Introduction
Acute myeloid leukemia (AML) is a highly
heterogeneous hematologic malignancy, which involves the
uncontrolled clonal proliferation of abnormal myeloid progenitor
cells in the bone marrow, peripheral blood and other tissues
(1,2).
The maintenance of hematopoietic cells involves three types of
genes, including genes that regulate cell differentiation, the cell
cycle and apoptosis (3). Changes in
these genes have previously been observed in the occurrence and
development of AML (4). Patients with
AML have previously benefited from personalized therapy, based on
their genetic background (5). Gene
hypermethylation may suppress specific gene expression by binding
certain proteins to methylated DNA, which induces alterations in
chromatin structures and a subsequent decreased affinity for the
binding of certain transcriptional factors to methylated
C-phosphate-G(CpG) sites (6).
Therefore, the inappropriate silencing of tumor suppressor genes
may contribute to cancer tumorigenesis, progression, pathologic
grade, invasion and metastasis (7,8). Reversing
DNA methylation may improve the sensitivity of leukemia cells to
chemotherapy drugs, and may facilitate the development of more
effective clinical treatments (9).
N-myc downstream-regulated gene 4 (NDRG4) is
a member of the N-myc down regulated gene family (9). NDRG4 is a hydrolase involved in
modulating cell proliferation, invasion, migration and angiogenesis
in certain types of cancer, and it may have a valuable role as a
molecular target for novel cancer treatments (10). N-myc overexpression is highly
oncogenic in myeloid cells and may contribute to human myeloid
leukemogenesis (11), suggesting that
the silencing of NDRG4 expression may have a role in the
development of AML.
The aim of the present study was to investigate the
methylation status of NDRG4 gene alterations during
chemotherapy treatment of AML.
Materials and methods
Patient and tissue sample
collection
Bone marrow samples from 30 patients with AML prior
to and following chemotherapy were obtained through bone marrow
puncture and collected from the Department of Hematology and
Oncology, Yuyao People's Hospital (Ningbo, China) between January
2013 and June 2014. All of the AML cases were assessed at first
onset prior to any treatment, and the methylation levels prior to
chemotherapy were used as the control. There were 13 male and 17
female patients with a mean age of 47.8±15.4 years (range, 19–76
years), among them; there were 22 patients in remission and 8
patients with poor prognosis. The AML subtype distribution was as
follows: three M1, eight M2, seven M3, six M4, three M5 and three
M6. The diagnosis of AML was determined in accordance with the
revised French-American-British classification system and
diagnostic criteria published in 2013 (12,13).
Clinical data, pathological data and chemotherapy regimens were
obtained from the medical records and pathology files of the
patients, and is described in our previous study (14). The ethical committee of the Yuyao
People's Hospital of Ningbo City provided ethical approval for this
study. All the patients involved in this study provided written
informed consent.
DNA methylation assay
DNA was extracted using the nucleic acid extraction
automatic analyzer (Lab-Aid 820; Zeesan Biotech, Xiamen, China) and
quantified as described previously using a NanoDrop 1000
spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA,
USA) (14–16). Genomic DNA was then chemically
modified using sodium bisulfite from the EpiTech Bisulfite kit
(Qiagen GmbH, Hilden, Germany) as described in our previous study
(17). Pyrosequencing was used to
evaluate the methylation levels of the cytosines in the amplified
DNA fragments, and the pyrosequencing procedure was conducted as
previously described (17): The
bisulfite modified DNA and NDRG4 primer were mixed and
amplified using polymerase chain reaction (PCR). The PCR products
were denatured to release the single strands, then the
single-stranded DNA template was hybridized to a sequencing primer
and sequencing was performed using a PyroMark Q24 system and Gold
Q96 reagent (both Qiagen GmbH) (17).
PyroMark Assay Design software (version 2.0.1.15, Qiagen GmbH) was
used for primer design. The primer sequences used were as follows:
forward, 5′-AGGGTTGGGGGTTTTAGA-3′; reverse,
5′-Biotin-CACCCTCTACCAAAAACTCAAAACTCAATT-3′; and sequencing,
5′-GGGGTTTTAGAGTGTAT-3′.
Statistical analysis
The DNA sequences following NDRG4 primer
addition were detected. Then the frequency of methylation at
specific sites on NDRG4 were analyzed using PyroMark Q24
software (Qiagen, Inc., Valencia, CA, USA). Methylation records of
specific sites of NDRG4 in patients with AML were reviewed.
The statistical analysis was performed using R software (version
3.1; GNU General Public License; Free Software Foundation, Boston,
MA, USA) or SPSS software (version 16.0; SPSS, Inc., Chicago, IL,
USA). A paired-sample t-test was used to compare
NDRG4 methylation levels prior to and following
chemotherapy. Wilcoxon signed-rank sum test was used to analyze
data that did not have a normal distribution. A Pearson's linear
regression analysis was used to determine the association between
the mean methylation and patient age, gender, prognosis and AML
subtypes. A two-tailed P<0.05 was considered to indicate a
statistically significant result.
Results
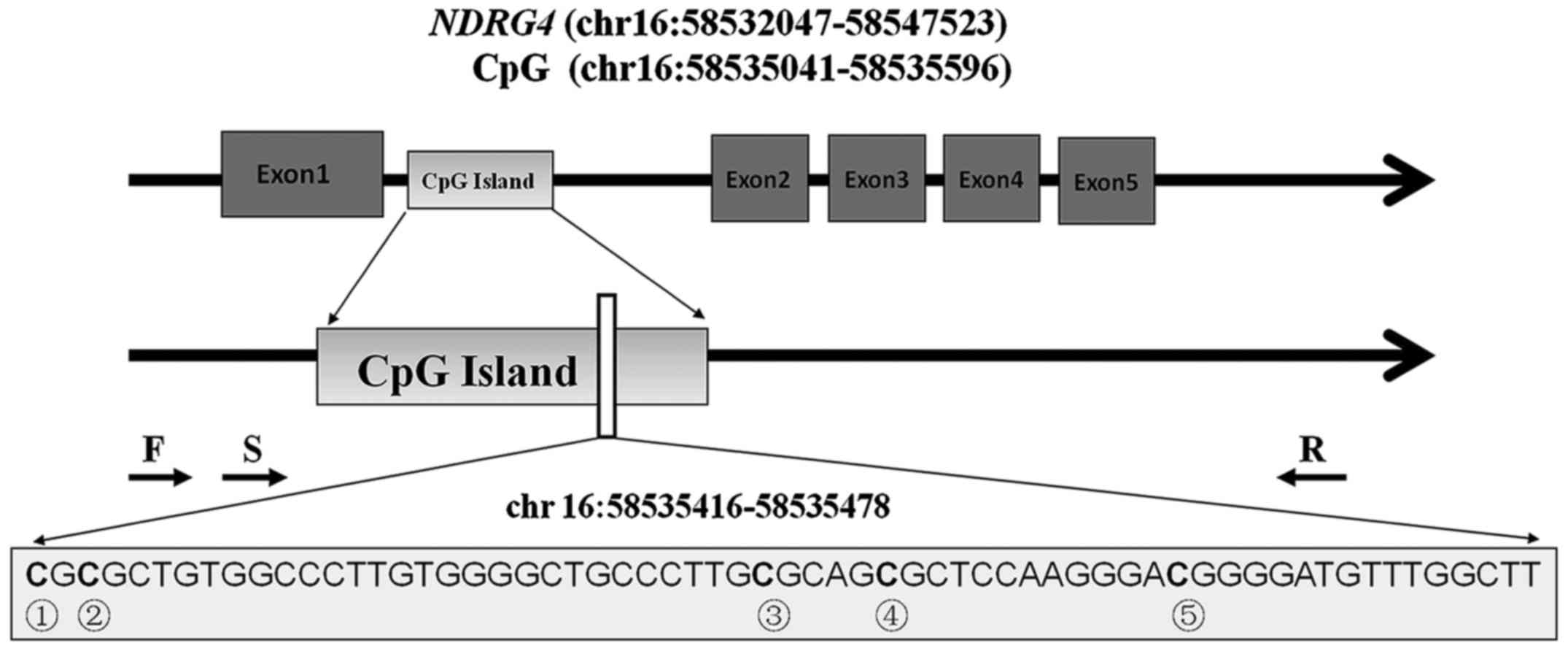
NDRG4 CpG sites
As presented in Fig.
1, a total of five CpG sites were included to represent the
methylation of the NDRG4 gene. These five CpG sites were
located in the gene body between exons 1 and 2. The DNA methylation
percentages of these five CpG sites were obtained using a bisulfite
pyrosequencing assay on a 63 bp fragment
(chr.16:58535416-58535478). The mean methylation levels were used
to compare the NDRG4 methylation level alterations in the
bone marrow DNA of the patients with AML prior to and following
chemotherapy (Fig. 2).
Changes in methylation status of NDRG4
in different subgroups
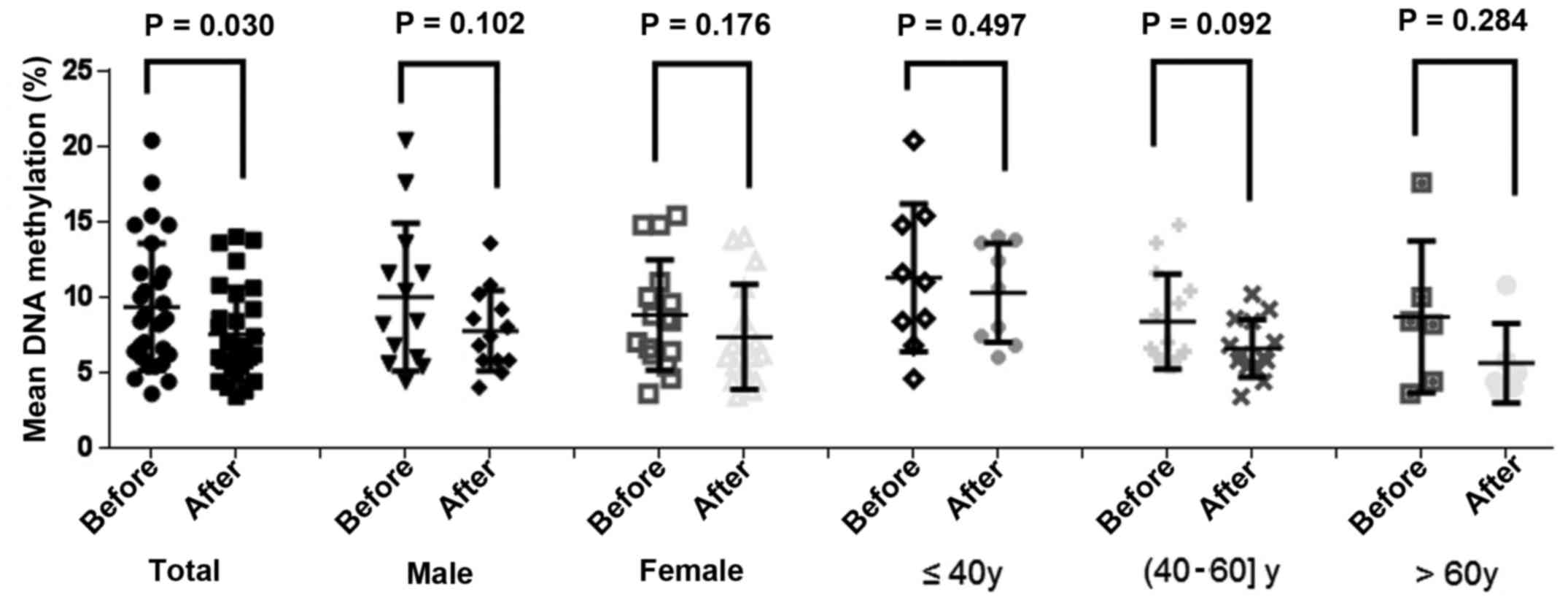
The results demonstrated a significant reduction in
NDRG4 methylation levels in the patients following
chemotherapy (prior to chemotherapy, 9.35±4.22%; following
chemotherapy, 7.54±3.11%; P=0.030; Fig.
2); however, no significant differences were identified between
gender and age, and methylation levels prior to, and following
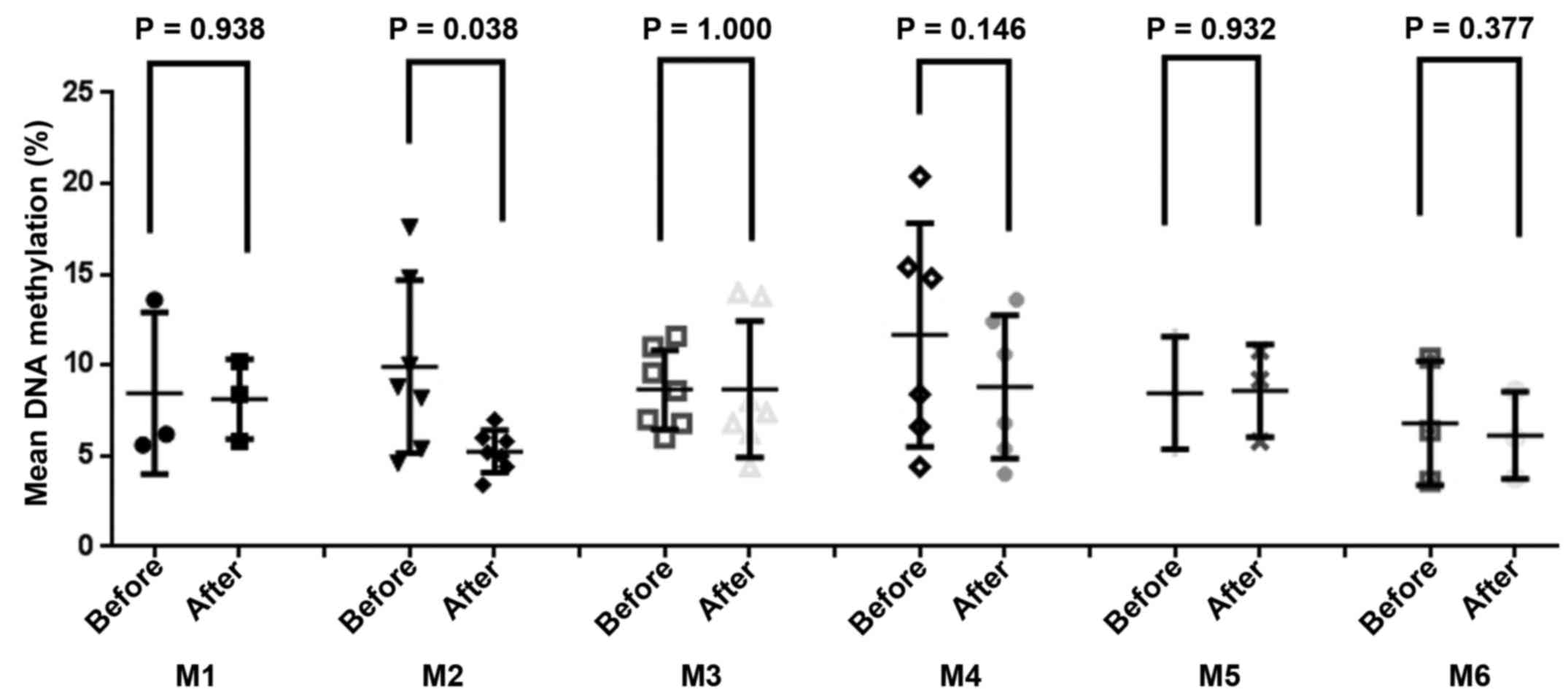
chemotherapy. A paired-sample t-test of the AML subtypes
indicated that patients with M2 subtype AML exhibited a significant
reduction of NDRG4 methylation following chemotherapy,
compared with patients with other subtypes (prior to chemotherapy,
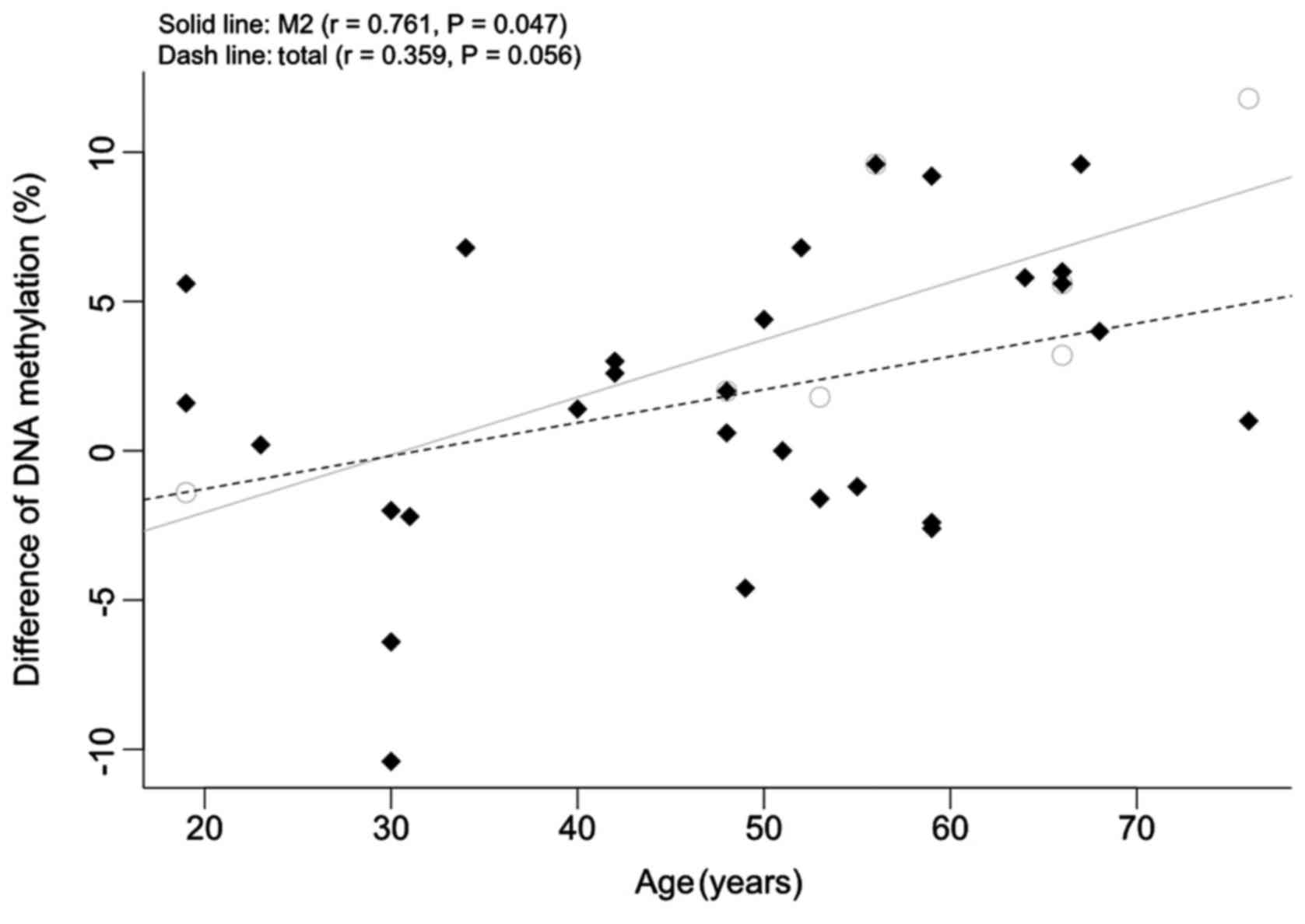
9.91±6.00%; following chemotherapy, 5.26±2.81%; P=0.038; Fig. 3). As presented in Fig. 4, the association between the
NDRG4 methylation changes and patient age was significant
for the patients with M2 subtype AML (r=0.761; P=0.047); however,
no significant association was observed in the entire patient group
(r=0.359; P=0.056).
Correlation between prognosis and
NDRG4 methylation
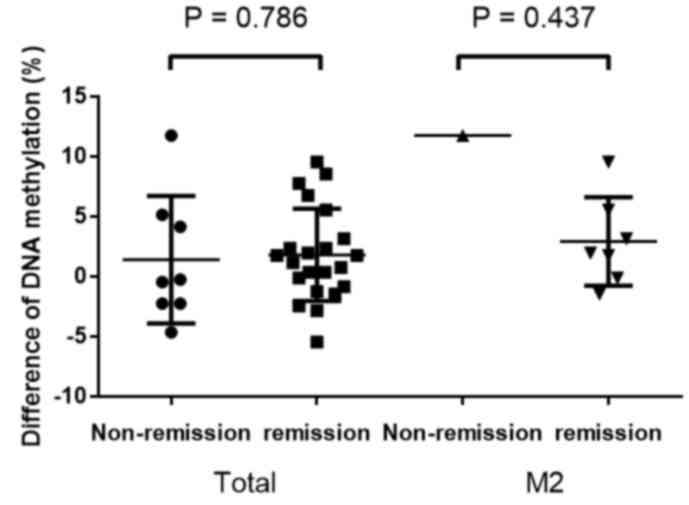
In addition, the present study investigated the
association between prognosis and NDRG4 methylation. The
results revealed that the level of chemotherapy-induced methylation
changes did not differ between patients with AML in remission and
those with a poor prognosis (P=0.786; Fig. 5). Concordantly, this was also observed
in patients with M2 subtype AML (P=0.437; Fig. 5). In addition, NDRG4
methylation prior to chemotherapy was not identified to be
associated with patient prognosis (P=0.274; data no shown).
Gender-based subgroup analysis revealed that the levels of
methylation changes in male patients did not differ from those in
female patients (male, 1.83±3.80%; female, 1.46±4.12%; P=0.806;
data no shown).
Discussion
In the current study, the NDRG4 methylation
level alterations in patients with AML were investigated. The
results demonstrated that NDRG4 methylation levels in
patients with AML were significantly reduced during chemotherapy,
particularly for patients with M2 subtype AML, indicating a
chemo-sensitive mechanism underlying NDRG4 methylation. The
NDRG4 methylation changes during chemotherapy were
positively associated with patient age in patients with M2 subtype
AML; further studies are required to fully elucidate the molecular
mechanisms underlying the chemotherapy-induced changes observed in
M2 subtype AML.
DNA methylation in the gene body has a role in the
silencing of genes, and is a therapeutic target of methylation
inhibitors (18). Yang et al
(18) identified that
5-aza-2′-deoxycytidine treatment reactivated tumor-suppressor genes
in addition to decreasing the overexpression of proto-oncogenes.
NDRG4 gene-body methylation may improve the sensitivity of a
certain type of leukemia cells to chemotherapeutic agents, although
the present study did not identify a significant association
between the altered levels of NDRG4 methylation and the
prognosis of patients with AML, possibly due to the small sample
size used.
Cytogenetics is considered a valuable prognostic
determinant for AML (19). AML with
t(8;21)(q22;q22) is the most common karyotypic abnormality observed
in AML, comprising ~15% of total cases (20). The incidence of t(8;21) in M2 was
24.1% (21), and it was typically
considered to be among the most favorable subtypes for predicting a
high response to treatment (~60%) (22). The M2 subtype has been frequently
associated with additional chromosome abnormalities (23), such as AML/RUNX1 translocation
partner 1 fusion gene, which has been detected in 40% of M2 cases
(24) and DEK
proto-oncogene-nucleoporin 214 fusion gene, which has been
associated with poor prognosis (25).
In addition, unique methylation signatures have previously been
associated with specific cytogenetic subtypes of AML (26). For example, the spalt like
transcription factor 4 hypomethylation rate was demonstrated to be
higher in patients with the M1 subtype compared with the M2 and
other subtypes (27). p15
methylation has been associated with the M2 subtype (28), whereas the aberrantly methylated
BMP/retinoic acid inducible neural specific 1 (DBC1) was
observed in cases with nucleophosmin mutations, and fms related
tyrosine kinase 3 aberrations were identified to be more prevalent
in the methylated DBC1 group (29). In the current study, patients with M2
subtype AML exhibited a significant reduction in NDRG4
methylation levels during chemotherapy. The incidence of AML is
frequently associated with increased age, but may occur at any age
(30–33). The clinical characteristics of elderly
patients with AML differ from those of younger patients, with
poorer survival and treatment outcomes (34). Surveillance, epidemiology and end
results data demonstrated that the overall 5-year survival was
<5% in patients aged >65 years of age (35); however, those patients who received
high-dose chemotherapy exhibited improved outcomes (34). In the current study, more changeable
hypomethylation was observed in elderly patients (particularly
patients with M2 subtype AML) than in younger patients, which may
facilitate the elucidation of age-associated variations in clinical
treatment methods.
There were certain limitations in the present study.
The patients enrolled were Chinese individuals from the city of
Ningbo; therefore, the significant association between NDRG4
gene-body methylation levels and AML may not be applicable to other
ethnic populations. Furthermore, the current study evaluated five
CpG sites, rather than the entire NDRG4 gene region.
Therefore, additional CpG sites must be investigated in further
studies. Alternate morphological types have previously been
demonstrated to have various effects on AML (36), and the heterogeneity of AML may have
influenced the results of the present study. Finally, the
conclusions presented were based on a moderate sample size, and
therefore further studies using a greater sample size are required
to investigate the association between NDRG4 methylation
levels and chemotherapeutic outcomes. In conclusion, the results of
the present study suggest there may be an age-dependent mechanism
underlying the induced methylation levels of the NDRG4 gene
body in patients with AML, particularly the M2 subtype, during
chemotherapy.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant no. 81371469), the Zhejiang
Provincial Natural Science Foundation (grant no. LR13H020003), the
Ningbo City Medical Science and Technology projects (grant no.
2014A20), the K. C. Wong Magna Fund of the Ningbo University and
the Zhejiang Province Student Science and Technology Innovation
Plan (grant no. 2015R405081).
Glossary
Abbreviations
Abbreviations:
|
AML
|
acute myeloid leukemia
|
|
NDRG4
|
N-myc downstream-regulated gene 4
|
|
CpG
|
cytosine-phosphate-guanine
dinucleotide
|
References
|
1
|
Estey EH: Acute myeloid leukemia: 2013
update on risk-stratification and management. Am J Hematol.
88:318–327. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Davis AS, Viera AJ and Mead MD: Leukemia:
An overview for primary care. Am Fam Physician. 89:731–738.
2014.PubMed/NCBI
|
|
3
|
Yi S, Wen L, He J, Wang Y, Zhao F, Zhao J,
Zhao Z, Cui G and Chen Y: Deguelin, a selective silencer of the
NPM1 mutant, potentiates apoptosis and induces differentiation in
AML cells carrying the NPM1 mutation. Ann Hematol. 94:201–210.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fang F and Zhu P: Analysis of gene
expression profiles to improve the treatment of leukemia. Zhongguo
Shi Yan Xue Ye Xue Za Zhi. 22:1735–1738. 2014.(In Chinese).
PubMed/NCBI
|
|
5
|
Schlenk RF and Döhner H: Genomic
applications in the clinic: Use in treatment paradigm of acute
myeloid leukemia. Hematology Am Soc Hematol Educ Program.
2013:324–330. 2013.PubMed/NCBI
|
|
6
|
Pogribny IP, Pogribna M, Christman JK and
James SJ: Single-site methylation within the p53 promoter region
reduces gene expression in a reporter gene construct: Possible in
vivo relevance during tumorigenesis. Cancer Res. 60:588–594.
2000.PubMed/NCBI
|
|
7
|
Ushijima T, Watanabe N, Okochi E, Kaneda
A, Sugimura T and Miyamoto K: Fidelity of the methylation pattern
and its variation in the genome. Genome Res. 13:868–874. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jones PA and Baylin SB: The epigenomics of
cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Itzykson R, Thépot S, Berthon C, Delaunay
J, Bouscary D, Cluzeau T, Turlure P, Prébet T, Dartigeas C,
Marolleau JP, et al: Azacitidine for the treatment of relapsed and
refractory AML in older patients. Leuk Res. 39:124–130. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tao YF, Xu LX, Lu J, Cao L, Li ZH, Hu SY,
Wang NN, Du XJ, Sun LC, Zhao WL, et al: Metallothionein III (MT3)
is a putative tumor suppressor gene that is frequently inactivated
in pediatric acute myeloid leukemia by promoter hypermethylation. J
Transl Med. 12:1822014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kawagoe H, Kandilci A, Kranenburg TA and
Grosveld GC: Overexpression of N-Myc rapidly causes acute myeloid
leukemia in mice. Cancer Res. 67:10677–10685. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bennett JM, Catovsky D, Daniel MT,
Flandrin G, Galton DA, Gralnick HR and Sultan C: Proposed revised
criteria for the classification of acute myeloid leukemia. A report
of the French-American-British Cooperative Group. Ann Intern Med.
103:620–625. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fasan A, Alpermann T, Haferlach C,
Grossmann V, Roller A, Kohlmann A, Eder C, Kern W, Haferlach T and
Schnittger S: Frequency and prognostic impact of CEBPA proximal,
distal and core promoter methylation in normal karyotype AML: A
study on 623 cases. PLoS One. 8:e543652013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hong Q, Chen X, Ye H, Zhou A, Gao Y, Jiang
D, Wu X, Tian B, Chen Y, Wang M, et al: Association between the
methylation status of the MGMT promoter in bone marrow specimens
and chemotherapy outcomes of patients with acute myeloid leukemia.
Oncol Lett. 11:2851–2856. 2016.PubMed/NCBI
|
|
15
|
Huang Y, Ye H, Hong Q, Xu X, Jiang D, Xu
L, Dai D, Sun J, Gao X and Duan S: Association of CDKN2BAS
polymorphism rs4977574 with coronary heart disease: A case-control
study and a meta-analysis. Int J Mol Sci. 15:17478–17492. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cheng C, Lingyan W, Yi H, Cheng Z, Huadan
Y, Xuting X, Leiting X, Meng Y and Shiwei D: Association between
TLR2, MTR, MTRR, XPC, TP73, TP53 genetic polymorphisms and gastric
cancer: A meta-analysis. Clin Res Hepatol Gastroenterol.
38:346–359. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang L, Ye H, Hong Q, Wang L, Wang Q, Wang
H, Xu L, Bu S, Zhang L, Cheng J, et al: Elevated CpG island
methylation of GCK gene predicts the risk of type 2 diabetes in
Chinese males. Gene. 547:329–333. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang X, Han H, De Carvalho DD, Lay FD,
Jones PA and Liang G: Gene body methylation can alter gene
expression and is a therapeutic target in cancer. Cancer Cell.
26:577–590. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Grimwade D, Walker H, Oliver F, Wheatley
K, Harrison C, Harrison G, Rees J, Hann I, Stevens R, Burnett A and
Goldstone A: The importance of diagnostic cytogenetics on outcome
in AML: Analysis of 1,612 patients entered into the MRC AML 10
trial. The medical research council adult and Children's Leukaemia
working parties. Blood. 92:2322–2333. 1998.PubMed/NCBI
|
|
20
|
Cheng CK, Li L, Cheng SH, Ng K, Chan NP,
Ip RK, Wong RS, Shing MM, Li CK and Ng MH: Secreted-frizzled
related protein 1 is a transcriptional repression target of the
t(8;21) fusion protein in acute myeloid leukemia. Blood.
118:6638–6648. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Byun JM, Kim YJ, Yoon HJ, Kim SY, Kim HJ,
Yoon J, Min YH, Cheong JW, Park J, Lee JH, et al: Cytogenetic
profiles of 2806 patients with acute myeloid leukemia-a
retrospective multicenter nationwide study. Ann Hematol.
95:1223–1232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nucifora G, Larson RA and Rowley JD:
Persistence of the 8;21 translocation in patients with acute
myeloid leukemia type M2 in long-term remission. Blood. 82:712–715.
1993.PubMed/NCBI
|
|
23
|
Shi HX, Jiang B, Qiu JY, Lu XJ, Fu JF,
Wang DB and Lu DP: Studies of treatment strategy and prognosis on
acute myeloid leukemia with chromosome 8 and 21 translocation.
Zhonghua Xue Ye Xue Za Zhi. 26:481–484. 2005.(In Chinese).
PubMed/NCBI
|
|
24
|
Hong Q, Ye H, Tang L, Jiang D, Ji H, Dai
D, Ouyang G and Duan S: Progress in DNA Methylation Research on
Genes Associated with Acute Myeloid Leukemia. Chinese J Cell Biol.
37:299–308. 2015.
|
|
25
|
Servitzoglou M, Grenzelia M, Baka M,
Harisi M, Pourtsidis A, Bouhoutsou D, Varvoutsi M, Doganis D, Dana
H, Divane A and Kosmidis H: A novel karyotype in acute myeloid
leukemia with basophilia. Pediatr Hematol Oncol. 31:149–156. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ho PA, Kutny MA, Alonzo TA, Gerbing RB,
Joaquin J, Raimondi SC, Gamis AS and Meshinchi S: Leukemic
mutations in the methylation-associated genes DNMT3A and IDH2 are
rare events in pediatric AML: A report from the Children's oncology
group. Pediatr Blood Cancer. 57:204–209. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ma JC, Qian J, Lin J, et al: Aberrant
hypomethylation of SALL4 gene is associated with intermediate and
poor karyotypes in acute myeloid leukemia. Clin Biochem.
46:304–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chim CS, Wong AS and Kwong YL: Epigenetic
inactivation of INK4/CDK/RB cell cycle pathway in acute leukemias.
Ann Hematol. 82:738–742. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Alvarez S, Suela J, Valencia A, et al: DNA
methylation profiles and their relationship with cytogenetic status
in adult acute myeloid leukemia. PLoS One. 5:e121972010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shah A, Andersson TM, Rachet B, Björkholm
M and Lambert PC: Survival and cure of acute myeloid leukaemia in
England, 1971–2006: A population-based study. Br J Haematol.
162:509–516. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dores GM, Devesa SS, Curtis RE, Linet MS
and Morton LM: Acute leukemia incidence and patient survival among
children and adults in the United States, 2001–2007. Blood.
119:34–43. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bhayat F, Das-Gupta E, Smith C, McKeever T
and Hubbard R: The incidence of and mortality from leukaemias in
the UK: A general population-based study. BMC Cancer. 9:2522009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Australian Institute of Health and
Welfare: Cancer survival and prevalence in Australia: Period
estimates from 1982 to 2010. Asia Pac J Clin Oncol. 9:29–39.
2013.PubMed/NCBI
|
|
34
|
Yi HG, Lee MH, Kim CS, Hong J, Park J, Lee
JH, Han BR, Kim HY, Zang DY, Kim SH, et al: Clinical
characteristics and treatment outcome of acute myeloid leukemia in
elderly patients in Korea: a retrospective analysis. Blood Res.
49:95–99. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Thein MS, Ershler WB, Jemal A, Yates JW
and Baer MR: Outcome of older patients with acute myeloid leukemia:
An analysis of SEER data over 3 decades. Cancer. 119:2720–2727.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gritsaev SV, Martynkevich IS, Ziuzgin IS,
Kariagina EV, Martynenko LS, Petrova EV, Tsybakova Nlu, Ivanova MP,
Kostroma II, Tiranova SA, et al: Heterogeneity of acute myeloid
leukemia with the translocation t(8;21)(q22;q22). Ter Arkh.
86:45–52. 2014.(In Russian). PubMed/NCBI
|