Introduction
Forkhead box O class (FOXO) proteins are
evolutionally conserved transcription factors. FOXO transcription
factors perform an important role in tumor suppression, longevity
and metabolism by up regulating target genes associated with
oxidative stress resistance, metabolism, apoptosis, cell cycle
arrest, aging and autophagy (1–5). There are
four FOXOs members in mammalian cells, FOXO1, 3, 4 and 6 (6). As presented in Fig. 1, all FOXO members consist of the
following four conserved regions (CRs): An N-terminal highly
conserved 100-residue-long forkhead DNA-binding domain (FH); a
nuclear localization signal (NLS); a nuclear export signal (NES);
and a C-terminal transactivation domain (CTD) (1,7–9). Crystal structure analysis revealed that
the winged helix H3 region of the FH is the primary element for DNA
recognition (7,8) and is capable of binding the consensus
DNA sequence, 5′-TTGTTTAC-3′, termed the forkhead response element
(FRE) (9,10).
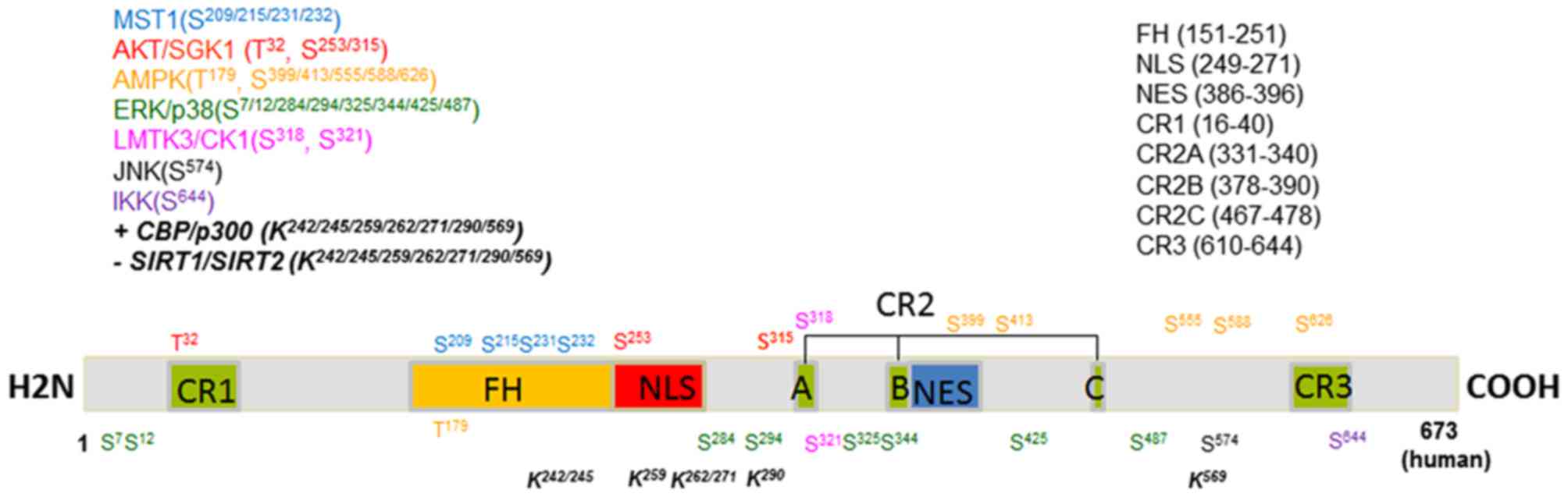 | Figure 1.Amino acid sites of phosphorylation
and acetylation of human FOXO3a. The different colors represent
different kinases for FOXO3a modification and different regions of
FOXO3a protein. A, B and C represent different sections of CR2.
MST1, mammalian sterile 20-like kinase 1; AKT, protein kinase B;
SGK, serum and glucocorticoid-induced kinase; AMPK, AMP-activated
protein kinase; ERK, extracellular signal-regulated kinase; LMTK,
lemur tyrosine kinase; CK1, casein kinase-1; JNK, c-Jun N-terminal
kinase; IKK, IκB kinase; CBP, cyclic AMP response element binding
protein; SIRT, silent information regulator; CR, conserved region;
FH, forkhead; NLS, nuclear localization signal; NES, nuclear export
signal; S, serine; T, threonine; K, lysine. |
There are two aspects of FOXO regulation;
subcellular localization and transcription activity. Multiple
post-translational modifications (PTMs) serve a major role in
determining FOXO subcellular localization and transcription
activity. As shown in Fig. 2, in
response to insulin/growth factor signaling, a number of different
protein kinases, including protein kinase B (AKT), serum and
glucocorticoid-induced kinase (SGK), extracellular signal-regulated
kinase (ERK), casein kinase-1 (CK1), and IkB kinase (IKK), are
capable of inducing translocation of FOXO into the cytoplasm
(11–13). By contrast, several upstream
regulators, such as c-Jun N-terminal kinase (JNK), mammalian
sterile 20-like kinase 1 (MST1), AMP-activated protein kinase
(AMPK) and lemur tyrosine kinase-3 (LMTK3) can phosphorylate FOXO
at specific residues, which in turn promotes nuclear localization
and transcriptional activity (11,12,14,15).
In addition to various phosphorylation modifications, acetylation
is another important regulator of FOXO proteins. The cyclic AMP
response element binding protein (CBP) and its associated protein
p300 form a coactivator complex (CBP/p300), which possesses histone
acetyl transferase (HAT) activity, interacts with FOXO and
regulates FOXO transcriptional activity (16,17).
Silent information regulator (SIRT) 1 and 2 are histone
deacetylases, which deacetylate FOXO and increase the expression of
specific stress resistance genes, including β-catenin and Gadd45
(18,19).
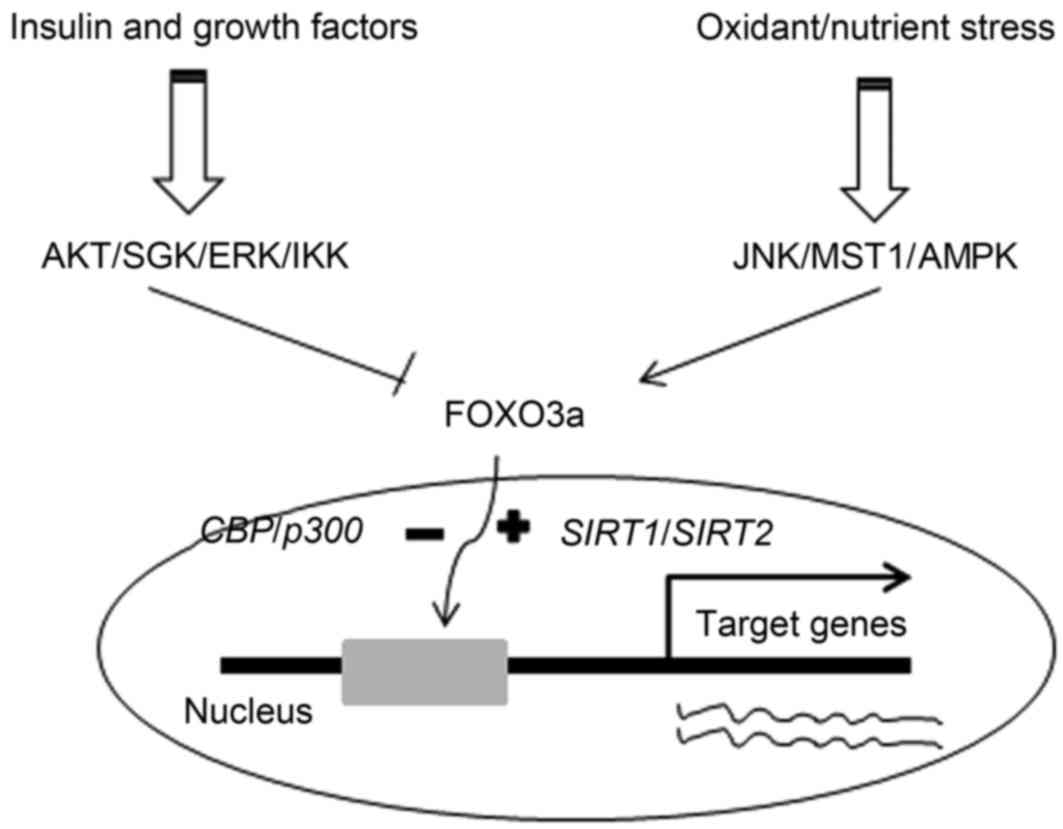 | Figure 2.Association between insulin/growth
factor signaling and oxidative/nutrient stress signaling in regards
to FOXO3a. AKT, protein kinase B; SGK, serum and
glucocorticoid-induced kinase; ERK, extracellular signal-regulated
kinase; IKK, IκB kinase; JNK, c-Jun N-terminal kinase; MST1,
mammalian sterile 20-like kinase 1; AMPK, adenosine
monophosphate-activated protein kinase; CBP, cyclic AMP response
element binding protein; FOXO3a, forkhead box class O 3a; SIRT,
silent information regulator. |
The effect of different types of phosphorylation on
the function of FOXO3a has been studied extensively (19). This has revealed that phosphorylation
and acetylation frequently occur simultaneously and affect one
other in certain conditions (19).
The present review summarized the phosphorylation and acetylation
sites of FOXO3a, in addition to analyzing the reciprocal regulation
of phosphorylation/acetylation and their effects on FOXO3a
subcellular localization and transcriptional activity.
Regulation of FOXO3a by phosphorylation
Phosphorylation modification regulates FOXO activity
via a cytoplasmic-nuclear shuttle mechanism. FOXO3a is targeted for
phosphorylation by numerous protein kinases. Several regulators
have been identified as kinases of FOXO3a in mammals (Table I). A summary of these regulatory
mechanisms are described below.
 | Table I.FOXO3a kinases, their phosphorylation
sites and effects. |
Table I.
FOXO3a kinases, their phosphorylation
sites and effects.
| Kinase | FOXO3a type | Sites of
phosphorylation | Localization | Effect |
|---|
| ERK/p38 | Human | S7,
S12, S284, S294, S325,
S344, S425, S487 | Cytoplasmic | Inhibition and
degradation |
|
| Rat | S7,
S12, S283, S293 | Cytoplasmic | Inhibition and
degradation |
|
| Mouse | S7,
S12, S283, S293,
S424 | Cytoplasmic | Inhibition and
degradation |
| IKK | Human |
S644 | Cytoplasmic | Inactivation and
degradation |
| AKT/SGK | Human | T32,
S253, S315 | Cytoplasmic | Inhibition |
|
| Rat | T32,
S252 | Cytoplasmic | Inhibition |
|
| Mouse | T32,
S252, S314 | Cytoplasmic | Inhibition |
| CK1 | Human | S318,
S321 | Cytoplasmic | Inhibition |
|
| Mouse | S317,
S320 | Cytoplasmic | Inhibition |
| DYRK1A/B | Human |
S325 | Cytoplasmic | Inhibition |
| AMPK | Human | T179,
S399, S413, S555, S588,
S626 | No effect |
Transactivation |
|
| Mouse |
S412 | No effect |
Transactivation |
| LMTK3 | Human | S318,
S321 | Nuclear |
Transactivation |
|
| Mouse | S317,
S320 | Nuclear |
Transactivation |
| MST1 | Human | S209,
S215, S231, S232 | Nuclear | Activation |
|
| Mouse |
S208 | Nuclear | Activation |
| JNK | Human |
S574 | Nuclear | Activation |
|
| Mouse |
S573 | Nuclear | Activation |
Negative effect of phosphorylation on
FOXO3a
As the major regulator of FOXO activation, AKT is
responsible for the phosphorylation of FOXO3a at threonine (Thr)32,
serine (Ser)253 and Ser315. This phosphorylation leads to the
exclusion of FOXO3a from the nucleus and the association of FOXO3a
with 14-3-3 proteins, which subsequently inhibits the
transactivation activity of FOXO3a (20,21). In
addition to phosphorylation and inhibition by AKT, FOXO3a is
phosphorylated by a number of other kinases. SGK is reported to
share the same FOXO phosphorylation sites as AKT, and also
decreases FOXO3a nuclear localization and DNA-binding activity
(11,22). ERKs have been demonstrated to inhibit
FOXO3a by directly phosphorylating FOXO3a at Ser294, Ser344 and
Ser425, and to induce the degradation of FOXO3a via the murine
double minute-2 signaling pathway, which consequently stimulates
cell survival and anti-apoptotic gene expression (23). In addition, IKK can phosphorylate
FOXO3a at Ser644, promoting cytoplasmic retention and
polyubiquitination-mediated degradation of FOXO3a (13).
CK1 has been identified to phosphorylate FOXO3a at
Ser318 and Ser321, leading to nuclear export and the inhibition of
FOXO3a activity (24). However, the
phosphorylation of FOXO3a at Ser315 by AKT is required for
CK1-dependent Ser318 and Ser321 phosphorylation (24). Dual-specificity tyrosine
phosphorylation and regulated kinase 1 (DYRK1) is also able to
phosphorylate FOXO proteins. It has been reported that DYRK1A
phosphorylates FOXO1 at Ser329 and FOXO3a at Ser325 (1,25). DYRK1B
serves an important role in ovarian cancer cell survival by
mediating the nuclear translocation of FOXO3a and the expression of
apoptotic genes (26). These data
suggest that the phosphorylation of FOXO3a by these kinases induces
cytoplasmic retention and degradation.
Positive effect of phosphorylation on
FOXO3a
FOXO-activating kinases can promote the nuclear
translocation and activity of FOXO3a. JNK is a member of the
oxidative stress-activated mitogen-activated protein kinase family.
Upon activation, JNK can phosphorylate Ser574 of FOXO3a, which
antagonizes the AKT signaling pathway and promotes the
transcription and nuclear translocation of FOXO3a (4,27). A
second kinase capable of promoting FOXO3a activation is AMPK, which
can phosphorylate FOXO3a at Thr179, Ser399, Ser413, Ser555, Ser588
and Ser626 (28). AMPK
phosphorylation increases the transcription activity of FOXO3a
without effecting the cytoplasmic-nuclear shuttling of FOXO3a
(28). The potential molecular
mechanism through which AMPK regulates FOXO is the recruitment of
additional proteins to transcriptional complexes or the direction
of FOXO to certain promoter regions. In response to doxorubicin,
p38 phosphorylates FOXO3a at Ser7, which promotes FOXO3 a nuclear
localization in breast carcinoma MCF-7 cells (29).
MST1 is also a positive regulator of FOXO activity.
Activated MST1 interacts with FOXO3a to phosphorylate Ser207, which
inhibits the association of FOXO3a with 14-3-3 proteins, and
promotes the nuclear translocation and transcription activity of
FOXO3a (15). LMTK3 is a
serine-threonine-tyrosine kinase that can inhibit protein kinase C
and AKT, thereby promoting the binding of FOXO3a to the estrogen
receptor 1 gene promoter element (30). In addition, upon silencing of LMTK3,
the level of AKT phosphorylation at Ser473 is increased, and the
frequency of FOXO3a phosphorylation by AKT at Ser318, Ser321 and
Thr32 is markedly decreased (30).
Cyclin-dependent kinase 1/2 (CDK1/2) has been demonstrated to
phosphorylate FOXO1 at conserved sites (31), although it remains unclear whether
they phosphorylate FOXO3a.
There is disagreement regarding the effect of CDK1/2
on FOXO1. CDK1 has been demonstrated to elicit cellular
proliferation, survival and tumorigenesis via the specific
phosphorylation of FOXO1 at Ser249, which inhibits the activity of
FOXO1 (31). In addition, CDK2 has
been reported to phosphorylate FOXO1 at Ser249 and Ser298, causing
cytoplasmic accumulation and inhibiting FOXO1 activity (32). However, a previous study demonstrated
that the phosphorylation of FOXO1 at Ser249 disrupted the binding
between FOXO1 and 14-3-3 proteins, thereby promoting the nuclear
accumulation of FOXO1 and FOXO1-dependent transcription, leading to
cell death in neurons (33).
Similarly to FOXO1, FOXO3a has been suggested as a potential target
of CDK1/2. Thus, studies investigating the effect of CDK1/2 on the
subcellular localization and transcription activity of FOXO3a in
response to certain stimuli are warranted.
Synergistic effect of multiple
phosphorylations of FOXO3a
The effects of multiple phosphorylation
modifications of FOXO3a by different kinases are not independent.
Specific residues of FOXO3a can be phosphorylated by a number of
kinases. As illustrated in Table I,
ERK and p38 share several conserved FOXO3a phosphorylation sites in
humans, rats and mice. Similarly, AKT and SGK1 have several of the
same phosphorylation sites on FOXO3a, and inhibit the activity of
FOXO3a. In humans, two conserved residues sites (Ser318 and Ser321)
can be phosphorylated by LMTK3 and CK1. However, LMTK3 and CK1 can
elicit opposite effects through phosphorylating the same sites.
LMTK3 phosphorylation of FOXO3a at Ser318/Ser321 in humans has been
demonstrated promote FOXO3a activity (30,31). By
contrast, the phosphorylation of these two sites by CK1 results in
the inhibition of FOXO3a activity (11,24).
Under certain conditions, kinases are likely to
simultaneously regulate one other. As described above, CK1
phosphorylates FOXO3a at Ser318/Ser321 following the
phosphorylation of Ser315 by AKT, which enabled the identification
of consensus sequences for CK1 phosphorylation (24). Under oxidative stress, JNK and MST1
participate in similar signaling pathways and act in a
complementary manner to activate FOXO3a. JNK directly
phosphorylates FOXO3a and also phosphorylates MST1 at Ser82, which
leads to enhanced MST1 activation (34). Accordingly, activated MST1
phosphorylates FOXO3 at Ser207 and promotes cell death (34). AKT inhibits MST1 through
phosphorylating it at Thr120, blocking MST1-mediated
phosphorylation and activation of FOXO3a (35). Additionally, MST1 has also been
demonstrated to be capable of binding to and inhibiting AKT,
resulting in an inhibition of cellular proliferation (36).
Regulation of FOXO3a by acetylation
Different PTMs change the subcellular localization
and/or DNA-binding activity of FOXO3a, allowing FOX3a to serve a
role in various cellular activities. Similarly to phosphorylation,
acetylation is another PTM that can regulate FOXO3a.
Dual effects of CBP/p300-mediate
acetylation of FOXO3a
In the majority of cases, acetylation aids the
transactivation of genes. The binding of the CBP/p300 coactivator
to FOXO proteins is essential for the transactivation of target
genes (37). CBP/p300, which
acetylates nucleosomal histones, can also acetylate FOXO3a.
Acetylation of nucleosomal histones stimulates transcription,
whereas the acetylation of FOXO3a has been reported to attenuate
its transcriptional activity (38,39). It is
possible that acetylated FOXO3a has a decreased DNA-binding
activity. In addition, acetylation has been observed to cause the
stimulation and depression of FOXO protein activity, dependent on
the FOXO isoform and binding partners present (38,39). A
previous study demonstrated that CBP/p300 acetylation of FOXO1 at
specific highly conserved sites decreased DNA-binding activity
(38). Additionally, under oxidative
stress CBP has been demonstrated to trigger the acetylation of
FOXO4 and inhibit its activity (39).
Furthermore, FOXO3a interacts with the HAT p300, the acetylation of
which causes cytoplasmic retention and induces the degradation of
FOXO3a via the proteasome (40,41). In
response to nutrient deprivation, dominant-negative p300 was
identified to induce FOXO3a nuclear translocation, while
transfection of CBP/p300 resulted in FOXO3a repression (41). In addition, AKT has been demonstrated
to phosphorylate acetylated FOXO proteins, which resulted in
cytoplasmic retention and 14-3-3 proteins binding (42).
Deacetylation of FOXO3a is induced by
sirtuins
The CBP/p300-mediated acetylation of FOXO3a
attenuates its transcriptional activity. By contrast, deacetylation
by SIRT1 increases the activity of FOXO3a. It has been reported
that SIRT1 deacetylates and activates FOXO3a in the nucleus
accumbens, leading to the expression of several targets genes
(43). However, the deacetylation of
FOXO3a by SIRT1 has also been reported to repress FOXO3a activity.
Motta et al (44) demonstrated
that SIRT1 deacetylates and decreases the activity of FOXO
proteins, including FOXO1, FOXO3a and FOXO4. A recent study also
demonstrated that SIRT1 could bind to and deacetylate FOXO3a, which
increased its ubiquitination level and protected against apoptosis
(45). SIRT2 also serves an important
role in FOXO3a deacetylation. SIRT2 deacetylates FOXO3, increasing
the DNA-binding activity of FOXO3 and thus increasing the
expression of its target genes, cyclin-dependent kinase inhibitor
1B (p27Kip1), manganese superoxide dismutase and BCL2 like 11 (Bim)
(18,19). Notably, in response to oxidative
stress, SIRT1 interacts with and deacetylates FOXO3a, which
decreases the expression of pro-apoptotic Bim, but increases the
expression of cell cycle arrest genes, including p27Kip1 and growth
arrest and DNA inducible α (16).
These results highlight the complex mechanisms by which FOXO is
regulated by acetylation and deacetylation. It is possible that the
interaction between acetylation and phosphorylation in FOXO
regulation may be the result of further mechanisms that have not
yet been elucidated.
Mechanisms of phosphorylation and
acetylation of FOXO3a
As shown in Fig. 1,
FOXO3a consists of the following CRs: A FH (residues 151–251); a
NLS (residues 249–271); a NES (residues 386–396); CR1 (residues
16–40); CR2A (residues 331–340); CR2B (residues 378–390); CR2C
(residues 467–478); and CR3 (residues 610–650) (1,7–10,19,46). FH is
a conserved DNA-binding domain, which is capable of binding to the
FRE and allowing transcription (1,7–9). MST1 (Ser209, Ser215, Ser231 and Ser232)
and AMPK (Thr179) can directly phosphorylate FOXO3a in the FH
region (15,34,43). These
conserved phosphorylation sites have been demonstrated to increase
the DNA-binding activity of FOXO3a. NLS and NES domains are
important for FOXO3a subcellular localization (15,34,43).
AKT/SGK phosphorylates FOX3a at Ser253, which is located in the NLS
and acts to disrupt NLS activity by introducing a negative charge
to the basic NLS region (19).
Phosphorylation of Ser315 reveals the NES of FOXO, increasing its
rate of nuclear export (38). The CR3
domain is a CTD that can interact with the FH region (7,10). It has
been demonstrated that the binding of FH to FRE releases CR3,
allowing it to bind to the KIX domain of CBP/p300, which can then
acetylate and inhibit the DNA binding capacity of FOXO3a (36). CR2C is also a transactivation domain,
and mediates the association between FOXO3a and CBP/p300 by binding
to the KIX domain (10,46).
Under certain conditions, coordinating the
regulation of phosphorylation and acetylation may synergistically
control the function of FOXO3a, including DNA binding activity,
subcellular localization and transactivation activity. The
processes of acetylation and phosphorylation of the CRs of FOXO3a
affects one other. It is known that MST1 (Ser209, Ser215, Ser231
and Ser232) and AMPK (Thr179) can directly phosphorylate FOXO3a in
the FH domain, which increases the DNA-binding affinity of FOXO3a.
Activated SIRT1 deacetylates Lysine (Lys)242 and Lys245 in the FH
domain and also enhances the DNA binding of FOXO3a (43). Previous studies have demonstrated that
SIRT1, MST1 and AMPK are activated in response to oxidative stress
(15,16,28,34,35).
This suggests that the DNA binding affinity of FOXO3a is controlled
by the interaction between acetylation and phosphorylation.
Certain acetylation sites in FOXO3a surround a
consensus site for AKT-induced Ser phosphorylation within the NLS
domain. Acetylation of FOXO3a by CBP/p300 has been suggested to
promote AKT-mediated phosphorylation at Ser253 in the NLS motif
(41,42). It is therefore possible that the
acetylation of Lys259, Lys262 and Lys271 in the NLS region of
FOXO3a changes the conformation of the NLS and results poor
affinity for the DNA, making the NLS prone to phosphorylation by
AKT (42). AKT phosphorylation of
FOXO3a at Thr32 in the CR1 region and Ser253 in the NLS region
increases the binding of FOXO3a to 14-3-3 proteins, and decreases
the interaction between FOXO3a and CBP/p300 (42). Phosphorylated FOXO3a (Thr32/Ser253) is
unable to bind to CBP/p300 and loses the ability for chromatin
remodeling. AMPK phosphorylation of FOXO3a at Ser626 in the CR3
domain increases the binding of CBP/p300 and transactivation
activity of FOXO3a (10). It has also
been demonstrated that AMPK enhances SIRT1 activity by increasing
cellular NAD+ levels, resulting in the deacetylation of
FOXO3a (47). The effect of
IKK-mediated Ser644 phosphorylation in the acetylated CR3 region of
FOXO3a remains unclear. Thus, the potential interaction between
acetylation and phosphorylation in the conserved PTM sites of
FOXO3a discussed in the present review is an important area to
investigate in future studies.
Conclusion
In conclusion, the effect of phosphorylation and
acetylation on the transactivation activity of FOXO3a represents a
complex balance between insulin/growth factor signaling and
oxidative stress signaling (Fig. 2).
Insulin and growth factors initiate a signaling cascade that
results in the activation of kinases, including AKT/SGK, ERK and
IKK (11–13). These kinases phosphorylate FOXO3a at
specific sites, inducing its cytoplasmic retention and/or
degradation, leading to inhibition of target genes transcription
(13,19–23). By
contrast, oxidative or nutrient stress signals induce activation of
JNK, MST1 and AMPK, which phosphorylate FOXO3a at the conserved
residue sites, resulting in nucleus localization of FOXO3a and
promoting the transcription of target genes (11,12,14,15).
Besides, stresses also increase SIRT1/SIRT2-FOXO3a interaction,
resulting in tightly binding of FOXO3a to DNA binding domain and
enhancing target genes transcription (18,19,45).
Although acetylation does not always correspond with
phosphorylation, phosphorylation and acetylation can cooperate to
regulate FOXO3a in response to different stimuli. Thus, the potent
functions of FOXO3a are tightly controlled by complex signaling
pathways under physiological conditions. Gaining better
understanding of the PTMs of FOXO3a remains an important area of
investigation and should be addressed in future studies.
Acknowledgements
The present review was supported by Hubei Province
Natural Science Foundation of China (grant no. 2016CFB180), Hubei
Province Health and Family Planning Scientific Research Project
(grant no. WJ2016Y07), Jingzhou Science and Technology Development
Planning Project (grant no. JZKJ15063) and the National Natural
Science Foundation of China (grant no. 31470072).
Glossary
Abbreviations
Abbreviations:
|
FOXO3a
|
forkhead box class O 3a
|
|
FH
|
forkhead DNA-binding domain
|
|
NLS
|
nuclear localization signal
|
|
NES
|
nuclear export signal
|
|
CTD
|
C-terminal transactivation domain
|
|
FRE
|
forkhead response element
|
|
PTM
|
post-translational modification
|
|
CK1
|
casein kinase-1
|
|
HAT
|
histone acetyltransferase
|
|
SGK
|
serum and glucocorticoid-induced
kinase
|
|
ERK
|
extracellular signal-regulated
kinase
|
|
AMPK
|
AMP-activated protein kinase
|
|
IKK
|
IκB kinase
|
|
MST1
|
mammalian sterile 20-like kinase 1
|
|
LMTK3
|
lemur tyrosine kinase-3
|
|
DYRK1
|
dual-specificity tyrosine
phosphorylated and regulated kinase 1
|
|
CDK1/2
|
cyclin-dependent kinase 1/2
|
|
SIRT
|
silent information regulator
|
|
CR
|
conserved region
|
References
|
1
|
Calnan DR and Brunet A: The FoxO code.
Oncogene. 27:2276–2288. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang J, Liu S, Yin Y, Li M, Wang B, Yang L
and Jiang Y: FOXO3-mediated up-regulation of Bim contributes to
rhein-induced cancer cell apoptosis. Apoptosis. 20:399–409. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kloet DE and Burgering BM: The PKB/FOXO
switch in aging and cancer. Biochim Biophys Acta. 1813:1926–1937.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang XW, Chen WR and Xing D: A pathway
from JNK through decreased ERK and Akt activities for FOXO3a
nuclear translocation in response to UV irradiation. J Cell
Physiol. 227:1168–1178. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhou J, Liao W, Yang J, Ma K, Li X, Wang
Y, Wang D, Wang L, Zhang Y, Yin Y, et al: FOXO3 induces
FOXO1-dependent autophagy by activating the AKT1 signaling pathway.
Autophagy. 8:1712–1723. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang M, Zhang X, Zhao H, Wang Q and Pan Y:
FoxO gene family evolution in vertebrates. BMC Evol Biol.
9:2222009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Clark KL, Halay ED, Lai E and Burley SK:
Co-crystal structure of the HNF-3/fork head DNA-recognition motif
resembles histone H5. Nature. 364:412–420. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Obsil T and Obsilova V: Structural basis
for DNA recognition by FOXO proteins. Biochim Biophys Acta.
1813:1946–1953. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang F, Marshall CB, Yamamoto K, Li GY,
Plevin MJ, You H, Mak TW and Ikura M: Biochemical and structural
characterization of an intramolecular interaction in FOXO3a and its
binding with p53. J Mol Biol. 384:590–603. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang F, Marshall CB, Yamamoto K, Li GY,
Gasmi-Seabrook GM, Okada H, Mak TW and Ikura M: Structures of KIX
domain of CBP in complex with two FOXO3a transactivation domains
reveal promiscuity and plasticity in coactivator recruitment. Proc
Natl Acad Sci USA. 109:6078–6083. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Boccitto M and Kalb RG: Regulation of
foxo-dependent transcription by post-translational modifications.
Curr Drug Targets. 12:1303–1310. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao Y, Wang Y and Zhu WG: Applications of
post-translational modifications of FoxO family proteins in
biological functions. J Mol Cell Biol. 3:276–282. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang
F, Yang JY, Zou Y, Bao S, Hanada N, Saso H, et al: IkappaB kinase
promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell.
117:225–237. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Essers MA, Weijzen S, de Vries-Smits AM,
Saarloos I, de Ruiter ND, Bos JL and Burgering BM: FOXO
transcription factor activation by oxidative stress mediated by the
small GTPaseRal and JNK. EMBO J. 23:4802–4812. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lehtinen MK, Yuan Z, Boag PR, Yang Y,
Villén J, Becker EB, DiBacco S, de la Iglesia N, Gygi S, Blackwell
TK and Bonni A: A conserved MST-FOXO signaling pathway mediates
oxidative-stress responses and extends life span. Cell.
125:987–1001. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brunet A, Sweeney LB, Sturgill JF, Chua
KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et
al: Stress-dependent regulation of FOXO transcription factors by
the SIRT1 deacetylase. Science. 303:2011–2015. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Daitoku H, Sakamaki J and Fukamizu A:
Regulation of FoxO transcription factors by acetylation and
protein-protein interactions. Biochim Biophys Acta. 1813:1954–1960.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nakagawa T and Guarente L: Sirtuins at a
glance. J Cell Sci. 124:833–838. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang F, Nguyen M, Qin FX and Tong Q: SIRT2
deacetylates FOXO3a in response to oxidative stress and caloric
restriction. Aging Cell. 6:505–514. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo
P, Hu LS, Anderson MJ, Arden KC, Blenis J and Greenberg ME: Akt
promotes cell survival by phosphorylating and inhibiting a Forkhead
transcription factor. Cell. 96:857–868. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dobson M, Ramakrishnan G, Ma S, Kaplun L,
Balan V, Fridman R and Tzivion G: Bimodal regulation of FoxO3 by
AKT and 14-3-3. Biochim Biophys Acta. 1813:1453–1464. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Brunet A, Park J, Tran H, Hu LS, Hemmings
BA and Greenberg ME: Protein kinase SGK mediates survival signals
by phosphorylating the forkhead transcription factor FKHRL1
(FOXO3a). Mol Cell Biol. 21:952–965. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang JY, Zong CS, Xia W, Yamaguchi H, Ding
Q, Xie X, Lang JY, Lai CC, Chang CJ, Huang WC, et al: ERK promotes
tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation.
Nat Cell Biol. 10:138–148. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rena G, Woods YL, Prescott AR, Peggie M,
Unterman TG, Williams MR and Cohen P: Two novel phosphorylation
sites on FKHR that are critical for its nuclear exclusion. EMBO J.
21:2263–2271. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Woods YL, Rena G, Morrice N, Barthel A,
Becker W, Guo S, Unterman TG and Cohen P: The kinase DYRK1A
phosphorylates the transcription factor FKHR at Ser329 in vitro, a
novel in vivo phosphorylation site. Biochem J. 355:597–607. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gao J, Yang X, Yin P, Hu W, Liao H, Miao
Z, Pan C and Li N: The involvement of FoxO in cell survival and
chemosensitivity mediated by Mirk/Dyrk1B in ovarian cancer. Int J
Oncol. 40:1203–1209. 2012.PubMed/NCBI
|
|
27
|
Tikhanovich I, Kuravi S, Campbell RV,
Kharbanda KK, Artigues A, Villar MT and Weinman SA: Regulation of
FOXO3 by phosphorylation and methylation in hepatitis C virus
infection and alcohol exposure. Hepatology. 59:58–70. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Greer EL, Oskoui PR, Banko MR, Maniar JM,
Gygi MP, Gygi SP and Brunet A: The energy sensor AMP-activated
protein kinase directly regulates the mammalian FOXO3 transcription
factor. J BiolChem. 282:30107–30119. 2007.
|
|
29
|
Ho KK, McGuire VA, Koo CY, Muir KW, de
Olano N, Maifoshie E, Kelly DJ, McGovern UB, Monteiro LJ, Gomes AR,
et al: Phosphorylation of FOXO3a on Ser-7 by p38 promotes its
nuclear localization in response to doxorubicin. J Biol Chem.
287:1545–1555. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Giamas G, Filipović A, Jacob J, Messier W,
Zhang H, Yang D, Zhang W, Shifa BA, Photiou A, Tralau-Stewart C, et
al: Kinome screening for regulators of the estrogen receptor
identifies LMTK3 as a new therapeutic target in breast cancer. Nat
Med. 17:715–719. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu P, Kao TP and Huang H: CDK1 promotes
cell proliferation and survival via phosphorylation and inhibition
of FOXO1 transcription factor. Oncogene. 27:4733–4744. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang H, Regan KM, Lou Z, Chen J and
Tindall DJ: CDK2-dependent phosphorylation of FOXO1 as an apoptotic
response to DNA damage. Science. 314:294–297. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yuan Z, Becker EB, Merlo P, Yamada T,
DiBacco S, Konishi Y, Schaefer EM and Bonni A: Activation of FOXO1
by Cdk1 in cycling cells and postmitotic neurons. Science.
319:1665–1668. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bi W, Xiao L, Jia Y, Wu J, Xie Q, Ren J,
Ji G and Yuan Z: c-Jun N-terminal kinase enhances MST1-mediated
pro-apoptotic signaling through phosphorylation at serine 82. J
Biol Chem. 285:6259–6264. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yuan Z, Kim D, Shu S, Wu J, Guo J, Xiao L,
Kaneko S, Coppola D and Cheng JQ: Phosphoinositide 3-kinase/Akt
inhibits MST1-mediated pro-apoptotic signaling through
phosphorylation of threonine 120. J Biol Chem. 285:3815–3824. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chao Y, Wang Y, Liu X, Ma P, Shi Y, Gao J,
Shi Q, Hu J, Yu R and Zhou X: Mst1 regulates glioma cell
proliferation via the AKT/mTOR signaling pathway. J Neurooncol.
121:279–288. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
van der Heide LP and Smidt MP: Regulation
of FoxO activity by CBP/p300-mediated acetylation. Trends Biochem
Sci. 30:81–86. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Daitoku H, Hatta M, Matsuzaki H, Aratani
S, Ohshima T, Miyagishi M, Nakajima T and Fukamizu A: Silent
information regulator 2 potentiates Foxo1-mediated transcription
through its deacetylase activity. Proc Natl Acad Sci USA.
101:10042–10047. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
van der Horst A, Tertoolen LG, de
Vries-Smits LM, Frye RA, Medema RH and Burgering BM: FOXO4 is
acetylated upon peroxide stress and deacetylated by the longevity
protein hSir2(SIRT1). J Biol Chem. 279:28873–28879. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bertaggia E, Coletto L and Sandri M:
Posttranslational modifications control FoxO3 activity during
denervation. Am J Physiol Cell Physiol. 302:C587–C596. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Senf SM, Sandesara PB, Reed SA and Judge
AR: p300 Acetyltransferase activity differentially regulates the
localization and activity of the FOXO homologues in skeletal
muscle. Am J Physiol Cell Physiol. 300:C1490–C1501. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Matsuzaki H, Daitoku H, Hatta M, Aoyama H,
Yoshimochi K and Fukamizu A: Acetylation of Foxo1 alters its
DNA-binding ability and sensitivity to phosphorylation. Proc Natl
Acad Sci USA. 102:11278–11283. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ferguson D, Shao N, Heller E, Feng J, Neve
R, Kim HD, Call T, Magazu S, Shen L and Nestler EJ: SIRT1-FOXO3a
regulate cocaine actions in the nucleus accumbens. J Neurosci.
35:3100–3111. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Motta MC, Divecha N, Lemieux M, Kamel C,
Chen D, Gu W, Bultsma Y, McBurney M and Guarente L: Mammalian SIRT1
represses forkhead transcription factors. Cell. 116:551–563. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang YQ, Cao Q, Wang F, Huang LY, Sang TT,
Liu F and Chen SY: SIRT1 protects against oxidative stress-induced
endothelial progenitor cells apoptosis by inhibiting FOXO3a via
FOXO3a ubiquitination and degradation. J Cell Physiol.
230:2098–2107. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang F, Marshall CB, Li GY, Yamamoto K,
Mak TW and Ikura M: Synergistic interplay between promoter
recognition and CBP/p300 coactivator recruitment by FOXO3a. ACS
Chem Biol. 4:1017–1027. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cantó C, Gerhart-Hines Z, Feige JN,
Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P and Auwerx
J: AMPK regulates energy expenditure by modulating NAD+ metabolism
and SIRT1 activity. Nature. 458:1056–1060. 2009. View Article : Google Scholar : PubMed/NCBI
|
















