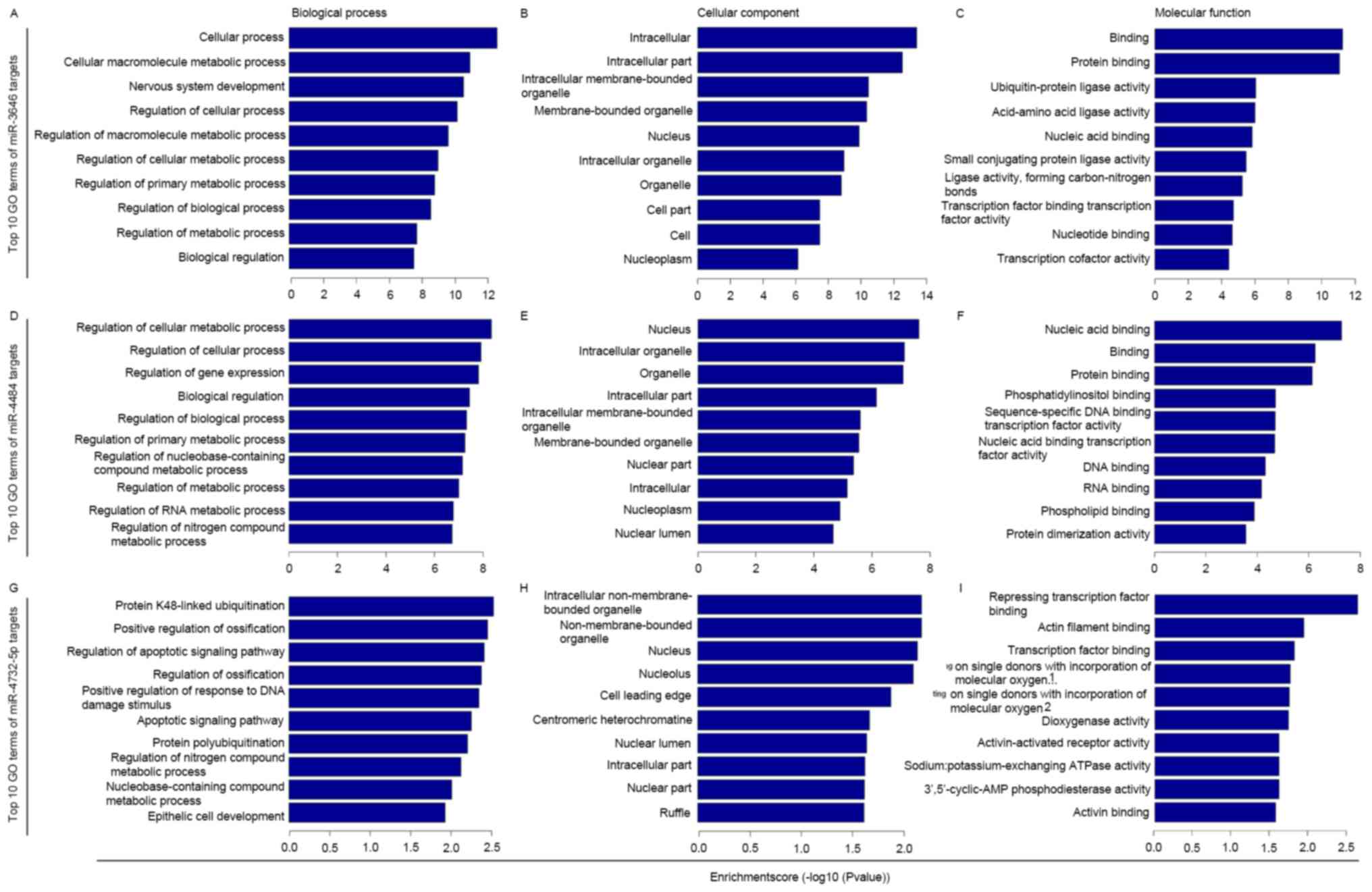
GO and KEGG enrichment analysis of the
putative targets of miR-3646,-4484 and-4732-5p
GO term enrichment analysis was performed for the
selected putative targets of miR-3646, −4484 and −4732-5p. The
results revealed that the targets were significantly involved in a
number of biological, cellular and molecular processes (P<0.05;
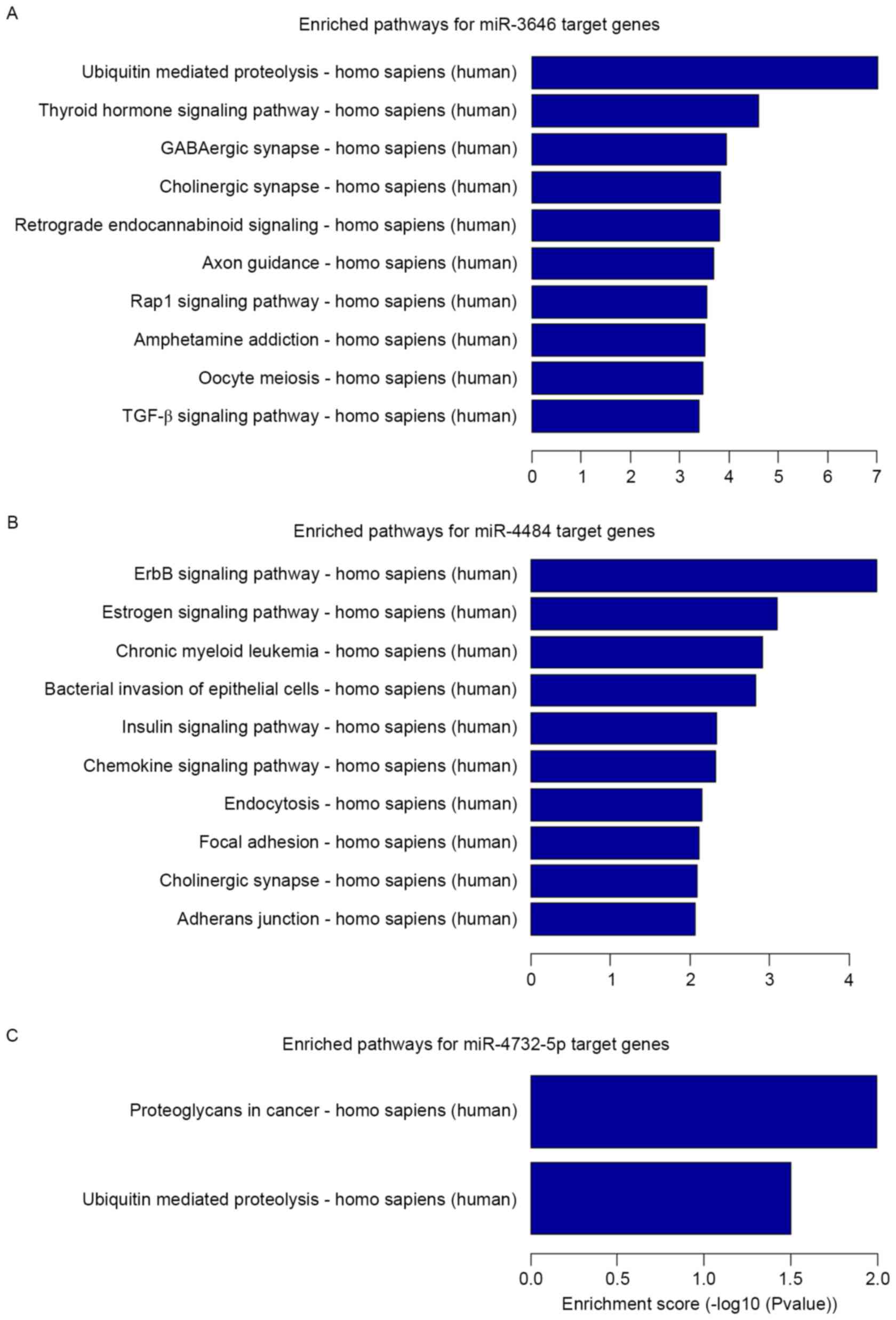
Fig. 2). KEGG pathway enrichment
analysis revealed that the miRNA targets were implicated in a
number of pathways that are associated with carcinogenesis and
tumor progression (Fig. 3). The
miR-3646 targets (P<0.05; Fig. 3A)
were significantly implicated in ubiquitin mediated proteolysis,
the thyroid hormone signaling pathway, the γ-aminobutyric acidergic
and cholinergic synapses, retrograde endocannabinoid signaling,
axon guidance, the Ras-related protein Rap1 signaling pathway,
amphetamine addiction, oocyte meiosis and the transforming growth
factor β (TGF-β) signaling pathway. The miR-4484 targets
(P<0.05; Fig. 3B) were
significantly implicated in epidermal growth factor receptor (ErbB)
signaling, estrogen signaling, chronic myeloid leukemia, bacterial
invasion of epithelial cells, insulin signaling, chemokine
signaling, endocytosis, focal adhesions, the cholinergic synapse
and adherens junction. For the miR-4732-5p targets (P<0.05;
Fig. 3C) the significantly enriched
pathways were proteoglycans in cancer and ubiquitin mediated
proteolysis.
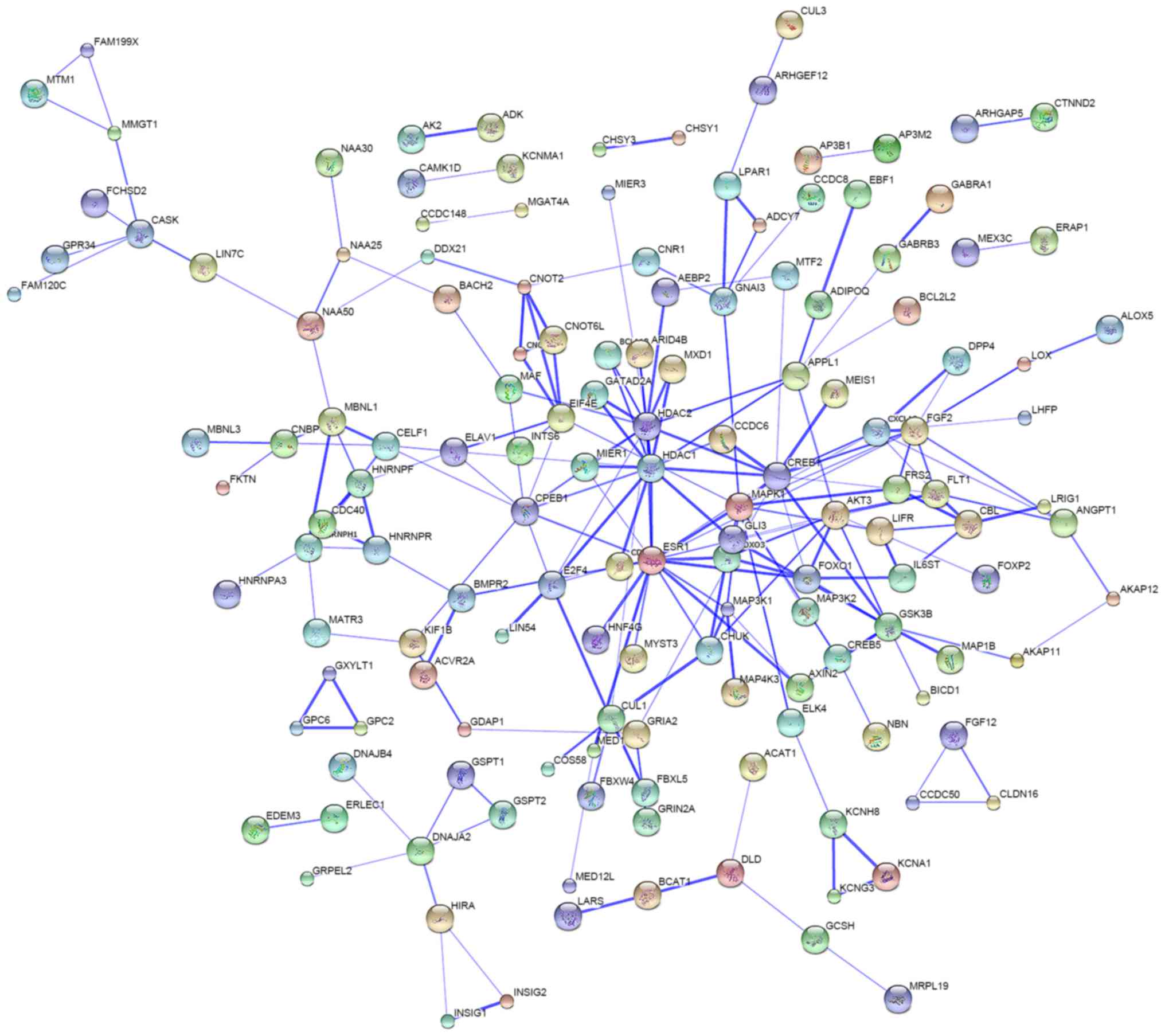
Protein-protein interaction
analysis
Protein interaction analysis was performed to
investigate the interactions of the proteins translated from the
mRNA targets of miR-3646, −4484 and −4732-5p. The data identified a
number of proteins that have a potential association with each
other (Fig. 4), suggesting that the
miRNA-regulated genes may form a crosstalk network in modulating
breast cancer development and progression.
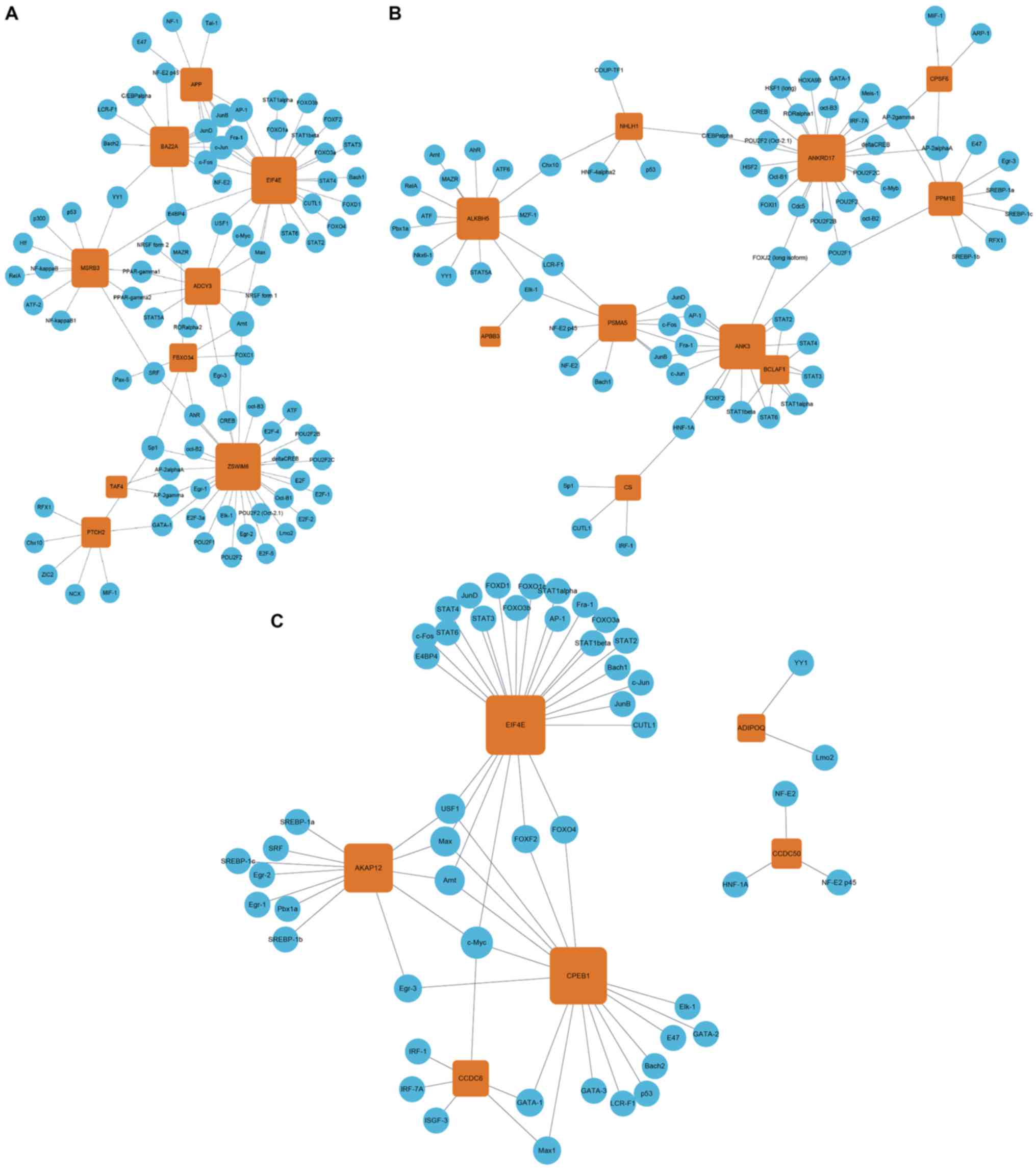
TF-mRNA regulatory network
construction
Following the identification of TF-mRNA pairs using
the UCSC genome browser, a TF-mRNA regulatory network was
constructed (Fig. 5). The data
suggested that the miRNA target genes are under the regulation of
numerous transcription factors, including cyclic AMP-dependent
transcription factor (ATF), cyclic AMP-responsive element-binding
protein 1 (CREB), CCAAT/enhancer-binding protein α (C/EBPα),
proto-oncogene c-Fos (c-Fos), transcription factor AP-1 (c-Jun),
myc proto-oncogene protein (c-Myc), transcription factor E2F1
(E2F), transcription factor JunB and JunD (JunB and JunD), cellular
tumor antigen p53 (p53), transcription factor SP1 (SP1), signal
transducer and activator of transcription 3 (STAT3), and
transcription factor p65 (NF-κβ). For miR-4484, ZSWIM6 was the
target gene regulated by the majority of TFs (n=28; Fig. 5A). For miR-4732-5p, ANKRD17 was the
target gene regulated by the largest number of TFs (n=24; Fig. 5B), and for miR-3646, EIF4E was the
target gene regulated by the largest number of TFs (n=25; Fig. 5C).
Discussion
Nipple discharge is one of the most common
complaints for patients with breast lesions (23). Identifying biomarkers in nipple
discharge that would allow breast cancers and benign breast lesions
to be distinguished, which traditional radiological and cytological
examinations cannot do, would be critical in improving diagnosis
(24,25). The expression signatures of miRNAs can
be informative in cancer diagnosis (13). The dysregulated expression of
miR-3646, −4484 and −4732-5p in the nipple discharge of patients
with breast cancer compared with that of patients with benign
breast lesions has previously been reported (26), implicating miRNAs as novel tumor
biomarkers for the diagnosis of breast cancer. In the present
study, bioinformatics analysis was performed to explore the
potential functions and underlying mechanisms of miR-3646, −4484
and −4732-5p in breast cancer.
The results of the present study revealed that
numerous genes associated with cell proliferation, apoptosis and
metastasis are potential targets of miR-3646, −4484 and −4732-5p.
For example, DNAJB4, a predicted target of miR-3646 in the present
study has been reported to be downregulated in colorectal carcinoma
and correlated with cancer cell metastasis, disease progression and
patient survival (27).
Simões-Correia et al (28)
demonstrated that the expression level of DNAJB4 was decreased in
human gastric cancer and inhibited cancer cell invasion in
vivo. DNAJB4 has also been identified as a tumor suppressor in
lung cancer, in which it inhibits cell proliferation,
anchorage-independent growth, tumorigenesis, cell motility and
invasion (29,30). Another target of miR-3646, ADIPOQ, has
been associated with breast cancer risk (31) and cell invasion in breast cancer
(32). More recently, Chen et
al (33) identified ADIPOQ as one
of the most downregulated genes in breast cancer in the Asian
population. CPEB1, another potential target of mIR-3646, is
downregulated in breast, ovarian and gastric cancers (34–36) and is
associated with cell growth, senescence and migration in glioma
(37,38). APP, a potential target of miR-4484,
has been implicated in several human malignancies and contributes
to the proliferation and migration of prostate cancer cells by
modulating the expression of metalloproteinases and
epithelial-mesenchymal transition-associated genes (39). The apoptosis-associated gene BCLAF1
(40), a potential target of
miR-4732-5p, has been associated with tumor recurrence in
esophageal squamous cell carcinoma (41).
Notably, EIF4E was identified as a shared target of
miR-3646 and miR-4484. EIF4E is deregulated in a number of human
malignancies, including lung, bladder, colon, breast, prostate,
cervical and ovarian cancer (42).
Pettersson et al (43)
suggested that EIF4E silencing could reduce the invasiveness and
metastatic capability of breast cancer cells. EIF4E is
overexpressed in invasive breast ductal carcinoma and is associated
with the occurrence and metastasis of breast cancer (44). Recently, high EIF4E expression has
been correlated with an increased risk of systemic metastasis in
patients with lymph node-positive breast cancer patients (45). There results indicate that miR-3646,
−4484 and −4732-5p may be associated with several essential
cellular processes, including cell proliferation, apoptosis and
metastasis, and serve critical roles in cancer initiation and
progression.
In order to investigate the underlying functional
mechanisms of miR-3646, −4484 and −4732-5p KEGG pathway enrichment
analysis was performed. As expected, the target genes of these
miRNAs were significantly enriched in several cancer-associated
signaling pathways, including those of Rap1, TGF-β, ErbB, estrogen,
focal adhesion and proteoglycans. Spanjaard et al (46) demonstrated that focal adhesion size,
sliding and intensity were regulated by the Rap1 signaling pathway
and in turn could influence tumor metastasis. High Rap1 activity is
correlated with poor differentiation and an advanced tumor stage in
serous ovarian cancer (47). The Rap1
signaling pathway has also been associated with breast tumor cell
migration and invasion, and breast cancer progression (48–51). TGF-β
signaling regulates diverse cellular processes, including cell
proliferation, differentiation, apoptosis, plasticity and
migration, and thus serves important roles in the development and
progression of various kinds of diseases, including human cancer
(52). TGF-β serves a tumorigenic
role, but it can also act as a tumor suppressor depending on the
cellular context and tumor stage (53). TGF-β signaling pathways have been
reported to be associated with epithelial-mesenchymal transition,
breast cancer stem cell maintenance and breast cancer metastasis
(54,55). Additionally, estrogen and ErbB
signaling pathways are associated with breast carcinogenesis,
invasion and cell growth (56).
Notably, agents co-targeting the estrogen and ErbB signaling
pathways have been used in the clinic and demonstrated a benefit
for breast cancer patients (56). The
results of the current study suggest that miR-3646, −4484 and
−4732-5p serve important roles in human cancers, including breast
cancer. However, this requires experimental validation and further
investigation.
To the best of our knowledge, there are few previous
studies investigating miR-3646, −4484 and −4732-5p. Rogler et
al (57) reported that miR-3646
exhibited strong expression in Caucasian bladder cancer cell lines
and tumor tissues. Only one report identified a clinical
significance of miR-4484 in human cancer; high serum expression of
miR-4484 was found to predict lymph node metastasis in patients
with early-stage cervical squamous cell carcinoma (58). Pouladi et al (59) predicted a binding site for miR-4732-5p
in the 5′ UTR of telomerase cajal body protein 1 mRNA. In addition,
miR-4732-5p has been revealed to be closely associated with
recurrence in gastric cancer patients treated with S-1 adjuvant
chemotherapy (60). Importantly, the
TF-mRNA regulatory network constructed in the present study
revealed that the target genes of the miRNAs investigated were
potentially regulated by numerous transcription factors, which
highlights the critical roles of these miRNAs and their targets in
physiological and pathological processes. Given the uncharacterized
roles and underlying molecular mechanisms of miR-3646, −4484 and
−4732-5p in human cancer, additional studies are required to fully
explore their functions. The results of the bioinformatics analysis
performed in the present study provide a theoretical basis for
further research.
In conclusion, the present study identified that
miR-3646, −4484 and −4732-5p are associated with breast cancer
development and progression, and in particular with certain
distinct signaling pathways within these processes. The target
genes of miR-3646, −4484 and −4732-5p were predicted in the current
study, with the findings indicating that miR-3646, −4484 and
−4732-5p are associated with signaling pathways important in
cancer, including breast carcinogenesis and progression. The
current study provides a comprehensive view of the functions and
molecular mechanisms of miR-3646, −4484 and −4732-5p from a
bioinformatics perspective, and will aid in future experimental
studies.
Acknowledgements
The present study was supported by the Shandong Key
Research and Development Plan (grant no. 2016GSF201128). The
authors thank Cloud-Seq Company (Shanghai, China) for the technical
assistance in bioinformatic analysis.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gülay H, Bora S, Kilicturgay S, Hamaloglu
E and Göksel HA: Management of nipple discharge. J Am Coll Surg.
178:471–474. 1994.PubMed/NCBI
|
|
3
|
Vargas HI, Vargas MP, Eldrageely K,
Gonzalez KD and Khalkhali I: Outcomes of clinical and surgical
assessment of women with pathological nipple discharge. Am Surg.
72:124–128. 2006.PubMed/NCBI
|
|
4
|
Jardines L: Management of nipple
discharge. Am Surg. 62:119–122. 1996.PubMed/NCBI
|
|
5
|
Murad TM, Contesso G and Mouriesse H:
Nipple discharge from the breast. Ann Surg. 195:259–264. 1982.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sabel MS, Helvie MA, Breslin T, Curry A,
Diehl KM, Cimmino VM, Chang AE and Newman LA: Is duct excision
still necessary for all cases of suspicious nipple discharge?
Breast J. 18:157–162. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kapenhas-Valdes E, Feldman SM and Boolbol
SK: The role of mammary ductoscopy in breast cancer: A review of
the literature. Ann Surg Oncol. 15:3350–3360. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
López-Ríos F, Vargas-Castrillón J,
González-Palacios F and de Agustín PP: Breast carcinoma in situ in
a male. Report of a case diagnosed by nipple discharge cytology.
Acta cytol. 42:742–744. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tsukamoto Y, Nakada C, Noguchi T, Tanigawa
M, Nguyen LT, Uchida T, Hijiya N, Matsuura K, Fujioka T, Seto M and
Moriyama M: MicroRNA-375 is downregulated in gastric carcinomas and
regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer
Res. 70:2339–2349. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang YW, Shi DB, Chen X, Gao C and Gao P:
Clinicopathological significance of microRNA-214 in gastric cancer
and its effect on cell biological behaviour. PloS one.
9:e913072014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fabbri M: miRNAs as molecular biomarkers
of cancer. Expert Rev Mol Diagn. 10:435–444. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang K, Zhang Y, Liu C, Xiong Y and Zhang
J: MicroRNAs in the diagnosis and prognosis of breast cancer and
their therapeutic potential (Review). Int J Oncol. 45:950–958.
2014.PubMed/NCBI
|
|
14
|
Zhong Z, Dong Z, Yang L, Chen X and Gong
Z: Inhibition of proliferation of human lung cancer cells by green
tea catechins is mediated by upregulation of let-7. Exp Ther Med.
4:267–272. 2012.PubMed/NCBI
|
|
15
|
Takamizawa J, Konishi H, Yanagisawa K,
Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y,
et al: Reduced expression of the let-7 microRNAs in human lung
cancers in association with shortened postoperative survival.
Cancer Res. 64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stahlhut C and Slack FJ: Combinatorial
ction of microRNAs let-7 and miR-34 effectively synergizes with
erlotinib to suppress non-small cell lung cancer cell
proliferation. Cell Cycle. 14:2171–2180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cui L, Zhang X, Ye G, Zheng T, Song H,
Deng H, Xiao B, Xia T, Yu X, Le Y and Guo J: Gastric juice
microRNAs as potential biomarkers for the screening of gastric
cancer. Cancer. 119:1618–1626. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang K, Zhao S, Wang Q, Yang HS, Zhu J
and Ma R: Identification of microRNAs in nipple discharge as
potential diagnostic biomarkers for breast cancer. Ann Surg Oncol.
22:(Suppl 3). S536–S544. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43:D146–D152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gene Ontology Consortium, Blake JA, Dolan
M, Drabkin H, Hill P, Li N, Sitnikov D, Bridges S, Burgess S, Buza
T, et al: Gene ontology annotations and resources. Nucleic Acids
Res. 41:D530–D535. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kanehisa M, Goto S, Sato Y, Furumichi M
and Tanabe M: KEGG for integration and interpretation of
large-scale molecular data sets. Nucleic Acids Res. 40:D109–D114.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vargas HI, Romero L and Chlebowski RT:
Management of bloody nipple discharge. Curr Treat Options Oncol.
3:157–161. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dinkel HP, Trusen A, Gassel AM, Rominger
M, Lourens S, Müller T and Tschammler A: Predictive value of
galactographic patterns for benign and malignant neoplasms of the
breast in patients with nipple discharge. Br J Radiol. 73:706–714.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sauter ER, Ehya H, Babb J, Diamandis E,
Daly M, Klein-Szanto A, Sigurdson E, Hoffman J, Malick J and
Engstrom PF: Biological markers of risk in nipple aspirate fluid
are associated with residual cancer and tumour size. Br J Cancer.
81:1222–1227. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang K, Zhao S, Wang Q, Yang HS, Zhu J
and Ma R: Identification of microRNAs in nipple discharge as
potential diagnostic biomarkers for breast cancer. Ann Surg Oncol.
22:(Suppl 3). S536–S544. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Y, Zhou J, Zhang C, Fu W, Xiao X, Ruan
S, Zhang Y, Luo X and Tang M: HLJ1 is a novel biomarker for
colorectal carcinoma progression and overall patient survival. Int
J Clin Exp Pathol. 7:969–977. 2014.PubMed/NCBI
|
|
28
|
Simões-Correia J, Silva DI, Melo S,
Figueiredo J, Caldeira J, Pinto MT, Girão H, Pereira P and Seruca
R: DNAJB4 molecular chaperone distinguishes WT from mutant
E-cadherin, determining their fate in vitro and in vivo. Hum Mol
Genet. 23:2094–2105. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen HW, Lee JY, Huang JY, Wang CC, Chen
WJ, Su SF, Huang CW, Ho CC, Chen JJ, Tsai MF, et al: Curcumin
inhibits lung cancer cell invasion and metastasis through the tumor
suppressor HLJ1. Cancer Res. 68:7428–7438. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tsai MF, Wang CC, Chang GC, Chen CY, Chen
HY, Cheng CL, Yang YP, Wu CY, Shih FY, Liu CC, et al: A new tumor
suppressor DnaJ-like heat shock protein, HLJ1 and survival of
patients with non-small-cell lung carcinoma. J Natl Cancer Inst.
98:825–838. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mantzoros C, Petridou E, Dessypris N,
Chavelas C, Dalamaga M, Alexe DM, Papadiamantis Y, Markopoulos C,
Spanos E, Chrousos G and Trichopoulos D: Adiponectin and breast
cancer risk. J Clin Endocrinol Metab. 89:1102–1107. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Libby Falk E, Liu J, Li YI, Lewis MJ,
Demark-Wahnefried W and Hurst DR: Globular adiponectin enhances
invasion in human breast cancer cells. Oncol Lett. 11:633–641.
2016.PubMed/NCBI
|
|
33
|
Chen L, Ye C, Huang Z, Li X, Yao G, Liu M,
Hu X, Dong J and Guo Z: Differentially expressed genes and
potential signaling pathway in Asian people with breast cancer by
preliminary analysis of a large sample of the microarray data. Nan
Fang Yi Ke Da Xue Xue Bao. 34:807–812. 2014.(In Chinese).
PubMed/NCBI
|
|
34
|
Nairismägi ML, Vislovukh A, Meng Q,
Kratassiouk G, Beldiman C, Petretich M, Groisman R, Füchtbauer EM,
Harel-Bellan A and Groisman I: Translational control of TWIST1
expression in MCF-10A cell lines recapitulating breast cancer
progression. Oncogene. 31:4960–4966. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hansen CN, Ketabi Z, Rosenstierne MW,
Palle C, Boesen HC and Norrild B: Expression of CPEB, GAPDH and
U6snRNA in cervical and ovarian tissue during cancer development.
APMIS. 117:53–59. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Caldeira J, Simões-Correia J, Paredes J,
Pinto MT, Sousa S, Corso G, Marrelli D, Roviello F, Pereira PS,
Weil D, et al: CPEB1, a novel gene silenced in gastric cancer: A
Drosophila approach. Gut. 61:1115–1123. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xiaoping L, Zhibin Y, Wenjuan L, Zeyou W,
Gang X, Zhaohui L, Ying Z, Minghua W and Guiyuan L: CPEB1, a
histone-modified hypomethylated gene, is regulated by miR-101 and
involved in cell senescence in glioma. Cell Death Dis. 4:e6752013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kochanek DM and Wells DG: CPEB1 regulates
the expression of MTDH/AEG-1 and glioblastoma cell migration. Mol
Cancer Res. 11:149–160. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Miyazaki T, Ikeda K, Horie-Inoue K and
Inoue S: Amyloid precursor protein regulates migration and
metalloproteinase gene expression in prostate cancer cells. Biochem
Biophys Res Commun. 452:828–833. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yoshitomi T, Kawakami K, Enokida H,
Chiyomaru T, Kagara I, Tatarano S, Yoshino H, Arimura H, Nishiyama
K, Seki N and Nakagawa M: Restoration of miR-517a expression
induces cell apoptosis in bladder cancer cell lines. Oncol Rep.
25:1661–1668. 2011.PubMed/NCBI
|
|
41
|
Chen Y, Wang Y, Song H, Wang J, Yang H,
Xia Y, Xue J, Li S, Chen M and Lu Y: Expression profile of
apoptosis-related genes potentially explains early recurrence after
definitive chemoradiation in esophageal squamous cell carcinoma.
Tumour Biol. 35:4339–4346. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Carroll M and Borden KL: The oncogene
eIF4E: using biochemical insights to target cancer. J Interferon
Cytokine Res. 33:227–238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pettersson F, Del Rincon SV, Emond A, Huor
B, Ngan E, Ng J, Dobocan MC, Siegel PM and Miller WH Jr: Genetic
and Pharmacologic inhibition of eIF4E reduces breast cancer cell
migration, invasion and metastasis. Cancer Res. 75:1102–1112. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hu A, Sun M, Yan D and Chen K: Clinical
significance of mTOR and eIF4E expression in invasive ductal
carcinoma. Tumori. 100:541–546. 2014.PubMed/NCBI
|
|
45
|
Yin X, Kim RH, Sun G, Miller JK and Li BD:
Overexpression of eukaryotic initiation factor 4E is correlated
with increased risk for systemic dissemination in node-positive
breast cancer patients. J Am Coll Surg. 218:663–671. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Spanjaard E, Smal I, Angelopoulos N,
Verlaan I, Matov A, Meijering E, Wessels L, Bos H and de Rooij J:
Quantitative imaging of focal adhesion dynamics and their
regulation by HGF and Rap1 signaling. Exp Cell Res. 330:382–397.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Che YL, Luo SJ, Li G, Cheng M, Gao YM, Li
XM, Dai JM, He H, Wang J, Peng HJ, et al: The C3G/Rap1 pathway
promotes secretion of MMP-2 and MMP-9 and is involved in serous
ovarian cancer metastasis. Cancer lett. 359:241–249. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Alemayehu M, Dragan M, Pape C, Siddiqui I,
Sacks DB, Di Guglielmo GM, Babwah AV and Bhattacharya M:
β-Arrestin2 regulates lysophosphatidic acid-induced uman breast
tumor cell migration and invasion via Rap1 and IQGAP1. PloS one.
8:e561742013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ahmed SM, Thériault BL, Uppalapati M, Chiu
CW, Gallie BL, Sidhu SS and Angers S: KIF14 negatively regulates
Rap1a-Radil signaling during breast cancer progression. J Cell
Biol. 199:951–967. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
McSherry EA, Brennan K, Hudson L, Hill AD
and Hopkins AM: Breast cancer cell migration is regulated through
junctional adhesion molecule-A-mediated activation of Rap1 GTPase.
Breast Cancer Res. 13:R312011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Itoh M, Nelson CM, Myers CA and Bissell
MJ: Rap1 integrates tissue polarity, lumen formation, and
tumorigenic potential in human breast epithelial cells. Cancer Res.
67:4759–4766. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhao B and Chen YG: Regulation of TGF-β
Signal Transduction. Scientifica (Cairo).
2014:8740652014.PubMed/NCBI
|
|
53
|
Zarzynska JM: Two faces of TGF-beta1 in
breast cancer. Mediators Inflamm. 2014:1417472014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kotiyal S and Bhattacharya S: Breast
cancer stem cells, EMT and therapeutic targets. Biochem Biophys Res
Commun. 453:112–116. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Imamura T, Hikita A and Inoue Y: The roles
of TGF-β signaling in carcinogenesis and breast cancer metastasis.
Breast Cancer. 19:118–124. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Mehta A and Tripathy D: Co-targeting
estrogen receptor and HER2 pathways in breast cancer. Breast.
23:2–9. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Rogler A, Hoja S, Socher E, Nolte E, Wach
S, Wieland W, Hofstädter F, Goebell PJ, Wullich B, Hartmann A and
Stoehr R: Role of two single nucleotide polymorphisms in secreted
frizzled related protein 1 and bladder cancer risk. Int J Clin Exp
Pathol. 6:1984–1998. 2013.PubMed/NCBI
|
|
58
|
Chen J, Yao D, Li Y, Chen H, He C, Ding N,
Lu Y, Ou T, Zhao S, Li L and Long F: Serum microRNA expression
levels can predict lymph node metastasis in patients with
early-stage cervical squamous cell carcinoma. Int J Mol Med.
32:557–567. 2013.PubMed/NCBI
|
|
59
|
Pouladi N, Kouhsari SM, Feizi MH, Gavgani
RR and Azarfam P: Overlapping region of p53/wrap53 transcripts:
Mutational analysis and sequence similarity with microRNA-4732-5p.
Asian Pac J Cancer Prev. 14:3503–3507. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Omura T, Shimada Y, Nagata T, Okumura T,
Fukuoka J, Yamagishi F, Tajika S, Nakajima S, Kawabe A and Tsukada
K: Relapse-associated microRNA in gastric cancer patients after S-1
adjuvant chemotherapy. Oncol Rep. 31:613–618. 2014.PubMed/NCBI
|