Introduction
Cholangiocarcinoma (CCA; a fatal bile duct cancer)
is a major public health concern in the Lower Mekong Basin and
South East Asia, which are areas endemic for Opisthorchis
viverrini liver fluke infection (1). The prognosis of CCA is principally poor
because the majority of patients with CCA are diagnosed at an
advanced stage, therefore they are inoperable and there are no
effective treatments available (2).
Additionally, CCA is prone to developing multidrug chemoresistance
(3,4).
Therefore, there is a requirement to investigate novel targeted
therapies and strategies to enhance chemosensitivity of CCA.
We previously demonstrated that the alteration of
cytoprotective enzymes or derangement of intracellular redox
balance and the signaling system were involved in the
chemoresistance of CCA (5–8). NAD(P)H:quinone oxidoreductase 1 (NQO1;
EC 1.6.5.2), one of the detoxifying enzymes with antioxidant
properties, has been proposed to be associated with the
chemotherapeutic response of CCA (5,8). NQO1 is
generally recognized as a ‘cell protector’, its induction in
response to various noxious stimuli provides protection for cells
against oxidative damage and oxidative stress-associated
pathological conditions including cancer (9,10).
Conversely, an increasing number of studies revealed abnormal
increases in NQO1 expression levels in solid tumors of the adrenal
gland, breast, colon, lung, ovary, pancreas, thyroid, skin and
bladder (9–16). High-level expression of NQO1 may be
associated with cancer progression and it was suggested to be a
poor prognostic marker of these types of cancer (14,16,17).
Upregulation of NQO1 during carcinogenesis may provide cancer cells
with a growth advantage and protection against extreme oxidative
stress environments (10,11). Considering the function of NQO1, an
increased NQO1 expression level may be associated with
disappointing outcomes to certain cancer treatment modalities,
including chemotherapy and radiotherapy, which induces cancer cell
death by the generation of free radicals and oxidative damage
(5,8).
The roles of NQO1 during carcinogenesis and
chemotherapeutic response have been demonstrated by numerous
previous studies (11,18,19).
Inhibition of NQO1 by a pharmacological inhibitor, dicoumarol,
suppressed urogenital and pancreatic cancer cell growth and also
potentiated cytotoxicity of cisplatin and doxorubicin (18,20).
Similarly, the roles of NQO1 in CCA have been previously
demonstrated (5,8,17,21). Significant association between high
NQO1 expression level in CCA tissues and short survival time of
patients was observed (17), implying
NQO1 is an independent predictor associated with prognosis of CCA.
Furthermore, dicoumarol was able to enhance gemcitabine-induced
cytotoxicity in CCA cells with increased NQO1 activity (5). In addition, knockdown of NQO1 expression
levels enhanced the cytotoxicity of chemotherapeutic agents;
conversely, overexpression of NQO1 protected the cells from
chemotherapeutic agents (8). These
results suggested roles for NQO1 in CCA chemotherapy; however, the
biological role of NQO1 in CCA cells has not yet been clearly
demonstrated.
The aim of the present study was to investigate the
biological role of NQO1 in CCA cells. The effects of NQO1 knockdown
on cell proliferation, cell cycle and migration were assessed in
KKU-100 CCA cells, which notably expressed NQO1. Furthermore, the
molecular events associated with NQO1 small interfering RNA
(siRNA)-induced inhibition of cell proliferation, inducing cell
cycle arrest and inhibiting migration of CCA cells were
investigated.
Materials and methods
Human cell line and cell culture
KKU-100 cells with high expression levels of NQO1
were provided by Professor Banchob Sripa (Department of Pathology,
Faculty of Medicine, Khon Kaen University, Khon Kaen, Thailand).
KKU-100 cells were established, characterized and derived from CCA
tissues (22). Cells were routinely
cultured in Ham's F-12 complete medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 10 mM
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (pH 7.3), 100
U/ml penicillin G and 100 µg/ml streptomycin, and maintained under
an atmosphere of 5% CO2 at 37°C. Cells were passaged
every 3 days using 0.25% trypsin-EDTA (2).
NQO1 siRNA transfection
The transfection of NQO1 siRNA was performed using
four sequences of predesigned NQO1 siRNA (siGENOME SMARTpool of
siRNA M-005133-02-0010; Dharmacon Inc., Lafayette, CO, USA), as
described previously (8). The NQO1
siRNA and the negative control siRNA (siGENOME non-targeting siRNA
pool#2 D-001206-14-20) at a final concentration of 100 nM siRNA
were introduced to the cells using Lipofectamine™ 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) as a transfection reagent,
according to the manufacturer's protocol. The efficiency of the
NQO1 knockdown by transient NQO1 siRNA transfection was confirmed
by evaluating levels of gene expression by performing the reverse
transcription-quantitative polymerase chain reaction (RT-qPCR),
protein expression levels by performing western blot analysis and
NQO1 enzyme activity using an enzymatic assay.
Clonogenic survival assay
Cells with NQO1 siRNA were used in the colony
formation assay. KKU-100 cells were transfected with NQO1 siRNA
then seeded in 6-well plates at densities of 300 and 600 cells/well
in Ham's F-12 complete medium. Two cell densities were used to
confirm the effect of NQO1 siRNA on replicative
potential of KKU-100 cells. Cells were cultured at 37°C in a 5%
CO2 incubator for another 10 days to obtain cell
colonies and the Ham's F-12 complete medium was freshly renewed
every 3 days. On the last experiment day, the cell colonies were
fixed with absolute methanol at room temperature (25°C) for 20 min,
stained with 0.5% crystal violet solution at room temperature
(25°C) for 15 min and the number of cell colonies was counted
(23).
Cell cycle analysis using flow
cytometry
KKU-100 cells were seeded in a 6-well plate with a
density of 150,000 cells/well in Ham's F-12 complete medium,
transfected with NQO1 siRNA for 24 h and then starved for 12 h
using serum-free Ham F-12 medium at 37°C and 5% CO2. At
24 h post-starvation, the cell cycle distribution was evaluated
using flow cytometric analysis. On the experimental day, cells were
harvested, washed with PBS and fixed overnight with 70% cold
ethanol at 4°C. Subsequently, the cell suspension was maintained at
−20°C for 3 h, followed by the addition of 0.02 mg/ml propidium
iodide and 0.02 mg/ml RNase A (Ameresco, Inc., Framingham, MA,
USA). The cell suspension was incubated further for 1 h at 4°C in
the dark. The cell cycle stage content was evaluated using a BD
FACSCanto II flow cytometer and determined using BD FACSDiva™
software v6.1.3 (BD Biosciences, San Jose, CA, USA). The service
was provided by the Research Instrument Center, Khon Kaen
University, Thailand.
Cell motility by wound healing and
Transwell migration assays
Cells with NQO1 siRNA were used in the wound-healing
and Transwell migration assays. For the wound-healing assay,
KKU-100 cells transfected with NQO1 siRNA were seeded in a 24-well
plate at a cell density of 150,000 cells/well in Ham's F-12
complete medium at 37°C with 5% CO2 overnight to obtain
90–100% cell confluence. On the next day, cells were scratched to
create a straight wound using a sterile 200-µl pipette tip.
Following scratching, the cells were washed twice with PBS to
remove any detached cells and further incubated for 72 h at 37°C
and 5% CO2. The width of the wound outline was monitored
under a phase-contrast microscope at designed times; 0, 3, 6, 12,
24, 48 and 72 h.
For the Transwell migration assay, KKU-100 cells
transfected with NQO1 siRNA were seeded onto a 6.5 mm Transwell
insert (Corning Incorporated, Corning, NY, USA) at a cell density
of 75,000 cells/well in serum-free Ham's F-12 medium at 37°C with
5% CO2, while the lower parts of the chambers were
filled with Ham's F-12 complete medium. Following a 24-h
incubation, cells were fixed with absolute methanol for 20 min and
stained with 0.05% crystal violet solution for 15 min. The number
of migrated cells on the lower surface of the filters was counted
under a light microscope (Eclipse Ni-U; Nikon Corporation, Tokyo,
Japan).
Western blot analysis
In order to determine the expression levels of p21,
cyclin D1 and cyclin A protein, western blot analysis was
performed. KKU-100 cells were seeded in a 6-well plate at a cell
density of 150,000 cells/well in Ham's F-12 complete medium and
transfected with NQO1 siRNA for 24 h. Then cells were serum-starved
for 12 h using serum-free Ham F-12 medium at 37°C in at atmosphere
containing 5% CO2 and grown in Ham's F-12 complete
medium for a further 24 h. Following this, the cells were harvested
for western blot analysis. Cells were washed twice with PBS and
lysed with radioimmunoprecipitation assay cell lysis buffer
supplemented with dithiothreitol, phenylmethylsulfonyl fluoride
(PMSF) and protease inhibitor cocktail (Ameresco, Inc.) at 4°C for
15 min. The concentration of extracted proteins was determined
using Bradford reagent, according to the manufacturer's protocol
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). Whole cell lysate
containing 30 mg protein was separated by SDS-PAGE using a 10%
polyacrylamide gel. The separated proteins were then transferred
onto polyvinylidene difluoride membranes (EMD Millipore, Billerica,
MA, USA). The membranes were blocked for 1 h at room temperature
with 5% non-fat milk in PBS supplemented with 0.1% Tween-20. They
were then probed with rabbit polyclonal anti-human cyclin D1
(dilution, 1:1,000; #sc-718), rabbit polyclonal anti-human cyclin A
(dilution, 1:2,000; #sc-751), mouse monoclonal anti-human p21
(dilution, 1:500; #sc-56335) or horseradish peroxidase
(HRP)-conjugated goat polyclonal anti-human β-actin (dilution,
1:5,000; #sc-1616) primary antibodies for 12 h at 4°C (all
antibodies were from Santa Cruz Biotechnology, Inc., Dallas, TX,
USA). The membranes were then incubated with HRP-conjugated goat
anti-mouse IgG (dilution, 1:5,000; #sc-2005) or HRP-conjugated goat
anti-rabbit IgG (dilution, 1:5,000; #sc-2004) secondary antibodies
for 2 h at room temperature (both from Santa Cruz Biotechnology
Inc.). The membranes were incubated with enhanced chemiluminescence
(ECL) substrate solution (Amersham™ ECL™ Prime Western Blotting
detection reagent; GE Healthcare Life Sciences, Chalfont, UK) for 1
min at room temperature (25°C). The optical densities of the
protein bands were determined using ImageQuant™ LAS4000 (GE
Healthcare Life Sciences) (8).
RT-qPCR
In order to determine the mRNA expression levels of
matrix metalloproteinases (MMPs) and tissue inhibitors of
metalloproteinases (TIMPs), RT-qPCR was performed. KKU-100 cells
were seeded in a 6-well plate with density of 150,000 cells/well in
Ham's F-12 complete medium, transfected with NQO1 siRNA for 24 h
and then starved for 12 h using serum-free Ham F-12 medium at 37°C
and 5% CO2. At 24 h post-starvation, total RNA was
extracted from CCA cells using TRIzol® reagent was
obtained from Thermo Fisher Scientific, Inc., according to a
previously described method (8). A
cDNA synthesis mixture consisting of 1 µg total RNA and 4 µl 5x
iScript™ Reverse Transcription Supermix for RT-qPCR (170–8841;
Bio-Rad Laboratories, Inc.) was mixed with RNase-free water in a
total volume of 20 µl. cDNA synthesis was performed using a C1000™
thermal cycler (Bio-Rad Laboratories, Inc.). The cDNA synthesis
conditions included priming for 5 min at 25°C and reverse
transcription for 30 min at 42°C, and the reaction was stopped by
incubation for 15 min at 70°C. The reverse transcription products
served as templates for qPCR. PCR amplification was performed using
specific primers for the MMP2, MMP3, MMP9, TIMP1, TIMP2 and the
internal control β-actin. The primer sequences were as follows: i)
MMP2 (NM_004530) (24), forward
primer 5′-AGCTCCCGGAAAAGATGGATG-3′ and reverse primer
5′-CAGGGTGCTGGCTGAGTAGAT-3′; ii) MMP3 (NM_002422) (25), forward primer
5′-GCGTGGATGCCGCATATGAAGTTA-3′ and reverse primer
5′-AAACCTAGGGTGTGGATGCCTCTT-3′; iii) MMP9 (NM_002422) (26), forward primer
5′-GAAGATGCTGCTGTTCAGCG-3′ and reverse primer
5′-ACTTGGTCCACCTGGTTCAA-3′; iv) TIMP1 (NM_003254) (27), forward primer
5′-AGGCTCTGAAAAGGGCTTCCA-3′ and reverse primer
5′-GAGTGGGAACAGGGTGGACA-3′; v) TIMP2 (NM_003255) (28), forward primer
5′-GACGGCAAGATGCACATCAC-3′ and reverse primer
5′-GAGATGTAGCACGGGATCATGG-3′; vi) β-actin (NM_001101) (8), forward primer 5′-TGCCATCCTAAAAGCCAC-3′
and reverse primer 5′-TCAACTGGTCTCAAGTCAGTG-3′. The
reverse-transcription fluorescence PCR, based on
EvaGreen® dye, was performed in a final volume of 20 µl
of 1x SsoFast™ EvaGreen® Supermix (172–5201; Bio-Rad
Laboratories, Inc.), 0.5 µmol/l MMP2, MMP3, MMP9, TIMP1 or TIMP2
primer and 0.25 µmol/l β-actin primer. A negative control (no cDNA
template) was included in the experimental runs. The qPCR was
performed for each gene in duplicate on cDNA samples in 96-well
reaction plates using LightCycler® 480 Real-Time PCR
System (Roche Applied Science, Madison, WI, USA). The cycling
conditions for qPCR were: 95°C for 3 min, followed by 40 cycles of
95°C for 15 sec and 60°C for 31 sec. To verify the purity of the
products, a melting curve analysis was performed following each
run. The concentration of PCR products was evaluated on the basis
of an established standard curve derived from serial dilutions of
the positive control for MMP2, MMP3, MMP9, TIMP1, TIMP2 and
β-actin.
Statistical analysis
Data were expressed as the mean ± standard error of
the mean from three independent experiments (SigmaPlot Version
10.0; Systat Software, Inc., San Jose, CA, USA). Statistical
comparison between the control and treatment groups was performed
using Student's t-test. P<0.05 was considered to indicate a
statistically significant difference (SigmaStat Version 3.11;
Systat Software, Inc.).
Results
Knockdown of NQO1 expression
suppresses the cell proliferation of KKU-100 cells
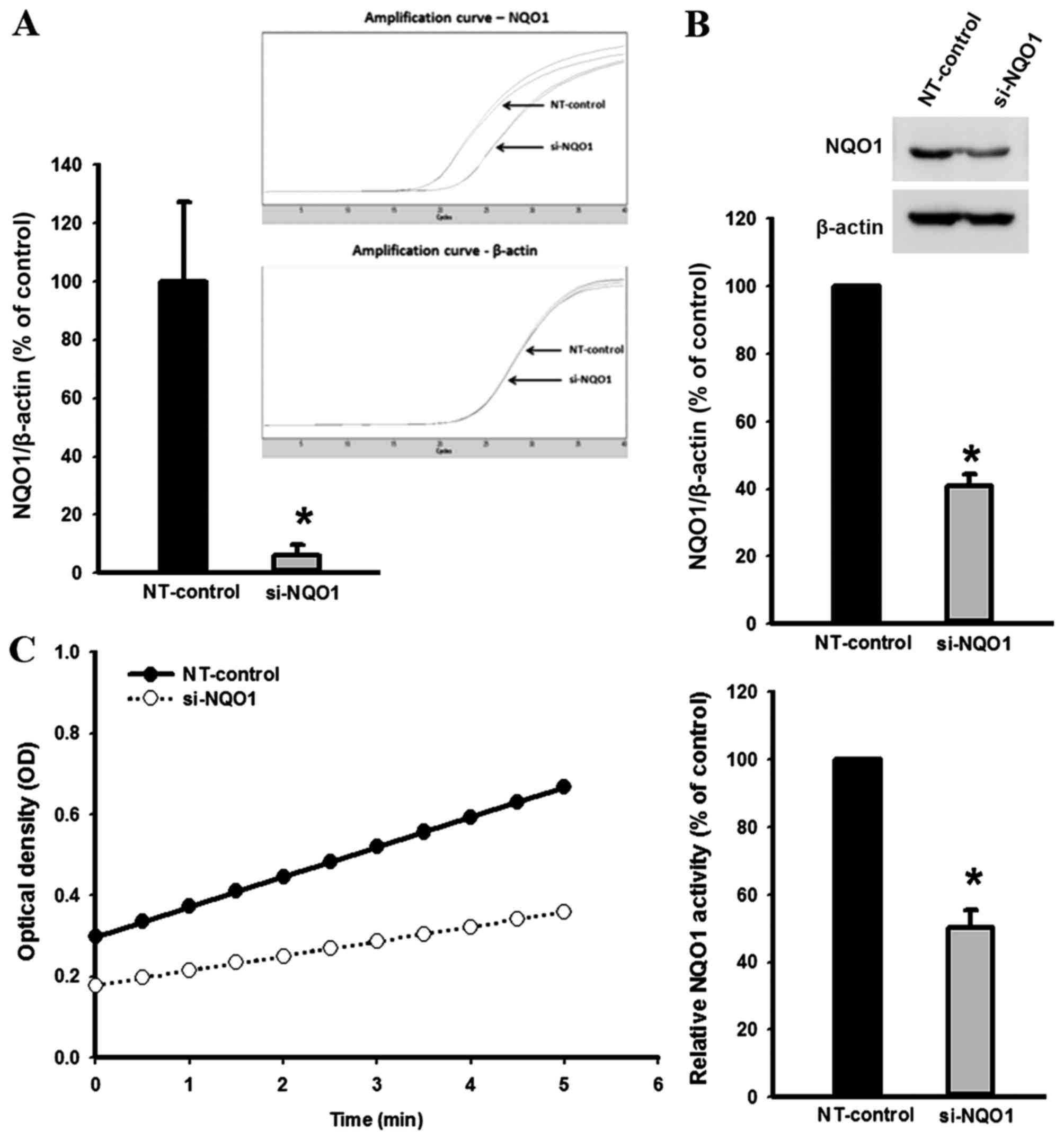
The efficiency of NQO1 siRNA transfection in KKU-100
cells was established using qPCR, western blot analysis and NQO1
enzymatic assay (Fig. 1). Knockdown
of NQO1 transcripts using siRNA transfection efficiently decreased
NQO1 expression at the mRNA and protein levels, and enzymatic
activity by ~90, ~60 and ~50%, respectively compared with the
non-targeting siRNA-transfected control cells (P<0.001; Fig. 1). To investigate the role of NQO1 on
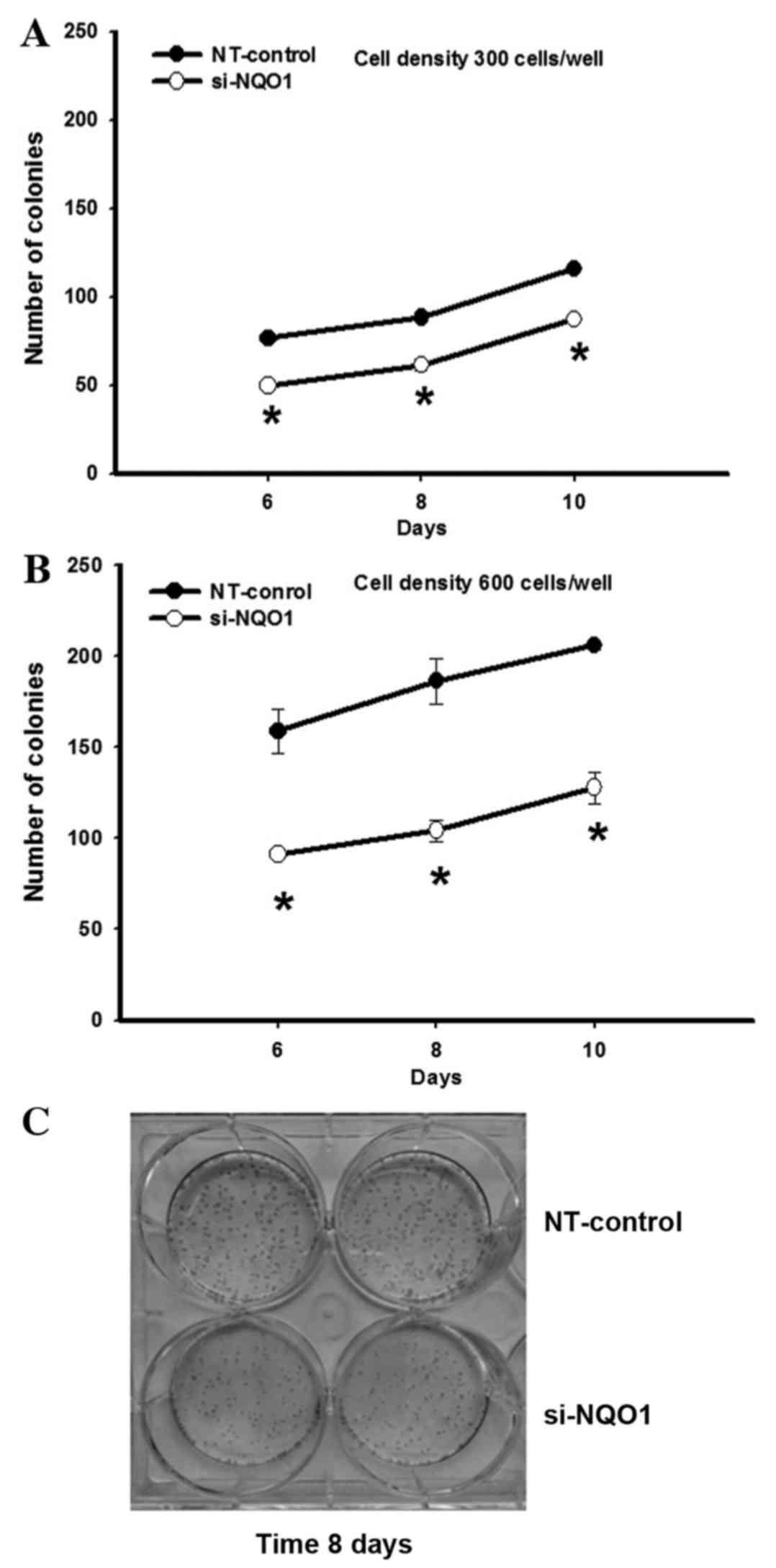
CCA cell proliferation, NQO1-knockdown cells using siRNA were
subjected to the clonogenic survival assay. The colony formation,
as an index of long-term cell proliferation, was observed by
plating the cells at low densities (300 and 600 cells/well;
Fig. 2). NQO1-knockdown CCA cells
exhibited a decreased number of colonies formed compared with
control cells (Fig. 2). A significant
antiproliferative effect of NQO1 siRNA was observed between days 6
and 10. At 10 days of culture, the NQO1 siRNA decreased colony
formation by ~24 and ~37% when cells were plated at 300 and 600
cells/well, respectively, compared with the non-targeting
siRNA-transfected control cells (P<0.05; Fig. 2). These results suggested that
knockdown of NQO1 transcripts using siRNA transfection decreases
the ability of CCA cells to grow and proliferate, resulting in a
decrease in colony formation. Therefore, these results implied that
NQO1 may serve a role in CCA cell growth and proliferation.
Knockdown of NQO1 expression arrests
the cell cycle progression of KKU-100 cells
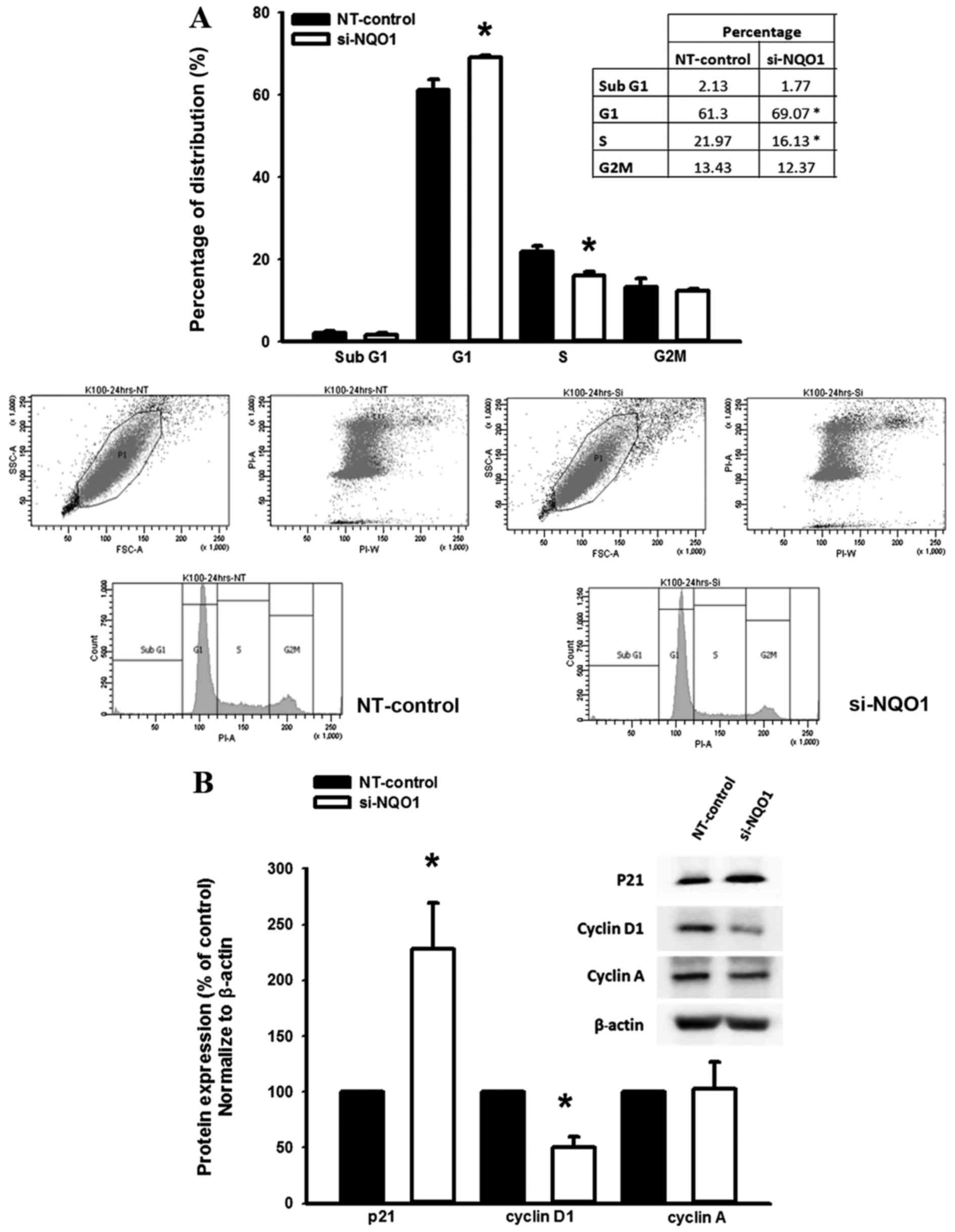
To investigate the role of NQO1 on the regulation of
the CCA cell cycle, NQO1-knockdown cells using siRNA were subjected
to cell cycle analysis using a flow cytometer. The results
demonstrated that NQO1-knockdown CCA cells significantly
accumulated at the G1 phase with a decreased proportion
of cells at the S phase compared with the non-targeting
siRNA-transfected control cells (P<0.05; Fig. 3A). Therefore, these results suggested
that NQO1 serves a role in cell cycle progression in CCA cells.
Knockdown of NQO1 expression
contributes to altered levels of p21 and cyclin D1 protein
The suppression of cell proliferation and arrestment
of cell cycle in KKU-100 cells by NQO1 siRNA transfection led to
further experiments investigating the influence of NQO1 knockdown
on the expression levels of proteins which are associated with cell
proliferation and cell cycle. The effect of NQO1 siRNA on the
expression levels of p21, cyclin A and cyclin D1 protein was
observed by western blot analysis. Knockdown of NQO1 transcripts
using siRNA transfection significantly increased p21 and decreased
cyclin D1 protein expression levels compared with in the
non-targeting siRNA-transfected control cells (P<0.05; Fig. 3B); however, cyclin A protein
expression levels remained the same. As p21 and cyclin D1 are
important cell cycle regulator proteins for G1 and S
phases, altered expression levels of p21 and cyclin D1 protein in
NQO1-knockdown cells may induce CCA cells arrest at the
G1 phase. These results provided evidence for the
biological role of NQO1 in cell proliferation and cell cycle
regulation of CCA cells.
Knockdown of NQO1 expression delays
CCA cell migration
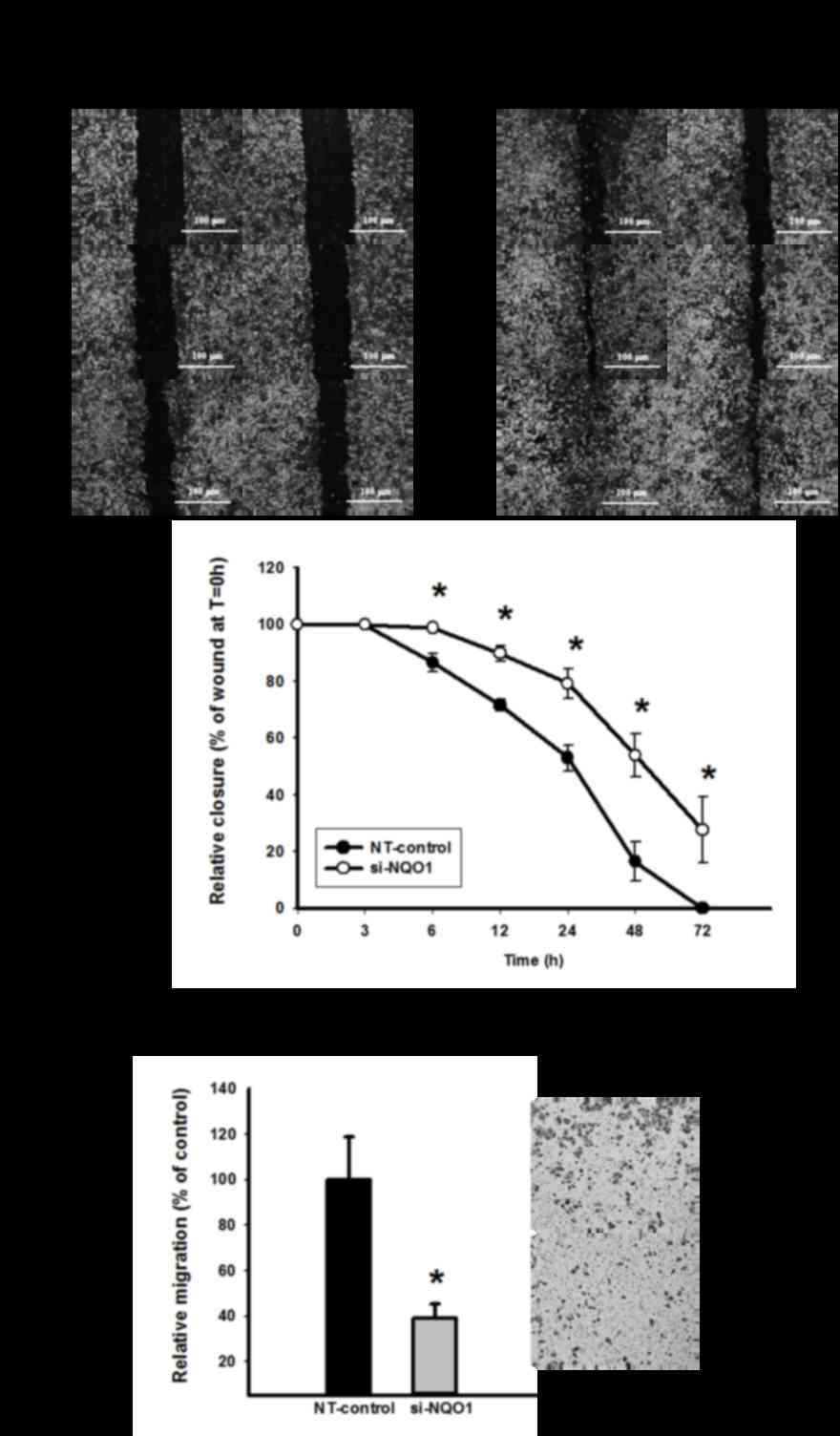
To investigate the role of NQO1 on CCA cell
migration, wound healing and Transwell migration assays were
performed. Knockdown of NQO1 transcripts using siRNA transfection
efficiently delayed cell migration, and this suppressive effect was
observed early at 6 h and continued until 72 h (P<0.05; Fig. 4A). To confirm the anti-migration
effect of NQO1 siRNA, a Transwell migration assay was additionally
performed. The results revealed that NQO1-knockdown CCA cells
significantly lost their migration ability, as presented in
Fig. 4B (P<0.05). The cell
migration of NQO1-knockdown CCA cells decreased by ~60% compared
with the non-targeting siRNA-transfected control cells. Therefore,
NQO1 siRNA exhibited anti-migration ability on CCA cells as
observed using wound healing and Transwell migration assays, and it
was concluded that NQO1 serves a role in the migration of CCA
cells.
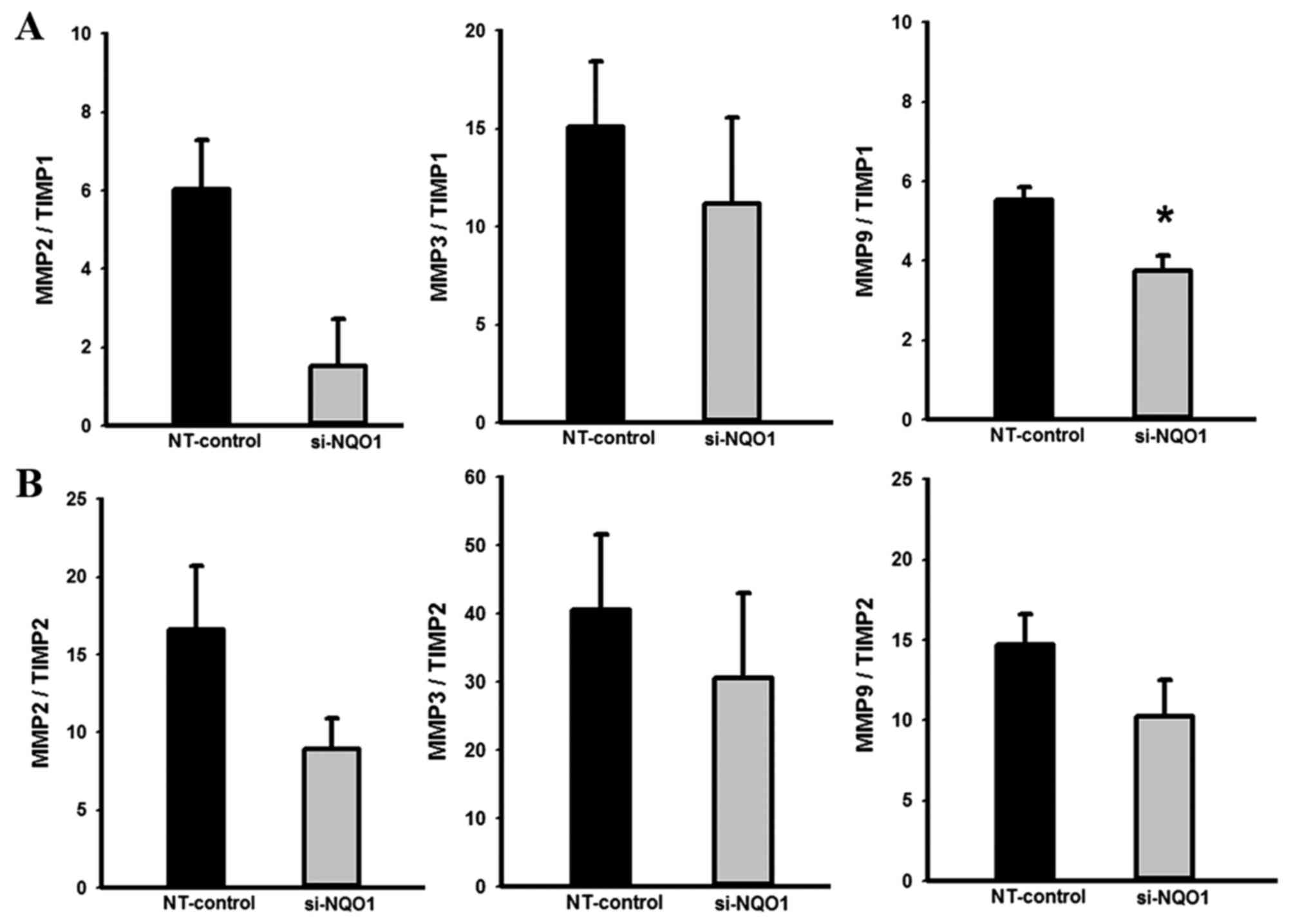
Knockdown of NQO1 expression
contributes to altered MMP/TIMP mRNA ratios
Further to demonstrating the biological role of NQO1
in CCA cell migration, the effects of NQO1 knockdown on expression
levels of migration-associated genes were investigated using
RT-qPCR. The RT-qPCR results revealed that knockdown of NQO1
transcripts using siRNA transfection did not alter expression
levels of MMP2, MMP3 and MMP9, but tended to increase level of
TIMP1 and significantly increased the expression level of TIMP2
mRNA compared with control cells (data not shown). The effects of
NQO1 knockdown on the MMP/TIMP mRNA ratios, which are generally
recognized as the index of the relative inhibition of MMP activity
(24,29), were analyzed. NQO1-knockdown CCA cells
demonstrated a decreased MMP9/TIMP1 ratio compared with in the
control cells (P<0.05; Fig. 5);
suggesting that NQO1 siRNA has an inhibitory effect on MMP
activity. The suppression of MMP activity in NQO1-knockdown cells
provided evidence of the biological role of NQO1 in the migration
of CCA cells.
Discussion
The involvement of NQO1 in various types of cancer
has previously been demonstrated (9–16). The
upregulation of NQO1 in certain types of solid tumor, including CCA
(17), was associated with poor
prognosis, thus NQO1 targeting has therapeutic potential (10,11). The
present study investigated the biological role of NQO1 in cell
proliferation, cell cycle and migration of CCA. The in vitro
experiments revealed that knockdown of NQO1 expression compromised
the proliferation and reproductive ability of cells, arrested cell
cycle progression and inhibited the motile capacity of CCA cells.
Therefore, suppression of NQO1 may be a potential target for the
treatment and/or strategy to enhance the chemosensitivity of CCA
and other types of cancer with increased NQO1 activity.
Previous studies have suggested possible
growth-inhibitory effects triggered by NQO1 suppression (11,18,19).
Dicoumarol, a potent inhibitor of the NQO1 enzyme, inhibited cell
growth of pancreatic cancer cells in a dose-dependent manner
(30), whereas a higher dose of
dicoumarol decreased colony formation on soft agar of pancreatic
cancer cells (18). For HeLa cells,
treatment with dicoumarol resulted in a marked decrease in the
viability of cells and proliferation rate (28). In accordance with these findings, the
results of the present study demonstrated that knockdown of NQO1 in
CCA cells significantly impaired cell proliferative ability. These
results provide evidence suggesting the role of NQO1 in maintaining
the ability of cell proliferation and replication.
The cell cycle, a series of events leading to cell
division and duplication, is a key regulatory process in cell
growth and proliferation (31). In
the present study, flow cytometric analysis revealed that NQO1
knockdown induced G1 phase arrest and decreased the
proportion of cells at the S phase, which may have contributed to
the inhibition of proliferation in the NQO1-knockdown cells. In
order to elucidate the possible molecular mechanism underlying
NQO1-mediated inhibition of the proliferation and reproductive
capacities of CCA cells, western blot analysis was performed to
identify the influence of NQO1 siRNA on the expression levels of
p21, cyclin A and cyclin D1 proteins. The results of the present
study demonstrated that the upregulation of p21 and downregulation
of cyclin D1 proteins were associated with the NQO1 expression
level, which indicated that these two proteins serve important
roles in the process of antiproliferation and cell cycle arrest
triggered by NQO1 siRNA. A similar observation was previously
revealed in melanoma cells, where knockdown of NQO1 decelerated the
transition of melanoma cells from G1 to G2-M
phase (32). Knockdown of NQO1 in
melanoma cells also induced downregulation of cyclins D1, A2 and B1
(32). Furthermore, treatment with
dicoumarol increased the number of cells that accumulated in the
sub-G1 phase (33).
Cell migration serves an important role in the
cancer progression (34). The ability
of NQO1 to drive human aortic vascular smooth muscle cell migration
and invasion was previously demonstrated (35). Treatment with dicoumarol or
transfection with NQO1 siRNA suppressed tumor necrosis factor
α-induced cell migration by inhibiting MMP9 protein and mRNA
expression (35,36). In the present study, NQO1 siRNA
significantly decreased the migratory ability of CCA cells, which
suggested a role for NQO1 in promoting the metastasis of CCA. In
order to elucidate the potential molecular mechanism underlying
NQO1-mediated inhibition of the motility of the CCA cells, RT-qPCR
analysis was performed to investigate the influence of NQO1 siRNA
on the expression levels of MMP2, MMP3, MMP9, TIMP1 and TIMP2 mRNA.
It was revealed that NQO1 siRNA exhibited an inhibitory effect on
MMP activity by altering the MMP/TIMP mRNA ratio, which indicated
that NQO1 serves a role in the process of cell migration of
CCA.
In conclusion, to the best of our knowledge, the
results of the present study provided evidence for the first time
that NQO1 serves a key role in cell proliferation, cell cycle and
migration of CCA cells. Furthermore, NQO1 siRNA demonstrated an
antiproliferative effect in KKU-100 cells by upregulation of p21
and downregulation of cyclin D1 proteins. Additionally, NQO1 siRNA
revealed an anti-migratory effect by decreasing the MMP9/TIMP1 mRNA
ratio. Therefore, the results of the present study highlighted the
biological role of NQO1 maintaining the ability of cell
proliferation and replication and migration in CCA. Thus, NQO1 may
be a potential molecular target to enhance CCA treatment and other
types of cancer with increased NQO1 activity.
Acknowledgements
This present study was supported by Khon Kaen
University Thailand Research Fund (grant no. RMU5380027), Faculty
of Medicine (grant no. I57206) and a scholarship from the National
Research University Project and Office of Higher Education
Commission through the Specific Health Problem in Greater Mekong
Sub-region project of Khon Kaen University to S.B.
References
|
1
|
Sripa B and Pairojkul C:
Cholangiocarcinoma: Lessons from Thailand. Curr Opin Gastroenterol.
24:349–356. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Khan SA, Davidson BR, Goldin RD, Heaton N,
Karani J, Pereira SP, Rosenberg WM, Tait P, Taylor-Robinson SD,
Thillainayagam AV, et al: Guidelines for the diagnosis and
treatment of cholangiocarcinoma: An update. Gut. 61:1657–1669.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Anderson CD, Pinson CW, Berlin J and Chari
RS: Diagnosis and treatment of cholangiocarcinoma. Oncologist.
9:43–57. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Patel T: Cholangiocarcinoma-controversies
and challenges. Nat Rev Gastroenterol Hepatol. 8:189–200. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Buranrat B, Prawan A, Kukongviriyapan U,
Kongpetch S and Kukongviriyapan V: Dicoumarol enhances
gemcitabine-induced cytotoxicity in high NQO1-expressing
cholangiocarcinoma cells. World J Gastroenterol. 16:2362–2370.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kongpetch S, Kukongviriyapan V, Prawan A,
Senggunprai L, Kukongviriyapan U and Buranrat B: Crucial role of
heme oxygenase-1 on the sensitivity of cholangiocarcinoma cells to
chemotherapeutic agents. PLoS One. 7:e349942012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Samatiwat P, Prawan A, Senggunprai L and
Kukongviriyapan V: Repression of Nrf2 enhances antitumor effect of
5-fluorouracil and gemcitabine on cholangiocarcinoma cells. Naunyn
Schmiedebergs Arch Pharmacol. 388:601–612. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zeekpudsa P, Kukongviriyapan V,
Senggunprai L, Sripa B and Prawan A: Suppression of NAD(P)H-quinone
oxidoreductase 1 enhanced the susceptibility of cholangiocarcinoma
cells to chemotherapeutic agents. J Exp Clin Cancer Res. 33:112014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ross D, Kepa JK, Winski SL, Beall HD,
Anwar A and Siegel D: NAD(P)H:Quinone oxidoreductase 1 (NQO1):
Chemoprotection, bioactivation, gene regulation and genetic
polymorphisms. Chem Biol Interact. 129:77–97. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dinkova-Kostova AT and Talalay P:
NAD(P)H:Quinone acceptor oxidoreductase 1 (NQO1), a multifunctional
antioxidant enzyme and exceptionally versatile cytoprotector. Arch
Biochem Biophys. 501:116–123. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Siegel D, Yan C and Ross D:
NAD(P)H:Quinone oxidoreductase 1 (NQO1) in the sensitivity and
resistance to antitumor quinones. Biochem Pharmacol. 83:1033–1040.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Basu S, Brown JE, Flannigan GM, Gill JH,
Loadman PM, Martin SW, Naylor B, Puri R, Scally AJ, Seargent JM, et
al: NAD(P)H:Quinone oxidoreductase-1 C609T polymorphism analysis in
human superficial bladder cancers: Relationship of genotype status
to NQO1 phenotype and clinical response to Mitomycin C. Int J
Oncol. 25:921–927. 2004.PubMed/NCBI
|
|
13
|
Cheng Y, Li J, Martinka M and Li G: The
expression of NAD(P)H:Quinone oxidoreductase 1 is increased along
with NF-kappaB p105/p50 in human cutaneous melanomas. Oncol Rep.
23:973–979. 2010.PubMed/NCBI
|
|
14
|
Cui X, Jin T, Wang X, Jin G, Li Z and Lin
L: NAD(P)H:Quinone oxidoreductase-1 overexpression predicts poor
prognosis in small cell lung cancer. Oncol Rep. 32:2589–2595.
2014.PubMed/NCBI
|
|
15
|
Kolesar JM, Pritchard SC, Kerr KM, Kim K,
Nicolson MC and McLeod H: Evaluation of NQO1 gene expression and
variant allele in human NSCLC tumors and matched normal lung
tissue. Int J Oncol. 21:1119–1124. 2002.PubMed/NCBI
|
|
16
|
Cui X, Li L, Yan G, Meng K, Lin Z, Nan Y,
Jin G and Li C: High expression of NQO1 is associated with poor
prognosis in serous ovarian carcinoma. BMC Cancer. 15:2442015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Buranrat B, Chau-in S, Prawan A, Puapairoj
A, Zeekpudsa P and Kukongviriyapan V: NQO1 expression correlates
with cholangiocarcinoma prognosis. Asian Pac J Cancer Prev. 13
Suppl:S131–S136. 2012.
|
|
18
|
Cullen JJ, Hinkhouse MM, Grady M, Gaut AW,
Liu J, Zhang YP, Weydert CJ, Domann FE and Oberley LW: Dicumarol
inhibition of NADPH: Quinone oxidoreductase induces growth
inhibition of pancreatic cancer via a superoxide-mediated
mechanism. Cancer Res. 63:5513–5520. 2003.PubMed/NCBI
|
|
19
|
Reigan P, Colucci MA, Siegel D, Chilloux
A, Moody CJ and Ross D: Development of indolequinone
mechanism-based inhibitors of NAD(P)H:Quinone oxidoreductase 1
(NQO1): NQO1 inhibition and growth inhibitory activity in human
pancreatic MIA PaCa-2 cancer cells. Biochemistry. 46:5941–5950.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Matsui Y, Watanabe J, Ding S, Nishizawa K,
Kajita Y, Ichioka K, Saito R, Kobayashi T, Ogawa O and Nishiyama H:
Dicoumarol enhances doxorubicin-induced cytotoxicity in p53
wild-type urothelial cancer cells through p38 activation. BJU Int.
105:558–564. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wakai T, Shirai Y, Sakata J, Matsuda Y,
Korita PV, Takamura M, Ajioka Y and Hatakeyama K: Prognostic
significance of NQO1 expression in intrahepatic cholangiocarcinoma.
Int J Clin Exp Pathol. 4:363–370. 2011.PubMed/NCBI
|
|
22
|
Sripa B, Leungwattanawanit S, Nitta T,
Wongkham C, Bhudhisawasdi V, Puapairoj A, Sripa C and Miwa M:
Establishment and characterization of an opisthorchiasis-associated
cholangiocarcinoma cell line (KKU-100). World J Gastroenterol.
11:3392–3397. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Aneknan P, Kukongviriyapan V, Prawan A,
Kongpetch S, Sripa B and Senggunprai L: Luteolin arrests cell
cycling, induces apoptosis and inhibits the JAK/STAT3 pathway in
human cholangiocarcinoma cells. Asian Pac J Cancer Prev.
15:5071–5076. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Figueira RC, Gomes LR, Neto JS, Silva FC,
Silva ID and Sogayar MC: Correlation between MMPs and their
inhibitors in breast cancer tumor tissue specimens and in cell
lines with different metastatic potential. BMC Cancer. 9:202009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim HA, Yeo Y, Kim WU and Kim S: Phase 2
enzyme inducer sulphoraphane blocks matrix metalloproteinase
production in articular chondrocytes. Rheumatology (Oxford).
48:932–938. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Onodera M, Zen Y, Harada K, Sato Y, Ikeda
H, Itatsu K, Sato H, Ohta T, Asaka M and Nakanuma Y: Fascin is
involved in tumor necrosis factor-alpha-dependent production of
MMP9 in cholangiocarcinoma. Lab Invest. 89:1261–1274. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pardo A, Gibson K, Cisneros J, Richards
TJ, Yang Y, Becerril C, Yousem S, Herrera I, Ruiz V, Selman M and
Kaminski N: Up-regulation and profibrotic role of osteopontin in
human idiopathic pulmonary fibrosis. PLoS Med. 2:e2512005.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang J, Zheng J, Wu L, Shi M, Zhang H,
Wang X, Xia N, Wang D, Liu X, Yao L, et al: NDRG2 ameliorates
hepatic fibrosis by inhibiting the TGF-β1/Smad pathway and altering
the MMP2/TIMP2 ratio in rats. PLoS One. 6:e277102011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Groblewska M, Siewko M, Mroczko B and
Szmitkowski M: The role of matrix metalloproteinases (MMPs) and
their inhibitors (TIMPs) in the development of esophageal cancer.
Folia Histochem Cytobiol. 50:12–19. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lewis A, Ough M, Li L, Hinkhouse MM,
Ritchie JM, Spitz DR and Cullen JJ: Treatment of pancreatic cancer
cells with dicumarol induces cytotoxicity and oxidative stress.
Clin Cancer Res. 10:4550–4558. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Alenzi FQ: Links between apoptosis,
proliferation and the cell cycle. Br J Biomed Sci. 61:99–102. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Garate M, Wani AA and Li G: The
NAD(P)H:Quinone Oxidoreductase 1 induces cell cycle progression and
proliferation of melanoma cells. Free Radic Biol Med. 48:1601–1609.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Scott KA, Barnes J, Whitehead RC,
Stratford IJ and Nolan KA: Inhibitors of NQO1: Identification of
compounds more potent than dicoumarol without associated off-target
effects. Biochem Pharmacol. 81:355–363. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Golias CH, Charalabopoulos A and
Charalabopoulos K: Cell proliferation and cell cycle control: A
mini review. Int J Clin Pract. 58:1134–1141. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee SO, Chang YC, Whang K, Kim CH and Lee
IS: Role of NAD(P)H:Quinone oxidoreductase 1 on tumor necrosis
factor-alpha-induced migration of human vascular smooth muscle
cells. Cardiovasc Res. 76:331–339. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Friedl P and Wolf K: Tumour-cell invasion
and migration: Diversity and escape mechanisms. Nat Rev Cancer.
3:362–374. 2003. View
Article : Google Scholar : PubMed/NCBI
|