Introduction
Cervical cancer is the second most common cancer in
females worldwide, ranking first in developing countries (1). In recent years, the incidence of
cervical adenocarcinoma and cervical cancer in young women has
increased significantly; therefore, early diagnosis is of high
importance (2–8). High-risk human papillomavirus (HR-HPV)
infection is an established cause of cervical cancer and this
disease is preventable and treatable. Although extensive screening
of cervical cancer enables early diagnosis and treatment in an
increasing number of patients, the detection of cervical cancer
using the HR-HPV ThinPrep cytology test may lead to overtreatment
and misdiagnosis, which may also impede prevention and therapeutic
intervention. Furthermore, the clinical outcomes of the same
treatment for similar pathological types of cervical cancer are
heterogeneous (9). Therefore, an
effective molecular marker of cervical lesions is required to
supplement screening for cervical cancer, in order to allow the
development of individual treatment regimens for various molecular
types and improve the prevention and treatment of cervical
cancer.
Ovarian cancer gene 1 (OVCA1) is a cancer suppressor
that may be correlated with the occurrence and development of
ovarian and cervical cancer, particularly in early lesions
(10). The inhibition of OVCA1 may be
associated with the regulation and control of the cell cycle,
leading to cell cycle arrest at the G1/S phase and,
thus, suppressing cell proliferation (11,12).
Aberrant expression of OVCA1 mRNA has been detected in cervical
cancer (13). However, the expression
levels of OVCA1, as well as its association with the clinical
pathology and its underlying mechanisms of action, remain to be
elucidated.
The current study aimed to evaluate molecular
biomarkers for the diagnosis and treatment of cervical lesions
through detection of the protein and mRNA expression levels of
OVCA1, cyclin D1 and p16 in cervical cancer, cervical
intraepithelial neoplasia (CIN) and normal cervix tissue. In
addition, the associations between OVCA1 expression and HR-HPV
infection were examined, in order to examine the potential
mechanisms underlying the involvement of OVCA1 in the development
of cervical lesions.
Materials and methods
Patients
In total, 130 female patients with cervical lesions
who were pathologically diagnosed between February 2014 and July
2014 at Liaoning Cancer Hospital and Institute (Shenyang, China)
were selected, including 66 cases of cervical cancer and 64 cases
of CIN. All pathological sections were reviewed by a pathologist
(Liaoning Cancer Hospital and Institute). In addition, 34 normal
cervix tissue specimens obtained from patients with hysteromyoma,
ovarian cysts and other non-malignant tumors with hysterectomy were
selected as controls. The present study was approved by Liaoning
Cancer Hospital Ethics Committee, and all patients provided
informed consent prior to biopsy surgery.
The clinical stages of cervical cancer were
classified according to the 2009 International Federation of
Gynecology and Obstetrics staging system (9,14). Of the
66 cases of cervical cancer, 20 cases were stage I, 8 were stage
II, 34 were stage III and 4 were stage IV. The cases included 60 of
squamous carcinoma and 6 of adenocarcinoma. The pathological grades
were as follows: 20 cases of high differentiation (G1), 42 cases of
moderate differentiation (G2) and 4 cases of low differentiation
(G3). In total, there were 31 cases with lymph node metastasis and
35 cases without lymph node metastasis. In addition, 64 cases of
CIN were graded according to the Richart Pathology criteria
(9,15–17) as
follows: 22 cases of CIN1, 22 cases of CIN2 and 20 cases of CIN3.
Among the 164 cervical cancer, CIN and control patients, HR-HPV
infection was detected in 103 cases and undetected in 61 cases. All
patients had no contraindications and received no radiotherapy or
chemotherapy prior to biopsy.
Two biopsy tissue specimens (3–8-mm thick) were
obtained for each patient with cervical cancer and each healthy
patient with normal cervical tissue. The first was fixed in 4%
paraformaldehyde for immunohistochemistry tissue microarray
analysis. The second tissue specimen was stored in at −80°C for
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) analysis. Following lymph node excision, lymph nodes of
>1 cm that had been deemed highly suggestive of metastasis
preoperatively using computed tomography/magnetic resonance imaging
were determined to be cases of lymph node metastasis. Prior to
treatment, the presence of HPV infection was determined from
cervical screening using hybrid capture II (HC-II; Qiagen, Inc.,
Valencia, CA, USA) and values of >1.0 relative light
units/cutoff were defined as HR-HPV-positive.
Reagents and instruments
The antibodies used were as follows: Mouse
anti-human OVCA1 (#ab54777; Abcam, Cambridge, UK), mouse anti-human
p16 (#ZM-0205; OriGene Technologies, Inc., Beijing, China) and
mouse anti-human cyclin D1 (#sc-8396; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA). The High-Capacity cDNA Reverse
Transcription kit was purchased from Thermo Fisher Scientific, Inc.
(Waltham, MA, USA) and the PrimeScript™ RT-PCR kit was purchased
from Takara Bio, Inc. (Otsu, Japan). TRIzol® RNA
Isolation reagent was purchased from Invitrogen (Thermo Fisher
Scientific, Inc.). Primers for OVCA1, cyclin D1, p16 and GAPDH
(internal control) were designed and synthesized by Invitrogen
(Thermo Fisher Scientific, Inc.). Immunohistochemistry imaging was
performed using NIS-Elements F3.0 software for image acquisition,
and image analysis utilized NIS-Elements Br3.0 software (Nikon
Corporation, Tokyo, Japan). The PCR instrument was produced by
Bio-Rad Laboratories, Inc. (Hercules, CA, USA).
Immunohistochemistry
The representative regions of normal cervix, CIN and
cervical cancer in paraffin-embedded specimens were labeled
according to the hematoxylin and eosin-stained sections.
Subsequently, tissue microarrays of cervical cancer, CIN and normal
cervical tissues were constructed, and all the representative
points were included in three arrays (10×8, 10×6 and 8×3). The
dilutions of primary antibodies were 1:50 (OVCA1), 1:100 (cyclin
D1) and 1:200 (p16). Immunohistochemical staining was performed
using a PicTure™ Two-Step Immunohistochemistry kit (OriGene
Technologies, Inc., Beijing, China) according to the manufacturer's
protocol. PBS was used to replace primary antibody as a negative
control, and cervical cancer tissue sections with established
cyclin D1- and p16-positive staining were used as positive
controls, whereas no positive control was established for OVCA1.
The presence of translucent brown granules in the nucleus and
cytoplasm was determined to indicate positivity for OVCA1, cyclin
D1 and p16. Abnormal OVCA1 and p16 proteins were detected using
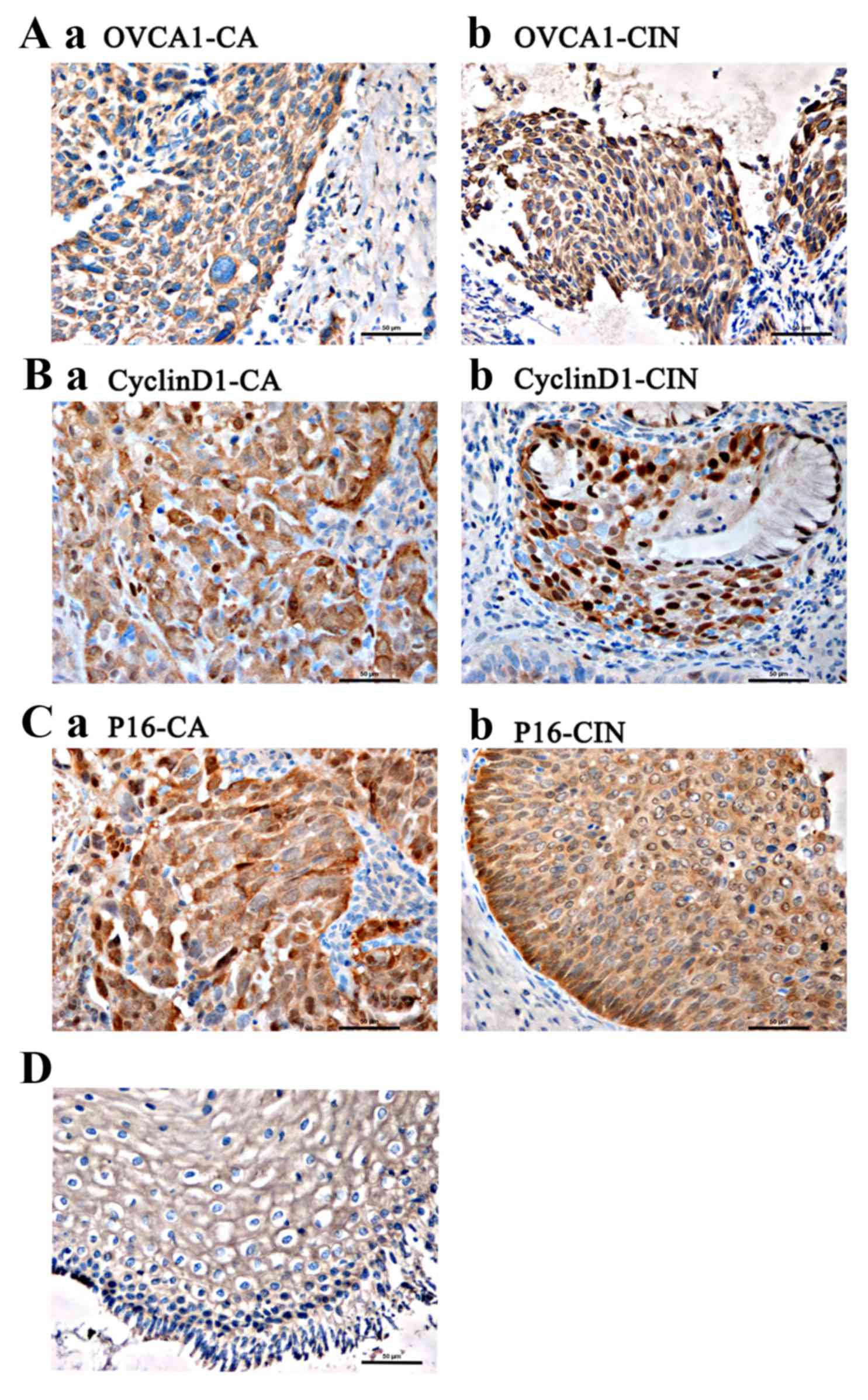
immunohistochemistry. Positive staining for OVCA1 was localized to
the nucleus and cytoplasm, primarily surrounding the nucleus.
CyclinD1 staining was primarily localized to the nucleus, as well
as being observed in the cytoplasm, whereas p16 staining was
localized to the nucleus and cytoplasm. In total, 5 representative
fields from each pathological section were examined under high
magnification using a scanning electron microscope (Nikon E800;
Nikon Corporation) to detect the average optical density (OD) and
color rendering area (S) of each field. The integral OD (IOD) was
calculated as ODxS, and the average IOD of each pathological
section was determined. The results were judged semi
quantitatively.
RT-qPCR
Total RNA was extracted from normal cervix and
cervical cancer tissue samples using TRIzol, and the OD of the RNA
samples, as detected using an ultraviolet spectrophotometer at an
absorbance of 260–280 nm, was ~1.8–2.0. The RNA bands were
separated using 1.5% agarose gel electrophoresis. The process of
cDNA synthesis was performed according to the manufacturer's
protocol. Briefly, reaction mixtures were prepared, which consisted
of 4 µl RNA samples treated with DNase (Takara Bio, Inc.), 2 µl of
5X PrimeScript® RT Master mix and 4 µl
diethylpyrocarbonate (DEPC)-treated water. The reactions were
incubated for 15 min at 37°C and 5 sec at 4°C, and the cDNA was
subsequently used as a template for RT-qPCR or stored at −70°C.
RT-qPCR reaction mixtures were composed of 10 µl
SYBR Green, 6 µl DEPC-treated water, 2.0 µl cDNA template and 2.0
µl of each 10 µM primers (total primer volume, 20 µl). The primer
sequences were as follows: OVCA1 forward,
5′-CTGAGGTGGATGTGTGGGTG-3′ and reverse, 5′-CCTCATAGGGTGTCAGCAGC-3′;
cyclin D1 forward, 5′-CCCTCGGTGTCCTACTTC-3′ and reverse,
5′-AGGAAGCGGTCCAGGTAGTT-3′; p16 forward, 5′-GTGGACCTGGCTGAGGAG-3′
and reverse, 5′-CTTTCAATCGGGGATGTCTG-3′; and GAPDH forward,
5′-AGGTGAAGGTCGGAGTCA-3′ and reverse, 5′-GGTCATTGATGGCAACAA-3′. The
thermocycling conditions consisted of an initial step for 5 min at
94°C, followed by 40 cycles of denaturation for 5 sec at 94°C and
annealing for 30 sec at 60°C. The results were analyzed using the
ΔΔCq relative quantification method (18), with GAPDH as the reference gene. The
ΔCq values for the experimental group and the control group were
calculated according to the formula: ΔCq=Cq (target gene)-Cq
(reference gene). Subsequently, according to the formula ΔΔCq=ΔCq
(experiment group)-ΔCq (control group), ΔΔCq was calculated, and
the difference in relative mRNA expression (2−ΔΔCq), was
obtained.
Statistical analysis
One-way analysis of variance, the Mann-Whitney U
rank sum test and Spearman's correlation analysis were applied to
analyze the data using SPSS version 19.0 (IBM SPSS, Armonk, NY,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
Protein expression of OVCA1, cyclin D1
and p16 in various cervical tissues
The IOD values of the OVCA1 (P<0.01), cyclin D1
(P<0.01) and p16 (P<0.01) protein bands increased from the
normal cervix to CIN and cervical cancer tissues. Following further
pairwise comparisons, a statistically significant difference was
observed in the OVCA1 and p16 expression levels between normal and
cancer tissues (P<0.01), normal and CIN tissues (P<0.01) and
CIN and cancer tissues (P<0.01), and in the protein expression
levels of cyclin D1 between normal and cancer tissues (P<0.01)
and CIN and cancer tissues (P<0.01). Conversely, no significant
difference in the expression levels of cyclin D1 was observed
between normal and CIN tissues (P=0.136; Table I; Fig.
1A).
 | Table I.Expression levels of OVCA1, cyclin D1
and p16 proteins in various cervical tissues (IOD value). |
Table I.
Expression levels of OVCA1, cyclin D1
and p16 proteins in various cervical tissues (IOD value).
| Group | n | IOD value,
OVCA1 |
F-value/P-value | IOD value, cyclin
D1 |
F-value/P-value | IOD value, p16 |
F-value/P-value |
|---|
| Normal tissue | 34 |
44,318.68 |
13.703a/<0.001a | 342,846.11 | 2.26a/0.136a |
42,779.59 |
19.53a/<0.001a |
| CIN | 64 |
71,011.03 |
73.078b/<0.001b |
41,502.09 | 11.88b/<0.001b |
88,331.77 | 126.11b/<0.001b |
| Cervical
cancer | 66 | 120,093.60 |
159.949c/<0.001c |
60,090.93 | 13.57c/<0.001c | 172,974.76 | 445.76c/<0.001c |
| Total | 164 |
85,229.99 |
73.914d/<0.001d |
47,486.97 | 10.84d/<0.001d | 112,951.67 | 130.82d/<0.001d |
Associations between the protein
expression levels of OVCA1, cyclin D1 and p16 and clinical
features
The protein expression levels of OVCA1 in high-level
CIN tissues (CIN2-3) were significantly higher compared with
low-level CIN tissues (CIN1; P=0.031, <0.05), whereas no
significant differences were observed in cyclin D1 and p16
expression levels between the high and low-level CIN groups
(Pcyclin D1=0.977; Pp16=0.056; Table II). The protein expression levels of
OVCA1, cyclin D1 and p16 in advanced-stage (II–IV) cervical cancer
were significantly higher compared with early-stage cervical cancer
(I; POVCA1=0.016; Pcyclin D1=0.001;
Pp16=0.005; Table
II).
 | Table II.Correlations between the protein
expression levels of OVCA1, cyclin D1 and p16 and various
clinicopathological features. |
Table II.
Correlations between the protein
expression levels of OVCA1, cyclin D1 and p16 and various
clinicopathological features.
| Group | n | IOD value,
OVCA1 | Z/F-value
P-value | IOD value, cyclin
D1 | Z/F-value
P-value | IOD value, p16 | Z/F-value
P-value |
|---|
| CIN level |
| CIN
1 | 22 |
51,995.00 | −2.941 | 42,772.36 | −0.280 |
69,577.02 | −1.908 |
| CIN
2–3 | 42 |
80,971.81 |
0.031a | 41,664.49 |
0.977 |
98,155.69 |
0.056 |
| FIGO clinical stage
of cervical cancer |
| Stage
I | 20 | 109,356.18 | −2.401 | 36,212.49 | −3.471 | 159,454.92 | −2.832 |
| Stage
II–IV | 46 | 124,762.04 |
0.016a | 70,472.35 |
<0.001b | 178,852.94 |
0.005b |
| Pathological
grade |
| G1 | 20 | 123,756.57 | 0.251 | 63,685.25 | 0.496 | 170,175.84 |
0.486 |
| G2 | 42 | 118,751.72 | 0.779 | 63,685.25 | 0.496 | 176,783.05 |
0.617 |
| G3 | 4 | 115,868.47 |
| 73,442.64 |
| 172,357.07 |
|
| Pathological
type |
|
Squamous carcinoma | 60 | 120,694.21 | −1.026 | 66,669.11 | −0.580 | 170,586.98 | −2.164 |
|
Adenocarcinoma | 6 | 114,087.48 |
0.305 | 64,282.41 | 0.562 | 196,852.47 |
0.030a |
| Lymph node
metastasis |
|
Positive | 31 | 123,897.14 | −1.774 | 70,232.35 | −2.211 | 173,387.03 | −0.084 |
|
Negative | 35 | 116,724.75 |
0.076 | 51,108.52 | 0.027 | 172,609.60 |
0.933 |
| HPV |
|
Positive | 103 |
94,994.85 | −3.934 | 49,858.21 | 0.864 | 127,905.46 | −3.479 |
|
Negative | 61 |
68,741.79 |
<0.01b | 43,483.07 | 0.387 | 87,701.82 |
0.001b |
The protein expression levels of OVCA1 and p16 in
the HR-HPV (+) group were significantly higher, as compared with
the HR-HPV (−) group (POVCA1<0.01;
P16<0.01; Table II).
The expression levels of p16 protein in adenocarcinoma were higher
compared with those detected in squamous carcinoma (P=0.030;
Table II). No statistically
significant differences (P=0.933) were identified in the protein
expression levels of cyclin D1 between the HR-HPV (+) and HR-HPV
(−) groups. In addition, no statistically significant differences
in OVCA1 and cyclin D1 protein expression levels were identified
between various pathological types, as well as in OVCA1, cyclin D1
and p16 protein expression levels between the groups with or
without lymph node metastasis and between pathological grades
(G1/G2/G3; POVCA1=0.799; Pcyclin D1=0.496;
Pp16=0.617; Table II;
Fig. 1B and C). Intense positive
staining was observed in the cytoplasm of the koilocytes in the
tissue samples, which was indicative of HPV infection (Fig. 1D).
Associations between the protein
expression levels of OVCA1, cyclin D1 and p16
The IOD values of the OVCA1, cyclin D1 and p16
proteins in the normal cervix, CIN and cervical cancer tissues
increased as the lesions progressed towards malignancy. Positive
staining for OCVA1 protein was significantly positively correlated
with that of cyclin D1 and p16 (POVCA1, cyclin
D1<0.01; POVCA, p16<0.01), and
staining for cyclin D1 and p16 were positively correlated
(Pp16 & cyclin D1<0.01; Table III).
 | Table III.Correlations between OVCA1, cyclin D1
and p16 protein expression levels. |
Table III.
Correlations between OVCA1, cyclin D1
and p16 protein expression levels.
|
| Cyclin D1 | p16 |
|---|
|
|
|
|
|---|
|
| r | P-value | r | P-value |
|---|
| OVCA1 | 0.249 | 0.001a | 0.618 |
<0.001a |
| Cyclin D1 | – | – | 0.336 |
<0.01a |
OVCA1, cyclin D1 and p16 mRNA
expression levels in cervical cancer and normal cervix tissues
The relative mRNA expression levels of OVCA1 and p16
in cervical cancer tissues were lower compared with that in normal
cervix tissues (POVCA1<0.01; Pp16=0.005).
In addition, the relative cyclin D1 mRNA expression levels were
higher in cervical cancer tissues compared with normal cervix
tissues (Pcyclin D1<0.01; Table IV).
 | Table IV.Relative expression levels of OVCA1,
cyclin D1 and p16 mRNA in normal cervix and cervical cancer
tissues. |
Table IV.
Relative expression levels of OVCA1,
cyclin D1 and p16 mRNA in normal cervix and cervical cancer
tissues.
| Group | n | OVCA1, ΔΔCq | cyclin D1,
ΔΔCq | p16, ΔΔCq |
|---|
| Normal tissue | 34 | 1.07 | 1.25 | 0.94 |
| Cervical
cancer | 66 | 0.64 | 1.98 | 0.72 |
| Z-value |
| −5.520 | −3.247 | −2.825 |
| P-value |
|
<0.001a | 0.001a | 0.005a |
Associations between the mRNA
expression levels of OVCA1, cyclin D1 and p16 and clinical
features
The mRNA expression levels of OVCA1 decreased
gradually from low- to moderately- and highly-differentiated
cervical cancer tissues (P=0.012) and were significantly lower in
the HR-HPV (+) group compared with the HR-HPV (−) group
(P<0.01). No significant differences were observed in the cyclin
D1 and p16 expression levels between various pathological grades
and between the HR-HPV (+) and HR-HPV (−) groups (Pcyclin
D1-pathological grades=0.359; Pcyclin
D1-HPV=0.250; Pp16-pathological grades=0.481;
Pp16-HPV=0.411). No significant differences were
identified in the mRNA expression levels of OVCA1, cyclin D1 and
p16 between the various stages and pathological types of cervical
cancer, and the presence or absence of lymph node metastasis
(Table V).
 | Table V.Correlations between OVCA1, cyclin D1
and p16 mRNA expression levels and various clinicopathological
features. |
Table V.
Correlations between OVCA1, cyclin D1
and p16 mRNA expression levels and various clinicopathological
features.
| Group | n | OVCA1, ΔΔCq | Z/F value
P-value | Cyclin D1,
ΔΔCq | Z/F value
P-value | p16, ΔΔCq | Z/F value
P-value |
|---|
| FIGO clinical stage
of cervical cancer |
| Stage
I | 20 | 0.71 | −0.671 | 1.78 | −0.614 | 0.80 | −1.173 |
| Stage
II–IV | 46 | 0.61 |
0.502 | 2.04 |
0.539 | 0.69 |
0.241 |
| Pathological
grade |
| G1 | 20 | 0.30 |
4.487 | 1.79 |
1.043 | 0.75 |
0.740 |
| G2 | 42 | 0.76 |
0.012a | 2.12 |
0.359 | 0.73 |
0.481 |
| G3 | 4 | 1.04 |
| 1.17 |
| 0.53 |
|
| Lymph node
metastasis |
|
Positive | 31 | 0.68 | −0.328 | 1.91 | −0.129 | 0.62 | −0.926 |
|
Negative | 35 | 0.60 |
0.743 | 2.00 |
0.898 | 0.81 |
0.355 |
| HPV |
|
Positive | 103 | 0.53 | −5.753 | 1.80 | −1.132 | 0.78 | −0.822 |
|
Negative | 61 | 1.15 |
<0.001b | 2.29 |
0.250 | 0.62 |
0.411 |
Associations between the mRNA
expression levels of OVCA1, cyclin D1 and p16
The relative mRNA expression levels of OVCA1 and p16
were decreased as the lesions progressed, and there was a positive
correlation between them (P=0.007). The relative expression levels
of cyclin D1 mRNA increased as lesions progressed; however, no
correlation was identified between OVCA1 and cyclin D1 and between
cyclin D1 and p16 mRNA expression levels (POVCA1 & cyclin
D1=0.845; Pp16 & cyclin D1=0.471; Table VI).
 | Table VI.Correlations between OVCA1, cyclin D1
and p16 mRNA expression levels. |
Table VI.
Correlations between OVCA1, cyclin D1
and p16 mRNA expression levels.
|
| Cyclin D1 | p16 |
|---|
|
|
|
|
|---|
|
| r | P-value | r | P-value |
|---|
| OVCA1 | 0.20 | 0.845 |
0.286 | 0.007a |
| Cyclin D1 | – | – | −0.073 | 0.471 |
Discussion
Although pathological detection remains the gold
standard for the diagnosis of cervical cancer, it is an invasive
examination, with a lack of consensus between pathologists, low
reproducibility rates for diagnosis and a high rate of misdiagnosis
of early or focal lesions (19,20). At
present, persistent HR-HPV infection is a recognized pathogenic
factor of cervical cancer, but HR-HPV infection alone is not
sufficient to cause cervical cancer, with few HR-HPV-infected
individuals developing cancer. The detection of HR-HPV lacks
specificity for the detection of cancer; therefore, medical
interventions to combat HR-HPV infection may cause patient distress
and overtreatment (19,20).
Advancements in molecular biology techniques have
allowed an improved understanding of the occurrence and development
of tumor sat the genetic level (21).
If molecular factors that are associated with tumor development are
identified, the accuracy of early diagnosis and severity evaluation
of cervical lesions may improve, providing a reference for the
prevention and treatment of cervical cancer.
First identified by Schultz et al (22) using positional cloning, the OVCA1 gene
is a tumor suppressor gene localized to17p13.3 that is widely
expressed in various human tissues. OVCA1 is transcribed as a 213
kb mRNA molecule, and translated to a protein encoding 443 amino
acids; its subcellular localization is surrounding the nucleus, and
its gene products in mammals are highly conserved (21). Increased OVCA1 protein is able to
inhibit the growth of tumor cells significantly, and small
alterations in its gene expression levels may lead to anomalies of
the cell cycle, promoting tumor development (10). A loss of heterozygosity in the OVCA1
gene has been detected in ovarian, breast and cervical cancer
(23–25), as well as other tumors (26–30),
indicating its potential role in the development of various
malignant tumors. In addition, previous studies have demonstrated
that OCVA1 is important in tumorigenesis, cell proliferation,
maintaining the normal growth of animals and inhibiting apoptosis,
metastasis and cell invasion (31,32).
In previous studies, the mRNA and protein expression
levels of the OVCA1 gene were decreased significantly or were lost
in ovarian cancer tissues and cell lines, as compared with normal
ovarian tissues, via an underlying mechanism that may be correlated
with the downregulation of cyclin D1 and upregulation of p16
expression (33–35).
High-frequency anomalies at the OVCA1 gene locus are
a feature of cervical cancer, and anomaly of number is as common as
that of structure. These abnormalities occur at an earlier stage
than p53 mutations, which are localized nearby (17p13.1). This
indicates that the deletion of OVCA1 in cervical cancer may be
correlated with cervical cancer progression, and appears at the
early-stage of disease (36,37). The detection of OVCA1 deletions may
contribute to the understanding of the molecular mechanisms
underlying the occurrence and development of cervical lesions at
the early stages. A previous study by Zhu (13) revealed that the rate of loss of all
exons of the OVCA1 gene in cervical cancer tissue was higher
compared with normal tissue, and that loss rates of the exons of
the OVCA1 gene in HPV16/18 positive individuals were higher
compared with HPV16/18 negative individuals. In addition, the
relative OVCA1 mRNA expression levels were higher in HPV16/18
positive cervical cancer tissues compared with HPV16/18 negative
samples; however, this difference was not significantly significant
(13). The relative OVCA1 mRNA
expression levels in cervical cancer tissues were significantly
decreased compared with those in normal tissues (13). Aberrant OVCA1 expression may occur at
the mRNA or protein level. To the best of our knowledge, no studies
have been conducted to examine the protein expression levels of
OVCA1 in cervical cancer and their association with cancer stage,
pathological grades, histological classification and HPV infection
status.
The current study revealed that the protein
expression levels of OVCA1 increased gradually in normal cervix,
CIN and cervical cancer tissues (P<0.05), whereas mRNA
expression decreased gradually (P<0.05). The protein and mRNA
expression levels of OVCA1 in cervical cancer exhibited
statistically significant differences between the early and
advanced stages (P<0.05), and the protein expression levels
exhibited statistically significant differences between CIN1 and
CIN2/3 (P<0.05), indicating that OVCA1 may function during the
process of cervical lesion development, particularly at the early
stages. There was a statistically significant difference between
OVCA1 protein and mRNA expression levels between the HR-HPV (+) and
HR-HPV (−) groups (P<0.05). Therefore, the aberrant expression
of OVCA1 in cervical lesions may be correlated with HR-HPV
infection, and HR-HPV infection may function during the
intermediate processes underlying cervical carcinogenesis. In
addition, the present study detected positive expression of OVCA1
in the HR-HPV (+) normal cervix tissue and CIN1 tissue, whereas
intense positive expression was observed in koilocytes, which
indicated the presence of HPV infection. Therefore, the current
study hypothesizes that, prior to triggering morphological changes
to cells, HR-HPV infection may promote the abnormal expression of
OVCA1 mRNA and protein. Furthermore, the detection of OVCA1
expression may aid the triage of diagnosis and the treatment of
HR-HPV-infected patients, as well as improving the detection rate
of CIN and cervical cancer at early stages. However, OVCA1 positive
expression was detected in HR-HPV (−) cases, which indicates that
the causes of abnormal OVCA1 expression may not be limited to
HR-HPV infection. The expression levels of OVCA1mRNA significantly
increased from the high to moderate to low differentiation cervical
cancer groups (P<0.05), and abnormal OVCA1 mRNA expression was
more common in well-differentiated cases at the early stage, but
there was no difference at the protein level. Therefore, further
studies with increased sample sizes are required.
Disorders of the cell cycle and the uncontrolled
growth of cells are primary mechanisms underlying tumorigenesis. As
a positive regulatory factor, cyclin is a convergent point of
numerous oncogenes and tumor suppressor genes, serving a key role
in the regulation of cell cycle events (38).
CyclinD1 is localized in the 11q13 chromosomal
region and is a regulatory nuclear protein, as well as being an
established potential oncogene, as its c-terminus has a PEST
sequence that is important in the cellular transformation process
(39). As a G1-stage cyclin, cyclin
D1 is important for G1/S-stage transformation of the
cell cycle. Following DNA synthesis, cyclinD1 forms a complex with
cyclin-dependent kinase (CDK) 6 or CDK4 at the G1 stage,
which combines with retinoblastoma protein (pRb) to promote DNA
transcription, allowing cells to pass the G1/S
checkpoint and triggers cell proliferation (39). Overexpression of cyclin D1 may shorten
the duration of the G1 stage and accelerate cell
proliferation, leading to tumorigenesis (39). The overexpression and amplification of
cyclin D1 are common features of cervical cancer and other
malignant tumors (40) and may
underlie the pathogenic process in ovarian cancer that involves the
loss of OVCA1 expression (34,41).
The p16 protein is an important tumor suppressor and
is a member of the cyclin-dependent kinase inhibitor family
(42–44). Through the competitive inhibition of
cyclin D1, p16 protein is able to function directly in the process
of cell cycle regulation to reduce cell proliferation (42–44).
Furthermore, p16 expression levels are negatively regulated by pRb
protein (45). Previous studies have
demonstrated that the abnormal expression of p16 is correlated with
the occurrence of cervical squamous carcinoma and CIN, and may
occur in part due to the combination of the E7 carcinogenic protein
of HR-HPV and pRB (46,47). A previous study suggested the
detection of p16 protein expression levels may be applied in the
differential diagnosis of high or low level CIN (48).
Preventing cell transformation and proliferation
from the G1 to S stage via upregulation of p16 and
inhibition of cyclin D1 expression may be a possible mechanism
underlying the effects of OVCA1 in ovarian cancer (21). The current study hypothesizes that, if
this mechanism was disrupted, cell cycle dysregulation may occur,
leading to uncontrolled proliferation of tumor cells. Therefore,
the detection of cyclin D1 and p16, in addition to examining the
correlations between the OVCA1 gene and cervical cancer
progression, may aid the elucidation of the potential underlying
mechanisms of abnormal OVCA1 expression in the development and
progression of cervical lesions, and the interaction of various
genes in the occurrence of cervical lesions.
In the current study, the protein and mRNA
expression levels of p16 in normal cervix, CIN and cervical cancer
tissues differed significantly (P<0.05). The protein expression
of p16 in early-stage cervical cancer in the HR-HPV negative group
was lower compared with cervical cancer in the advanced-stage and
HR-HPV positive group (P<0.05). In the case of HR-HPV infection,
p16 may function in the overall process of the occurrence and
development of cervical lesions together with OVCA1; however, the
present study observed no statistically significant differences in
p16 expression between low vs. high level CIN in the early events
of cervical lesions, which revealed that changes in OVCA1
expression may occur earlier compared withp16 expression. The
protein and mRNA expression levels of cyclin D1 in normal cervix
and cervical cancer tissue exhibited significant differences
(P<0.01), indicating its potential participation in the
development of cervical cancer. However, the protein expression of
cyclin D1 in normal cervix, CIN and low vs. high level CIN tissues
exhibited no significant differences (P>0.05), indicating that
cyclin D1 may function in cervical cancer at the advanced stage or
during cervical cancer metastasis. This result was consistent with
a previous study, which detected a correlation between cyclin D1
expression levels and the infiltration, metastasis and
deterioration of tumors, indicating that cyclin D1 may be an
important prognostic indicator (24).
In the current study, statistically significant
differences were identified between the protein expression levels
of p16 in cervical squamous carcinoma and adenocarcinoma tissues
(P<0.05); however, as the numbers of cases in the two groups
differed greatly, this may have led to bias and the results require
further study in an increased sample size. The correlations between
OVCA1, cyclin D1 and p16 and the lymph node metastasis of cervical
cancer exhibited no statistical significance. In addition, no
significant association was observed between cyclin D1 expression
and HR-HPV infection, which also requires further study with an
increased number of cases.
Positive correlations were observed in all pairwise
comparisons between OVCA1, cyclin D1 and p16 protein expression
(all P<0.05), with a more pronounced correlation between OVCA1
and p16 expression compared with OVCA1 and cyclin D1, as well as
cyclin D1 and p16. For mRNA expression, only OVCA1 and p16
exhibited a significantly positive correlation (P<0.01).
Therefore, in the occurrence and development of cervical lesions,
there may be interactions among OVCA1, cyclin D1, p16 and HR-HPV
infection, particularly for p16.
The occurrence and development of cervical cancer is
a multi-factor, multi-stage process involving numerous genes,
including a variety of oncogenes and tumor suppressors. Abnormal
OVCA1 gene and protein expression and its synergistic effects with
various cancer-associated genes (oncogenes, tumor suppressor genes
and other regulatory factors) may be important in the occurrence
and development of cervical cancer. Although the upregulation of
p16 expression levels and the inhibition of cyclin D1 expression
levels in ovarian cancer were possible mechanisms underlying the
effects of OCVA1, there may be varied mechanisms in cervical
cancer. HR-HPV infection maybe the initial factor underlying
changes in cyclin D1 expression, and its synergistic effect with
P16 was more pronounced than the associated functions of cyclin D1.
Therefore, cyclin D1 may function in the advanced stages of
cervical cancer. At present, there are few studies examining the
role of OCVA1 in cervical cancer, and the associated underlying
mechanisms by which OVCA1 is involved in internal functions of
cells and interacts with associated genes or factors, including p16
and HR-HPV infection remain to be elucidated. OCVA1 may be a
potential tumor marker and provide a novel basis for the clinical
diagnosis and treatment of cervical cancer.
Glossary
Abbreviations
Abbreviations:
|
HR-HPV
|
high-risk human papillomavirus
|
|
CIN
|
cervical intraepithelial neoplasia
|
References
|
1
|
Apostolidou S, Hadwin R, Burnell M, Jones
A, Baff D, Pyndiah N, Mould T, Jacobs IJ, Beddows S, Kocjan G and
Widschwendter M: DNA methylation analysis in liquid-based cytology
for cervical cancer screening. Int J Cancer. 125:2995–3002. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jiang BG, Chen AE and Li ZT: Clinical and
pathological characteristics of early cervical carcinoma in women
under 35 years of age. Mod Pract Med. 3:141–142. 2002.
|
|
3
|
Liu WX: Clinieal and prognostic analysis
of iuvasvie cevrical careinoma in women aged 40 and yunger
(unpublished PhD thesis). Tianjin Medical University. 2002.
|
|
4
|
Yu HY and Zhang X: Clinical
characteristics and prognosis of young women with cervical cancer.
China Health Care Nutrition. 08:793–794. 2012.
|
|
5
|
Bray F, Carstensen B, Moller H, Møller H,
Zappa M, Zakelj MP, Lawrence G, Hakama M and Weiderpass E:
Incidence trends of adenocarcinoma of the cervix in 13 European
countrines. Cancer Epidemiol Biomarkers Prev. 14:2181–2199. 2005.
View Article : Google Scholar
|
|
6
|
Wang SS, Sherman ME, Hildesheim A, Lacey
JV Jr and Devesa S: Cervical adenocarcinomal and squamous cell
carcinoma incidence trends among white women and black women in the
United States for 1976–2000. Cancer. 100:1035–1044. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Castellsagué X, Díaz M, de Sanjosè S,
Muñoz N, Herrero R, Franceschi S, Peeling RW, Ashley R, Smith JS,
Snijders PJ, et al: Worldwide human papillomavirus etiology of
cervical adenocanimoma and its cofactors: Impications for screening
and prevention. J Natl Cancer Inst. 98:303–315. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sasieni P, Castanon A and Cuzick J:
Screening and adenocarcinoma of the cervix. Int J Cancer.
125:525–529. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun JH, Cai SM and Gao YL: Some problems
in gynecologic oncology and training of Gynecologic Oncology.
Zhonghua Zhong Liu Za Zhi. 31:946–948. 2009.(In Chinese).
PubMed/NCBI
|
|
10
|
Boabang P, Kurbacher CM, Waida A and
Amo-Takyi BK: Detection of aberrations of chromosome 17 and p53
gene expression and their correlation to histologic grading and
prognosis in primary invasive squamous cell carcinoma of the
cervix. Gynecol Obstet Invest. 51:233–239. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bruening W, Prowse AH, Schultz DC,
Holgado-Madruga M, Wong A and Godwin AK: Expression of OVCA1, a
candidate tumor suppressor, is reduced in tumors and inhibits
growth of ovarian cancer cells. Cancer Res. 59:4973–4983.
1999.PubMed/NCBI
|
|
12
|
Kong FD, Zhao CY, Jia LY, Miao XY, Wei W,
Zhao XY, Yang G and Sun WP: Cloning of human OVCA1 gene and its in
vitro inhibitory effect on ovarian cancer cell growth. Int J Lab
Med. 16:1805–1806. 2011.
|
|
13
|
Zhu BZ: The study on the correlation of
tumor suppressor gene of OVCA1 and cervical cancer (unpublished PhD
thesis). Dalian Medlical University. 2010.
|
|
14
|
Pecorelli S: Revised FIGO staging for
caicinoma of the vulva, cervix, and endometrium. Int J Gynaecol
Obstet. 105:103–104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Richart RM: Cervical intraepithelial
neoplasia. Pathol Annu. 8:301–328. 1973.PubMed/NCBI
|
|
16
|
Cervical Intraepithelial Neoplasia (CIN)
Pipeline Review. H2 2015. Research and Markets. 2016.
|
|
17
|
Pierre PL Martin-Hirsch, Evangelos
Paraskevaidis, Andrew Bryant, Heather O Dickinson and Sarah L Keep:
Surgery for cervical intraepithelial neoplasia Pierre PL
Martin-Hirsch1. Cochrane Database Syst Rev CD001318. 2010.
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-timequantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kiaer SK, Frederiksen K, Munk C and Iftner
T: Long-term absolute risk of cervical intraepithelial neoplasia
grade 3 or worse following human papillomavirus infection: Role of
persistence. J Natl Cancer Inst. 102:1478–1488. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rodriguez AC, Schiffman M, Herrero R,
Hildesheim A, Bratti C, Sherman ME, Solomon D, Guillén D, Alfaro M,
Morales J, et al: Longitudinal study of human papillomavirus
persistence and cervical intraepithelial neoplasia grade 2/3:
Critical role of furation of infection. J Natl Cancer Inst.
102:315–324. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tong R, Yang Q and Wang CY: Research
advances on tumor suppressor gene OVCA1. Lett Biotechnology.
2:258–263. 2015.
|
|
22
|
Schultz DC, Vanderveer L, Berman DB,
Hamilton TC, Wong AJ and Godwin AK: Identification of two candidate
tumor uppressor genes on chromosome 17p13.3. Cancer Res.
56:1997–2002. 1996.PubMed/NCBI
|
|
23
|
Voeghtly LM, Mamula K, Campbell JL,
Shriver CD and Ellsworth RE: Molecular alterations associated with
breast cancer mortality. PLoS One. 7:e468142012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang GL, Yang H and Xu K: Loss of
heterozygosity on chromosome 17p13.3 in ovarian cancer and cervical
cancer. Zhonghua Zhong Liu Za Zhi. 19:401–403. 1997.(In Chinese).
PubMed/NCBI
|
|
25
|
Park SY, Kang YS, Kim BG, Lee SH, Lee ED,
Lee KH, Park KB and Lee JH: Loss of heterozygosity on the short arm
of chromosome 17 in uterine cervical carcinomas. Cancer Genet
Cytogenet. 79:74–78. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shikeeva AA, Kekeeva TV, Zavalishina LÉ,
Andreeva IuIu and Frank GA: Allelic imbalance in patients with
non-small cell lung cancer. Arkh Patol. 75:3–8. 2013.(In Russian).
PubMed/NCBI
|
|
27
|
Zhang B, Jia WH, Matsuda K, Kweon SS,
Matsuo K, Xiang YB, Shin A, Jee SH, Kim DH, Cai Q, et al:
Large-scale genetic study in East Asians identifies six new loci
associated with colorectal cancer risk. Nat Genet. 46:533–542.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Smardova J, Liskova K, Ravcukova B,
Kubiczkova L, Sevcikova S, Michalek J, Svitakova M, Vybihal V, Kren
L and Smarda J: High frequency of temperature-sensitive mutants of
p53 in glioblastoma. Pathol Oncol Res. 19:421–428. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kanamori M, Sano A, Yasuda T, Hori T and
Suzuki K: Array-based comparative genomic hybridization for
genomic-wide screening of DNA copy number alterations in aggressive
bone tumors. J Exp Clin Cancer Res. 31:1002012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ninomiya S, Tyybäkinoja A, Borze I, Räty
R, Saarinen-Pihkala UM, Usvasalo A, Elonen E and Knuutila S:
Integrated analysis of gene copy number, copy neutral LOH, and
microRNA profiles in adult acute lymphoblastic leukemia. Cytogenet
Genome Res. 136:246–255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen CM and Behringer RR: Ovcal regulates
cell proliferation, embryonic development, and tumorigenesis. Genes
Dev. 18:320–332. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kong FD, Liu L, Wei W, Zhao XY, Miao XY
and Zhao CY: Tumor suppressor gene OVCAl inhibits migration and
invasion of ovarian cancer cells A2780 in vitro. Chin J Cancer
Biother. 154:351–355. 2008.
|
|
33
|
Zhang JF: The expression of OVCA1 and P53
in ovarian serous Tumors (unpublished PhD thesis). Dalian Medlical
University. 2011.
|
|
34
|
Kong F, Tong R, Jia L, Wei W, Miao X and
Zhao C: OVCA1 inhibits the proliferation of epithelial ovarian
cancer cells by decreasing cyclin D1 and increasing p16. Mol Cell
Biochem. 354:199–205. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liang XX, Liang JF, Xiao H, Cheng CX, Zhao
YZ and Zheng HX: Expression and clinicopathological significance of
OVCA1 and HIC1 in ovarian epithelial tumors. Chinese Remedies
Clinics. 113:255–258. 2011.
|
|
36
|
Wiper DW and Zanotti KM: Analysis of
allelic imbalance on chromosome 17p13 in stage I and stage II
epithelial ovarian cancers. Gynecol Oncl. 71:77–82. 1998.
View Article : Google Scholar
|
|
37
|
Phillips NJ, Ziegler MR, Radford DM, Fair
KL, Steinbrueck T, Xynos FP and Donis-Keller H: Allelic deletion on
chromosome 17p13.3 in early ovarian cancer. Cancer Res. 56:606–611.
1996.PubMed/NCBI
|
|
38
|
Wang Q, Deng J and Jiang YX: Research
progress of cyclin D1. J Modern Oncol. 2:350–353. 2009.
|
|
39
|
Liang S, Mu K, Wang Y, Zhou Z, Zhang J,
Sheng Y and Zhang T: CyclinD1 a prominent prognostic marker for
endometrial diseases. Diagnostic Pathology. 8:1382013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lu S, Zhang BH and Wang Z: Expression of
survivin, cyclin D1, p21(WAF1), caspase-3 in cervical cancer and
its relation with prognosis. J Huazhong Univ Sci Technolog Med Sci.
25:78–81. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu XL: Expression of OVCAl gene and its
relation with the Expression of cyclin D1 in epithelial ovarian
tissue (unpublished PhD thesis). Dalian Medlical University.
2007.
|
|
42
|
Kamb A: Cell-cycle regulators and cancer.
Trends Genet. 11:136–140. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kamb A, Gruis NA, Weaver-Feldhaus J, Liu
Q, Harshman K, Tavtigian SV, Stockert E, Day RS III, Johnson BE and
Skolnick MH: A cell cycle regulator potentially involved in genesis
of many tumour types. Science. 264:436–440. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Velez Abreu AM and Howard MS:
Tumor-suppressor Genes, cell cycle regulatory checkpoints and the
skin. N Am J Med Sci. 7:176–188. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hao HB and XU PQ: Research progress of
relationship between tumor suppressor gene p16 and malignant tumor.
J Bengbu Medical College. 3:361–363. 2012.
|
|
46
|
Huang K, Li LA, Meng YG and Fu XY: p16
expression in patients with cervical cancer and its prognostic
significance: Meta-analysis of published literature. Eur J Obstet
Gynecol Reprod Biol. 183:64–69. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cai S and Han K: Research on expression
and importance of p53, p16 and VEGF-C in cervical cancer. J Gynecol
Obstet Biol Reprod (Paris). 44:639–645. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Krishnappa P, Mohamad IB, Lin YJ and Barua
A: Expression of P16 in high-risk human papillomavirus related
lesions of the uterine cervix in a government hospital, Malaysia.
Diagn Pathol. 202:2014.
|