Introduction
Pancreatic cancer is an aggressive type of cancer
with an extremely poor prognosis (1).
This type of tumor often metastasizes to multiple lymph nodes, with
liver and peritoneal sites the most common for distant spread
(2). The identification of molecular
pathways and novel candidates for therapeutic targeting is urgently
required to develop alternative treatments in order to improve
clinical outcomes. Chemokine receptors have been implicated in
pancreatic cancer metastasis (3). In
particular, C-X-C chemokine receptor 4 (CXCR4), a chemokine
G-protein-coupled receptor, is frequently expressed in pancreatic
cancer and affects pancreatic tumor cell growth, adhesion,
migration and invasion in cellulo and in clinical samples
(4–8).
The ligand for CXCR4, C-X-C chemokine ligand 12 (CXCL12; also known
as stromal cell-derived factor 1) is expressed at high levels in
tissues prone to pancreatic cancer spread, including the lymph
nodes, liver and lung (8,9). While the expression of CXCR4 is low or
absent in the majority of healthy tissues, its expression is found
in high frequency in pancreatic cancer specimens, ranging from
71.2–90% depending on the study (4,6,8,9); however,
the mechanism for CXCR4 misexpression is not well understood. A
limited number of factors are associated with the regulation of
CXCR4 expression in pancreatic cancer (10–12). For
example, glycogen synthase kinase 3 β enhances CXCR4 expression
(12), and CXCR4 expression is also
promoted through the hypoxia-driven expression of the microRNA
miRNA-150 (11). Additionally, the
Boswellia serrata plant extract boswellic acid has been
demonstrated to reduce CXCR4 levels in pancreatic cancer cells
(10). As the CXCR4 receptor promotes
pancreatic cancer growth, migration and metastasis, the targeting
of factors that can downregulate CXCR4 has the potential to inhibit
pancreatic cancer metastasis.
Our recent study revealed that paired box
transcription factor (PAX3) regulates CXCR4 expression in melanoma
(13). PAX6, a PAX3-associated
protein, is expressed in pancreatic adenocarcinoma cell lines and
tumor samples (14). PAX6 contains a
paired-type DNA-binding domain that is similar to the paired domain
of PAX3, and is able to bind to similar, but not identical, DNA
sequences (15,16). PAX6, like PAX3, interacts with the
promoter of the receptor tyrosine kinase gene MET and promotes gene
expression (17,18). While PAX6 is normally expressed in the
embryonic pancreatic bud and in pancreatic islet α-cells, the
expression of this factor is downregulated during exocrine
pancreatic development and is absent in mature exocrine tissue
(19). Studies investigating PAX6
function in pancreatic cancer have supported that PAX6 acts as a
transcription factor (20); however,
downstream effector genes of PAX6, other than MET (18), have not been reported.
PAX6 and CXCR4 have been previously identified to
have alternative splice variants (21,22). The
canonical PAX6 protein has two DNA-binding domains (a bipartite
paired domain and a paired-type homeodomain) and a C-terminal
transcriptional activation domain (23). A major splice variant of PAX6 is the
exon 5 alternative transcript variant PAX6(5a), which possesses a
14-amino-acid insertion into an N-terminal region of the paired
domain which alters the DNA-binding preference (21). For the human CXCR4 gene, two major
alternative splice variants have been identified: The canonical
CXCR4B transcript, and CXCR4A (22).
The two isoforms differ only in the furthest N-terminal region of
the protein, and this difference may affect how the receptor
interacts with its major ligand, CXCL12 (22,24). The
expression and consequences of expression of these variant isoforms
of PAX6 and CXCR4 in pancreatic cancer, and in cancer in general,
are virtually unexplored.
The present study reports the expression of CXCR4
and PAX6 in pancreatic cancer primary tissues and cell lines, and
indicates that there is a significant positive correlation of their
co-expression. In parallel with our previous findings with PAX3,
PAX6 is also sufficient to drive gene expression from the CXCR4
regulatory region, and this ability of PAX6 to promote
transcription is dependent on the presence of a highly conserved
intronic enhancer element. In addition, pancreatic cancer cell
lines express the common transcripts of PAX6 and CXCR4, as well as
the variant transcripts PAX6(5a) and CXCR4A. Canonical and/or
variant forms of PAX6 and CXCR4 may participate in a shared
molecular pathway in pancreatic cancer.
Materials and methods
Immunohistochemistry of primary tumor
samples
A total of 22 primary tissue samples verified as
late-stage pancreatic adenocarcinoma tumor specimens, obtained as
autopsy specimens from the Cooperative Human Tissue Network, as
previously described (25), were
utilized in the current study; their use was compliant with and
approved by the University of Chicago institutional review board
and Clinical Trials Committees. Slides holding the tissue sections
were boiled in 1X citrate buffer for antigen retrieval. Samples
were blocked in 1% normal goat serum, followed by overnight
incubation at 4°C with primary antibodies against PAX6 (mouse
monoclonal; dilution, 1:500; University of Iowa Hybridoma Bank,
Iowa City, IA, USA) and CXCR4 (rabbit polyclonal; cat. no. Ab1670;
dilution, 1:200; Abcam, Cambridge, MA, USA). Incubation for 1 h at
4°C with goat anti-rabbit fluorescein (cat. no. 31509; dilution,
1:1,000; Thermo Fisher Scientific, Inc., Waltham, MA, USA) or goat
anti-mouse dyl647 (cat no. 21235; dilution, 1:1,000; Thermo Fisher
Scientific, Inc) secondary antibodies, which enabled the
fluorescence-mediated detection of antigens. Samples were scored as
‘positive’ or ‘negative’ for each antigen, with a positive score
determined when >25% of the tumor tissue expressed the
antigen.
Cell culture and transfection
The pancreatic carcinoma cell lines AsPC-1, BxPC-3,
Capan-2, HPAF II (HPAF), MIA PaCa-2, PANC-1, CFPAC-1 as well as
control cells HEK-293T (human embryonic kidney) and 3T3
(fibroblast) were obtained from American Type Culture Collection
(Manassas, VA, USA), the pancreatic carcinoma cell lines COLO-537,
CD11 and SW979 were obtained from Dr Ruggeri from Allegeny
University (Philadelphia, PA, USA) who was supplied them by Dr
Batra from Eppley Institute for Research in Cancer (Omaha, NE, USA)
(25), and all the cell lines were
cultured as previously described (14,18). Cells
were transfected using Effectene Transfection Reagent (Qiagen,
Inc., Valencia, CA, USA), according to manufacturer's protocols. In
addition to the reporter and/or expression constructs, cells were
also transfected with a β-galactosidase-expressing construct (pCMV;
Clontech Laboratories, Inc., Mountain View, CA, USA) to serve as an
internal control for luciferase assays. The total amount of DNA
transfected per cell was maintained at a constant amount by the
addition of pBluescript plasmid (Stratagene; Agilent Technologies,
Santa Clara, CA, USA). Levels of luciferase and β-galactosidase
were measured using assay kits from Promega (Promega Corporation,
Madison, WI, USA) and each experiment was performed minimally in
triplicate.
Vectors
The pGL2-CXCR4pm393, pGL2-CXCR4pm393L and
pGL2-CXCR4pm393L-ΔISH vectors were constructed as described
previously (13). The pGL2-CXCR4pm393
construct contains a sequence of 393 bp 5′-proximal to exon 1 of
the CXCR4 gene, as well as the 5′ untranslated region (UTR) from
exon 1. The pGL2-CXCR4pm393L vector contains the entire sequence of
pGL2-CXCR4pm393 as well as the coding sequence from all of exon 1
and part of exon 2 cloned in-frame with the luciferase reporter
gene cassette and the 1,781-bp intron 1. The pGL2-CXCR4pm393L-ΔISH
construct is the same as pGL2-CXCR4pm393L with the exception of a
deletion of a 267-bp segment from intron 1 containing the island of
homology with the PAX binding site.
Western blot analysis
Whole cell lysates from pancreatic cancer cells were
isolated using RIPA buffer, loaded onto 4–15% Tris-Bis gels (50 µg
total protein per cell line) for electrophoresis, then transferred
onto nitrocellulose membranes. Membranes were probed with an
anti-PAX6 antibody and an anti-β-tubulin (cat no. E7) antibody
(University of Iowa Hybridoma Bank; dilutions, 1:100 and 1:400,
respectively). Immunoreactivity was detected using a WesternBreeze
Chemiluminescent kit (cat. no. WB7104; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. All incubations
were performed at 4°C and for 1 h for primary and secondary
antibody incubations and the blocking step. Secondary antibodies
utilized were from the WesternBreeze kit. Following secondary
antibody incubation the blots were washed at room temperature four
times for five min for each wash. Densitometric analyses of
resultant western blots were performed with ImageJ software (ImageJ
version 1.47 public domain software; National Institutes of Health,
Bethesda, MD, USA). For quantification, raw densitometry numbers
were recorded for each band and normalized against the β-tubulin
readings. The data presented are densitometric readings from three
independent western blot analyses. Background readings are
arbitrarily set at 0.5 U.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RNA from pancreatic cancer cell lines was isolated
using Trizol (Thermo Fisher Scientific, Inc.), DNase-treated with a
Ambion DNA-free DNA Removal Kit (Thermo Fisher Scientific, Inc.),
and used as a template for RT with the iScript cDNA Synthesis kit
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). The mRNA
expression levels of CXCR4 transcript variants A and B (CXCR4A and
CXCR4B, respectively), PAX6, PAX6(5a) and GAPDH were evaluated
using SYBR-Green Master Mix (Bio-Rad Laboratories, Inc.) and the
CFX Connect Real-Time System (Bio-Rad Laboratories, Inc.). The
template for each RT-qPCR sample was 2.5 µl of a 1:10 dilution of
cDNA derived from 1 µg of starting template RNA. Cycling conditions
included 40 cycles of 95°C for 10 sec, annealing at 58°C for 10 sec
and a 30 sec 72°C extension. The results are presented as the mean
and standard error of the mean of three independent experiments,
normalized against GAPDH expression and compared to the MIA PaCa-2
cell line for relative expression using the Pfaffl method (26). MIA PaCa-2 was chosen as the cell line
of comparison due to this cell line having significant levels of
all four transcripts. Primer sequences are presented in Table I.
 | Table I.Primers utilized for the reverse
transcription-quantitative polymerase chain reaction analysis. |
Table I.
Primers utilized for the reverse
transcription-quantitative polymerase chain reaction analysis.
| Gene/primer
name | Sequence
(5′-3′) |
|---|
| CXCR4B forward |
CCGAGGGCCTGAGTGCTCCAG |
| CXCR4A forward |
GCAGAGGAGTTAGCCAAGATG |
| CXCR4 reverse |
ATCCATTGCCCACAATGCCAG |
| PAX6 and PAX6(5a)
forward |
TTCAGAGCCCCATATTCGAG |
| PAX6 reverse |
GTTGGACACCTGCAGAAT |
| PAX6(5a)
reverse |
TGCATGGGTCTGCAGAAT |
Statistical analysis
GraphPad Prism statistical software (version 5.0;
GraphPad Software, Inc., La Jolla, CA, USA) was utilized to
determine whether the findings were significant. Correlation
coefficients between PAX6-expressing and CXCR4-expressing tumor
samples were determined with two-tailed Fisher's exact probability
tests, with the analysis of nominal data as either positive or
negative expression. The significance of the differences between
the groups was determined with Student's t-test and χ2
analysis, with a confidence interval of 95%. All values stated as
significant have P-values of ≤0.05, unless indicated. All
experiments were performed in triplicate.
Results
PAX6 and CXCR4 are co-expressed in
primary pancreatic adenocarcinoma samples
We have previously described PAX6 expression in
pancreatic cancer established cell lines and primary tumor samples
(14,18). PAX6 is frequently co-expressed with
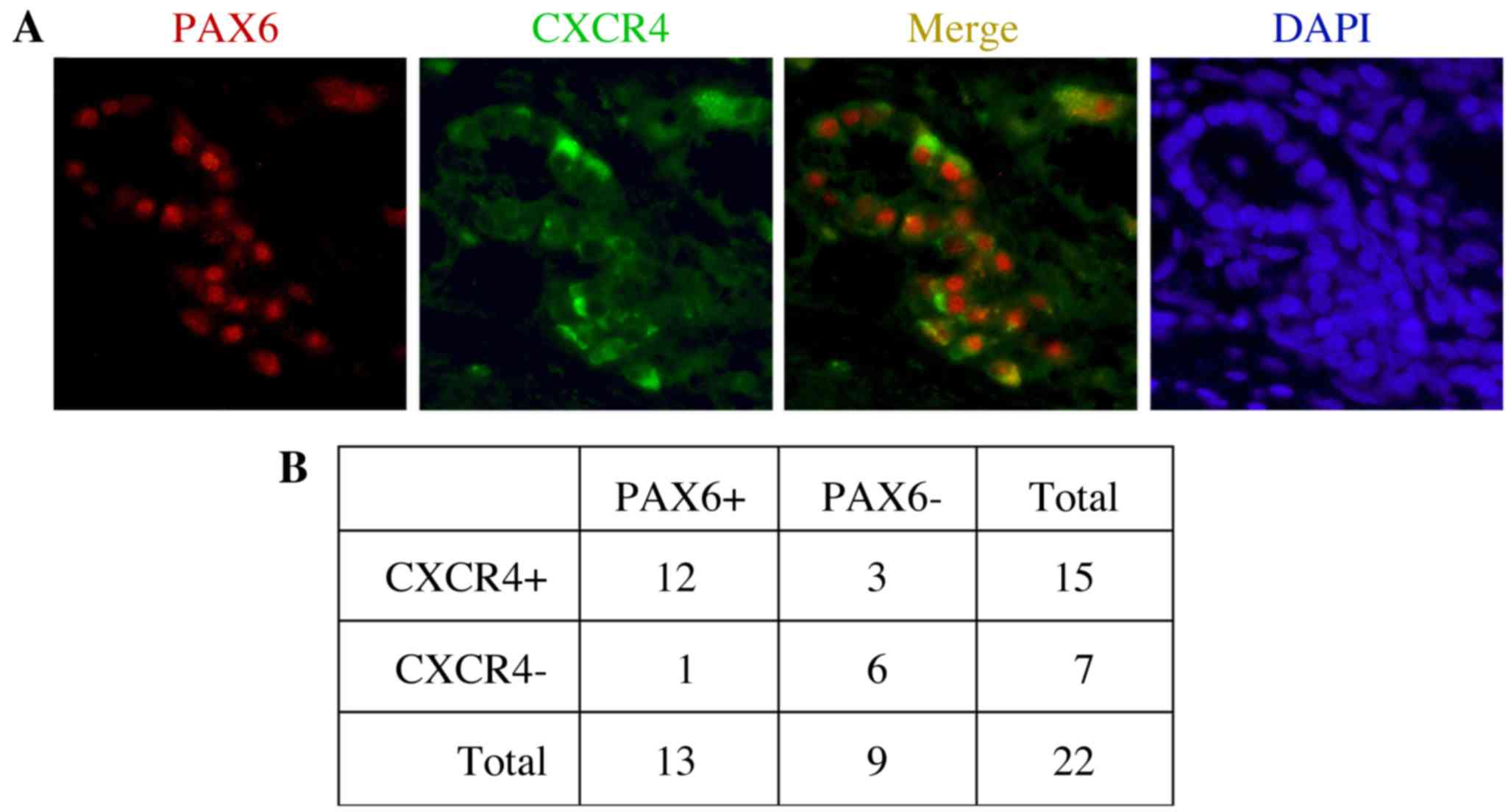
the receptor protein CXCR4 (Fig. 1).
In the present study, expression of PAX6 and CXCR4 was measured in
22 primary tumor tissues. Representative immunofluorescent staining
results indicating the expression of PAX6 and CXCR4, as well as
their co-expression, are shown in Fig.
1A. In the present study, 13/22 samples (59.1%) and 15/22
samples (68.2%) were positive for PAX6 and CXCR4 expression,
respectively (Fig. 1B). The
expression statuses of PAX6 and CXCR4 were positively correlated
when compared using Fisher's exact probability tests (P=0.0066). Of
the PAX6-expressing tumor samples, the majority (12/13; 92.3%) also
co-expressed CXCR4.
PAX6 activates a reporter containing
CXCR4 gene elements
We previously demonstrated that the PAX
transcription factor PAX3 promotes CXCR4 expression in melanoma
through a highly conserved enhancer in the CXCR4 gene (13). The PAX enhancer element is located
within the first intron of the CXCR4 gene, immediately 3′-proximal
to exon 1 (Fig. 2A). The element
(Fig. 2A, black box) is located in a
52-bp sequence that is highly conserved between mammals [Fig. 2A, grey box (13)]. PAX3 was found to bind to this PAX
element. The PAX6 paired domain binds to a sequence with a TT(A/C)
ACGC(A/T) core, first identified through in vitro site
selection assays (15) and recently
supported through in cellulo chromatin
immunoprecipitation-sequencing and systematic evolution of ligands
by exponential enrichment studies (27,28). The
PAX site in the CXCR4 enhancer region closely matches the preferred
PAX6 site, and is a more ideal site for PAX6 over PAX3 due to the
5′ TT rather than GT sequence (15,16). To
examine whether PAX6 is also capable of driving expression from
this element, vectors containing regions of the CXCR4 locus were
transfected into 293T cells with or without PAX6 and/or PAX6(5a),
an alternative splice version of PAX6. Three constructs were
tested: pGL2-CXCR4pm393, pGL2-CXCR4pm393L and
pGL2-CXCR4pm393L-ΔISH. The pGL2-CXCR4pm393 vector contains 393 bp
of sequence from 5′-proximal to exon 1 and the 5′-UTR. The
pGL2-CXCR4pm393L vector contains all the sequence of pGL2-CXCR4pm
with the addition of the first intron and part of the coding
sequence cloned in-frame with the luciferase gene cassette. The
pGL2-CXCR4pm393L-ΔISH vector has the same elements as
pGL2-CXCR4pm393L, but with a deletion of the island of sequence
homology. These vectors are shown schematically in Fig. 2B. PAX6 or PAX6(5a) did not drive
reporter expression from the pGL2-CXCR4pm393 vector (Fig. 2C). However, PAX6 and PAX6(5a) induced
significant reporter expression from the pGL2-CXCR4pm393L construct
containing an intact intron 1 [2.8±0.69-fold (P=0.011) and
3.8±0.63-fold (P=0.0017) for PAX6 and PAX6(5a), respectively,
relative to the reporter vector alone]. The combined addition of
PAX6 and PAX6(5a) produced the highest level of reporter expression
(8.1±0.25-fold). Deletion of the enhancer element within the intron
completely eliminated the ability of either of the PAX6 protein
variants to drive luciferase expression.
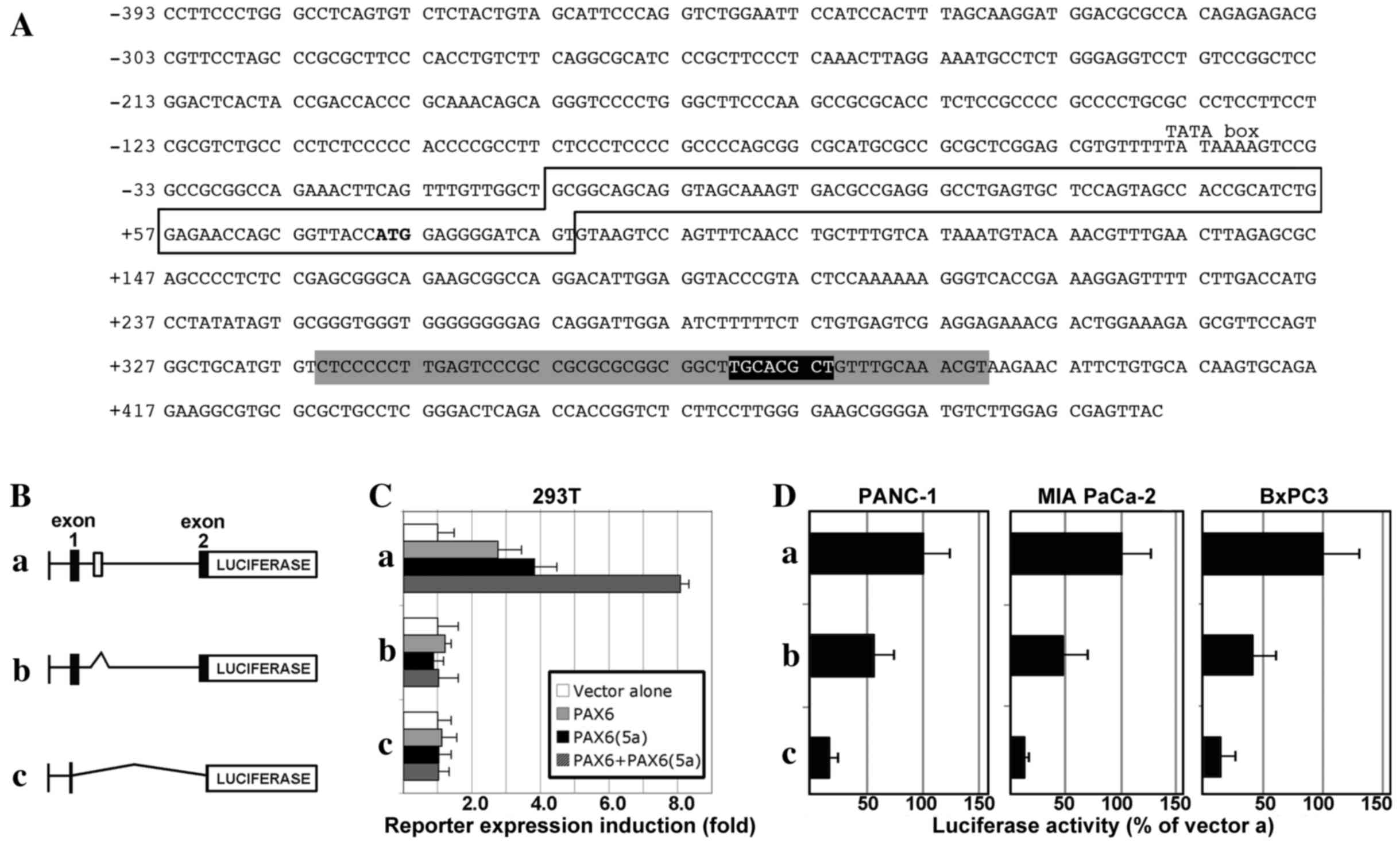 | Figure 2.The CXCR4 locus contains a conserved
enhancer element within intron 1 that is PAX6-responsive and active
in pancreatic cancer cells. (A) Sequence of a section of the human
CXCR4 locus, containing 393 bp 5′-proximal to exon 1, exon 1 (box)
with the translational start codon (ATG in bold), and partial
intron 1 sequence. The intronic sequence shown contains a 52-bp
island of high conservation of homology between mammals (grey
highlight), as well as a previously identified PAX site (black
highlight, white letters) (13)
within this homologous region. (B) Schematic of CXCR4 gene
expression constructs: (a) pGL2-CXCR4pm393L vector containing 393
bp of the sequence 5′-proximal to exon 1 of the CXCR4 gene, the
5′-untranslated region from exon 1 and the coding sequence from all
of exon 1 and part of exon 2, and the 1,781-bp intron 1; (b)
pGL2-CXCR4pm393L-ΔISH construct is the same as pGL2-CXCR4pm393L
except for a deletion of a 267-bp segment from intron 1, containing
the island of homology shown in the grey shaded region in (A); (c)
pGL2-CXCR4pm393 construct contains only the 393-bp sequence
5′-proximal to exon 1, and none of the other regions present in the
pGL2-CXCR4pm393L vector. (C) PAX6 proteins promote gene expression
through a highly conserved enhancer in the CXCR4 intron. The
different reporter constructs (a, pGL2-CXCR4pm393L; b,
pGL2-CXCR4pm393L-ΔISH; or c, pGL2-CXCR4pm393) were transfected into
293T cells with a reporter vector alone or with expression
constructs expressing canonical PAX6, PAX6(5a), or both PAX6 and
PAX6(5a) isoforms. Luciferase induction is shown as the fold
difference relative to the reporter vector alone, which was set at
1-fold. (D) Loss of the conserved intronic enhancer element from
the CXCR4 gene diminishes the activity of CXCR4 reporter constructs
in pancreatic cancer cell lines. Reporter vectors (a,
pGL2-CXCR4pm393L; b, pGL2-CXCR4pm393L-ΔISH; or c, pGL2-CXCR4pm393)
were transfected into PANC-1, MIA PaCa-2 and BxPC-3 cells. Light
levels measured following transfection with vector a
(pGL2-CXCR4pm393L) were set at 100%. CXCR4, C-X-C chemokine
receptor 4; PAX6, paired box transcription factor 6; PAX6(5a), PAX6
exon 5 alternative transcript variant. |
In order to determine whether the enhancer site
containing the PAX element drives expression in pancreatic cancer
cells, the three vectors were transfected into PANC-1, MIA PaCa-2,
and BxPC-3 cell lines. All cell lines exhibited luciferase activity
from the pGL2-CXCR4pm393L vector (set at 100%), and activity was
significantly decreased when the intronic enhancer element was
removed (pGL2-CXCR4pm393L-ΔISH): 57.09±17.80% in PANC-1 cells
(P=0.035), 47.7±23.11% in MIA PaCa-2 cells (P=0.034), and
42.39±19.18% in BxPC-3 cells (P=0.026; Fig. 2D). The expression of luciferase
reporter was significantly lower compared with pGL2-CXCR4pm393L,
when only the proximal promoter vector was utilized
(pGL2-CXCR4pm393) in all cell lines: PANC-1, 16.32±8.48%
(P=0.0025); MIA PaCa-2, 11.82±5.25% (P=0.0031); and BxPC-3,
15.53±11.74% (P=0.0059). These data support the hypothesis that
PAX6 proteins drive the CXCR4 gene through a conserved intronic
element, and that the loss of this site leads to a significant loss
of enhancer activity in pancreatic cancer cells.
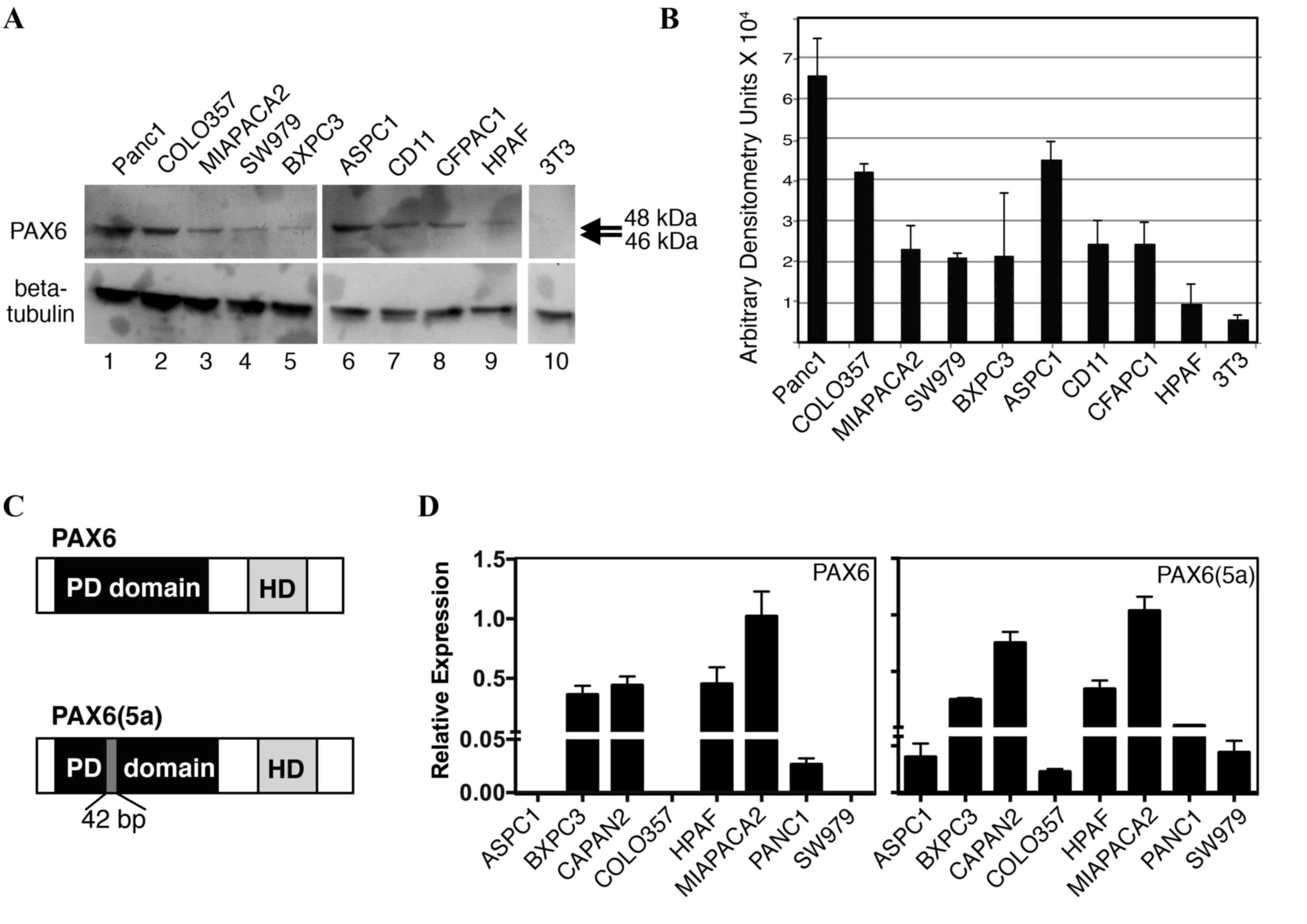
Pancreatic cancer cell lines express
PAX6
We have previously demonstrated that PAX6 is
expressed in pancreatic carcinoma primary tumors and cell lines
(14). Furthermore, unexpectedly, the
predominant PAX6 protein expressed in SW979, PANC-1, and MIA PaCa-2
cells was found to be the alternatively spliced 48-kDa version,
PAX6(5a), rather than the canonical 46 kDa protein (18). In the present study, PAX6 levels were
evaluated in nine independent pancreatic cell lines (Fig. 3A and B). All the cell lines expressed
measurable levels of PAX6 protein, except for the low or
undetectable levels in HPAF cells. As demonstrated in our
aforementioned results, the predominant band was indicated to be
~48 kDa, corresponding to the alternative rather than canonical
form of PAX6 (Fig. 3C). Bands at 46
kDa were absent or faint. To determine PAX6 transcript levels,
RT-qPCR analysis utilizing transcript-specific primers were
performed. The PAX6(5a) transcript was expressed in all eight of
the cell lines tested, while 5/8 lines produced the canonical
transcript (Fig. 3D); the cell lines
AsPC-1, COLO-357, and SW979 did not have measureable levels of PAX6
canonical transcript. No correlation was identified between the
levels of transcript and the detected levels of protein. However,
HPAF cells expressed significant levels of the two transcripts
while producing low or no PAX6 protein.
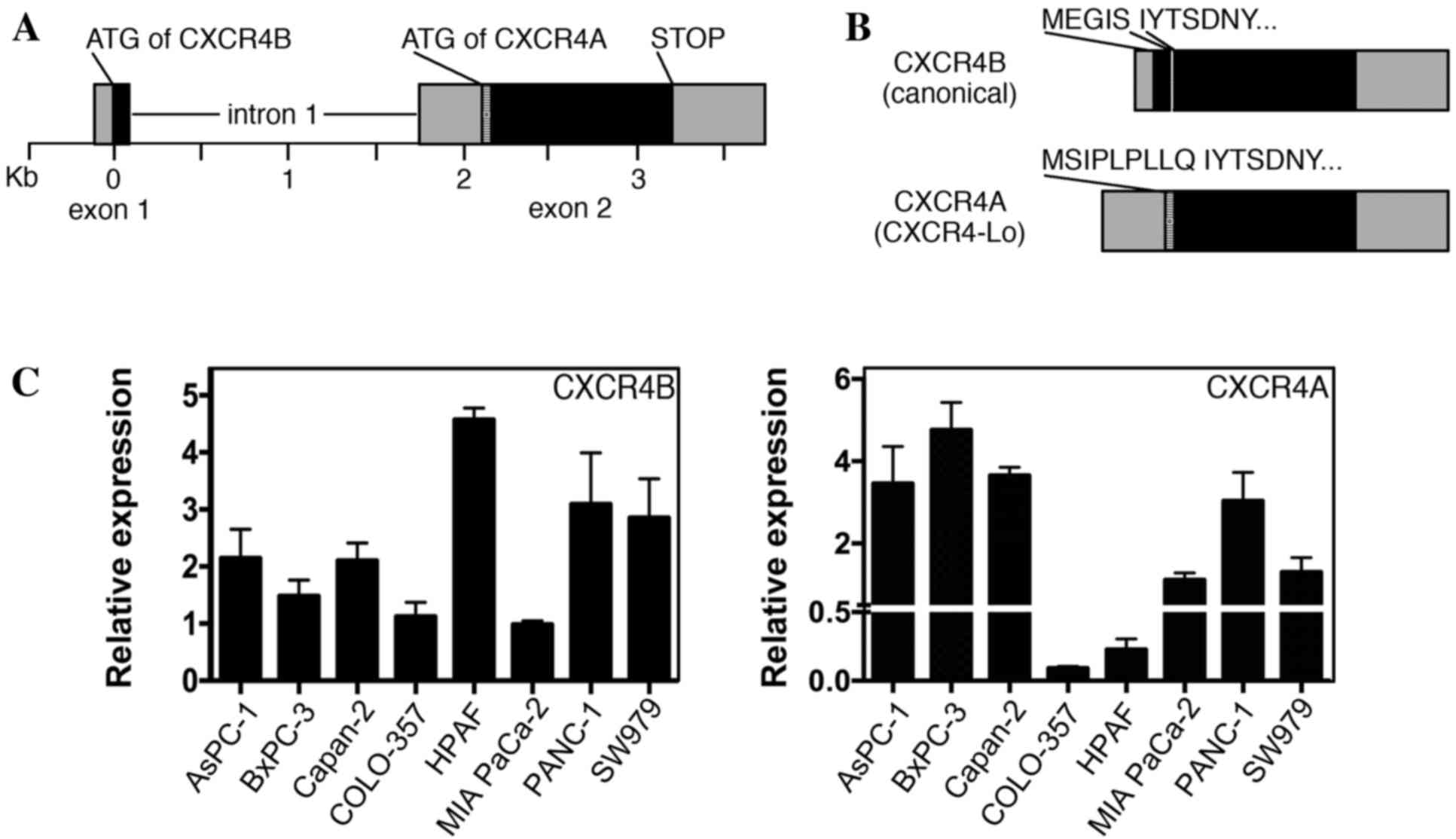
Pancreatic cancer cell lines express
variable levels of CXCR4A and CXCR4B transcripts
CXCR4 protein is widely expressed in pancreatic
cancer, with significant levels detected in various pancreatic
cancer cell lines, including all lines used in the present study
(4–6,29,30). Previous reports have identified CXCR4
expression in pancreatic cell lines by RT-PCR, western blot
analysis and immunohistochemistry; however, these methods do not
differentiate between the two major versions of CXCR4 (canonical
CXCR4 protein/CXCR4B transcript and CXCR4-Lo protein/CXCR4A
transcript). The human CXCR4 gene comprises two exons and two
alternative start codons (Fig. 4A)
(22). The two alternatively spliced
transcripts (CXCR4B and CXCR4A) differ from one another at the
N-terminal ends. CXCR4B (also known as CXCR4 variant 2) encodes the
more common CXCR4 protein utilizing codons from exons 1 and 2,
while CXCR4A (also referred to as CXCR4 variant 1) produces the
longer CXCR4-Lo protein and is encoded entirely from exon 2
(Fig. 4B). The canonical protein
encoded from the CXCR4B transcript is expressed in a wide array of
tissues, while CXCR4-Lo expression is normally restricted to
peripheral blood lymphocytes and spleen cells (22). The proteins differ only in the
furthest six or nine (CXCR4 or CXCR4-Lo, respectively) amino acids
of the N-terminal ends and, although the functional consequence of
this difference is unknown, there is evidence of a differential
response to ligand binding (22,24). Our
previous study revealed that the canonical CXCR4B transcript was
the dominant transcript in melanoma, with expression in all seven
lines analyzed (13). However, there
was measurable expression of the CXCR4A transcript in three of the
seven melanoma lines, albeit to significantly lower levels than
those of the canonical transcript. In the present study, pancreatic
cancer cell lines were found to express measurable levels of the
CXCR4A and CXCR4B transcripts in all eight cell lines measured
(Fig. 4C).
Discussion
The present study identified that, while both of the
PAX6 transcripts are expressed in pancreatic cancer cell lines, the
PAX6(5a) variant form is expressed in all eight cell lines tested
and produces the majority of the PAX6 protein (Fig. 3). It is not known why PAX6 is
aberrantly expressed in pancreatic cancer, or why the PAX6(5a)
variant is the predominant form expressed. The two proteins are
identical with the exception of a 14-amino-acid insertion into the
N-terminal of the paired DNA-binding domain (22). This insertion alters the DNA-binding
specificity of the domain, inducing the two PAX6 proteins to have
various DNA binding site preferences and affinities. The PAX6 and
PAX6(5a) paired domains bind to associated but distinct core DNA
elements, with lower affinities for the preferred DNA binding site
of the other protein (15,21,31). In
the present study, no difference in gene expression between PAX6
and PAX6(5a) proteins was measured in CXCR4 gene reporter assays
(Fig. 2C). In a previous study,
expressing PAX6 or PAX6(5a) in a cell line without endogenous PAX6
expression (mouse fibroblast 3T3 cells) identified genes that were
regulated by either of the PAX6 proteins or were subject to
transcript-specific gene regulation (32). In addition, isoform-specific PAX6
mouse mutants have overlapping but different phenotypes (33,34), and
this is in parallel with human studies wherein a specific PAX6(5a)
mutation has been shown to have similar abnormalities but does not
phenocopy other PAX6 mutations (35).
These findings support the hypothesis that PAX6 proteins have
redundant and unique functions.
While canonical PAX6 was found to be expressed in
five of the eight pancreatic cancer cell lines (Fig. 3D), the major PAX6 product expressed
was the larger PAX6(5a) protein [Fig.
3A and our previous study (18)].
In certain normal tissues and during development, the two isoforms
are expressed together and can functionally interact (36). The transcriptional function of the two
isoforms is influenced by the ratio of PAX6:PAX6(5a); depending on
cell type and developmental stage, the optimal balance ranges from
8:1-3:1 in the brain (depending on developmental stage) to 1:1 in
the developing and maturing retina (31,37–39). In
the present study, the two variants promoted the expression of a
reporter gene containing CXCR4 gene elements (Fig. 2B). While the expression of the
reporter was significantly increased with the addition of both
isoforms, it is not clear whether expression would be altered or
optimized utilizing the protein ratios found in pancreatic cancer
cells. Notably, in these cancer cells, the majority of the protein
is the variant form, which is the opposite of what is found in
normal tissues wherein the canonical is the dominant form. What
remains unknown is how the interaction of the two isoforms affects
gene expression, whether this is purely due to DNA site binding
specificity or due to changes in interactions with protein
co-factors, and how this impacts pancreatic cancer selection,
survival or progression.
This present study provides evidence that PAX6 is in
a common molecular pathway with CXCR4, since the two proteins are
expressed together (Fig. 1), and that
PAX6 proteins are sufficient to drive expression through an
intronic element within the CXCR4 gene (Fig. 2). Loss of this conserved intronic
enhancer led to a decrease in gene expression in three pancreatic
cancer cell lines (Fig. 2D). It is
not clear whether inhibition of one or both of the PAX6 isoforms
would lead to alteration of CXCR4 expression in pancreatic cancer,
or if PAX6 drives expression of CXCR4A and CXCR4B equally or if
there is specificity. The present results identified the expression
of the canonical CXCR4B transcript as well as the CXCR4A variant
(Fig. 4). While expression of the
CXCR4B transcript is well documented in many types of human cancer,
the presence of the alternative CXCR4A variant is not well studied.
Indeed, the expression of CXCR4A even in normal mature tissues is
highly restricted to peripheral blood lymphocytes and spleen cells
(22). In cancer cells, our group
identified expression in a subset of melanoma cells (13). In addition, the two transcripts are
expressed in Ewing sarcoma primary tissue and cell lines (40). It is not clear how common the
expression of CXCR4A transcript is in cancer in general, and what
is the potential impact of its expression. However, since the
extracellular domain differs between the receptors, this variance
may be exploited in the development of targeted therapies specific
for the more cell-type restricted CXCR4A.
In conclusion, the present study revealed that PAX6
and CXCR4 are co-expressed in pancreatic cancer, may be part of a
shared pathway, and are each expressed as their respective
canonical and variant transcripts. These present findings suggest
that PAX6 and CXCR4 are good candidates for therapies, and the
presence of the non-canonical proteins may provide novel targets
for future therapeutics.
Acknowledgements
This study was supported in whole or in part by the
National Institutes of Health (grant nos. NIH R01CA130202, NIH
R01CA184001 and NIH P30-CA014599), the American Cancer Society
(grant no. RSG-CSM-121505) and the Friends of
Dermatology-Chicago.
Glossary
Abbreviations
Abbreviations:
|
CXCL12
|
C-X-C chemokine ligand 12
|
|
CXCR4
|
C-X-C chemokine receptor 4
|
|
CXCR4A
|
CXCR4 transcript variant A
|
|
CXCR4B
|
CXCR4 transcript variant B
|
|
kDa
|
kilodaltons
|
|
PAX6
|
paired box transcription factor 6
|
|
PAX6(5a)
|
PAX6 exon 5 alternative transcript
variant
|
References
|
1
|
Ilic M and Ilic I: Epidemiology of
pancreatic cancer. World J Gastroenterol. 22:9694–9705. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wolfgang CL, Herman JM, Laheru DA, Klein
AP, Erdek MA, Fishman EK and Hruban RH: Recent progress in
pancreatic cancer. CA Cancer J Clin. 63:318–348. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marchesi F, Grizzi F, Laghi L, Mantovani A
and Allavena P: Molecular mechanisms of pancreatic cancer
dissemination: The role of the chemokine system. Curr Pharm Des.
18:2432–2438. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Koshiba T, Hosotani R, Miyamoto Y, Ida J,
Tsuji S, Nakajima S, Kawaguchi M, Kobayashi H, Doi R, Hori T, et
al: Expression of stromal cell-derived factor 1 and CXCR4 ligand
receptor system in pancreatic cancer: A possible role for tumor
progression. Clin Cancer Res. 6:3530–3535. 2000.PubMed/NCBI
|
|
5
|
Mori T, Doi R, Koizumi M, Toyoda E, Ito D,
Kami K, Masui T, Fujimoto K, Tamamura H, Hiramatsu K, et al: CXCR4
antagonist inhibits stromal cell-derived factor 1-induced migration
and invasion of human pancreatic cancer. Mol Cancer Ther. 3:29–37.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wehler T, Wolfert F, Schimanski CC, Gockel
I, Herr W, Biesterfeld S, Seifert JK, Adwan H, Berger MR, Junginger
T, et al: Strong expression of chemokine receptor CXCR4 by
pancreatic cancer correlates with advanced disease. Oncol Rep.
16:1159–1164. 2006.PubMed/NCBI
|
|
7
|
Marchesi F, Monti P, Leone BE, Zerbi A,
Vecchi A, Piemonti L, Mantovani A and Allavena P: Increased
survival, proliferation, and migration in metastatic human
pancreatic tumor cells expressing functional CXCR4. Cancer Res.
64:8420–8427. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gebauer F, Tachezy M, Effenberger K, von
Loga K, Zander H, Marx A, Kaifi JT, Sauter G, Izbicki JR and
Bockhorn M: Prognostic impact of CXCR4 and CXCR7 expression in
pancreatic adenocarcinoma. J Surg Oncol. 104:140–145. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Marechal R, Demetter P, Nagy N, Berton A,
Decaestecker C, Polus M, Closset J, Deviére J, Salmon I and Van
Laethem JL: High expression of CXCR4 may predict poor survival in
resected pancreatic adenocarcinoma. Br J Cancer. 100:1444–1451.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Park B, Sung B, Yadav VR, Cho SG, Liu M
and Aggarwal BB: Acetyl-11-keto-β-boswellic acid suppresses
invasion of pancreatic cancer cells through the downregulation of
CXCR4 chemokine receptor expression. Int J Cancer. 129:23–33. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun JS, Zhang XL, Yang YJ, Nie ZG and
Zhang Y: Hypoxia promotes C-X-C chemokine receptor type 4
expression through microRNA-150 in pancreatic cancer cells. Oncol
Lett. 10:835–840. 2015.PubMed/NCBI
|
|
12
|
Ying X, Jing L, Ma S, Li Q, Luo X, Pan Z,
Feng Y and Feng P: GSK3β mediates pancreatic cancer cell invasion
in vitro via the CXCR4/MMP-2 Pathway. Cancer Cell Int. 15:702015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kubic JD, Lui JW, Little EC, Ludvik AE,
Konda S, Salgia R, Aplin AE and Lang D: PAX3 and FOXD3 promote
CXCR4 expression in melanoma. J Biol Chem. 290:21901–21914. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lang D, Mascarenhas JB, Powell SK,
Halegoua J, Nelson M and Ruggeri BA: PAX6 is expressed in
pancreatic adenocarcinoma and is downregulated during induction of
terminal differentiation. Mol Carcinog. 47:148–156. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Epstein J, Cai J, Glaser T, Jepeal L and
Maas R: Identification of a Pax paired domain recognition sequence
and evidence for DNA-dependent conformational changes. J Biol Chem.
269:8355–8361. 1994.PubMed/NCBI
|
|
16
|
Goulding MD, Chalepakis G, Deutsch U,
Erselius JR and Gruss P: Pax-3, a novel murine DNA binding protein
expressed during early neurogenesis. Embo J. 10:1135–1147.
1991.PubMed/NCBI
|
|
17
|
Mascarenhas JB, Littlejohn EL, Wolsky RJ,
Young KP, Nelson M, Salgia R and Lang D: PAX3 and SOX10 activate
MET receptor expression in melanoma. Pigment Cell Melanoma Res.
23:225–237. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mascarenhas JB, Young KP, Littlejohn EL,
Yoo BK, Salgia R and Lang D: PAX6 is expressed in pancreatic cancer
and actively participates in cancer progression through activation
of the MET tyrosine kinase receptor gene. J Biol Chem.
284:27524–27532. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
St-Onge L, Sosa-Pineda B, Chowdhury K,
Mansouri A and Gruss P: Pax6 is required for differentiation of
glucagon-producing alpha-cells in mouse pancreas. Nature.
387:406–409. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Epstein JA, Lam P, Jepeal L, Maas RL and
Shapiro DN: Pax3 inhibits myogenic differentiation of cultured
myoblast cells. J Biol Chem. 270:11719–11722. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Epstein JA, Glaser T, Cai J, Jepeal L,
Walton DS and Maas RL: Two independent and interactive DNA binding
subdomains of the PAX6 paired domain are regulated by alternative
splicing. Genes Dev. 8:2022–2034. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gupta SK and Pillarisetti K: Cutting edge:
CXCR4-Lo: Molecular cloning and functional expression of a novel
human CXCR4 splice variant. J Immunol. 163:2368–2372.
1999.PubMed/NCBI
|
|
23
|
Glaser T, Walton DS and Maas RL: Genomic
structure, evolutionary conservation and aniridia mutations in the
human PAX6 gene. Nat Genet. 2:232–239. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Duquenne C, Psomas C, Gimenez S, Guigues
A, Carles MJ, Barbuat C, Lavigne JP, Sotto A, Reynes J, Guglielmi
P, et al: The two human CXCR4 isoforms display different HIV
receptor activities: Consequences for the emergence of X4 strains.
J Immunol. 193:4188–4194. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ruggeri BA, Huang L, Berger D, Chang H,
Klein-Szanto AJ, Goodrow T, Wood M, Obara T, Heath CW and Lynch H:
Molecular pathology of primary and metastatic ductal pancreatic
lesions: Analyses of mutations and expression of the p53, mdm-2 and
p21/WAF-1 genes in sporadic and familial lesions. Cancer.
79:700–716. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun J, Rockowitz S, Xie Q, Ashery-Padan R,
Zheng D and Cvekl A: Identification of in vivo DNA-binding
mechanisms of Pax6 and reconstruction of Pax6-dependent gene
regulatory networks during forebrain and lens development. Nucleic
Acids Res. 43:6827–6846. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bryne JC, Valen E, Tang MH, Marstrand T,
Winther O, da Piedade I, Krogh A, Lenhard B and Sandelin A: JASPAR,
the open access database of transcription factor-binding profiles:
New content and tools in the 2008 update. Nucleic Acids Res.
36(Database Issue): D102–D106. 2008.PubMed/NCBI
|
|
29
|
Heinrich EL, Lee W, Lu J, Lowy AM and Kim
J: Chemokine CXCL12 activates dual CXCR4 and CXCR7-mediated
signaling pathways in pancreatic cancer cells. J Transl Med.
10:682012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Singh S, Srivastava SK, Bhardwaj A, Owen
LB and Singh AP: CXCL12-CXCR4 signalling axis confers gemcitabine
resistance to pancreatic cancer cells: A novel target for therapy.
Br J Cancer. 103:1671–1679. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kozmik Z, Czerny T and Busslinger M:
Alternatively spliced insertions in the paired domain restrict the
DNA sequence specificity of Pax6 and Pax8. Embo J. 16:6793–6803.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kiselev Y, Eriksen TE, Forsdahl S, Nguyen
LH and Mikkola I: 3T3 cell lines stably expressing Pax6 or
Pax6(5a)-a new tool used for identification of common and isoform
specific target genes. PLoS One. 7:e319152012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Duncan MK, Kozmik Z, Cveklova K,
Piatigorsky J and Cvekl A: Overexpression of PAX6(5a) in lens fiber
cells results in cataract and upregulation of (alpha)5(beta)1
integrin expression. J Cell Sci. 113:3173–3185. 2000.PubMed/NCBI
|
|
34
|
Singh S, Mishra R, Arango NA, Deng JM,
Behringer RR and Saunders GF: Iris hypoplasia in mice that lack the
alternatively spliced Pax6(5a) isoform. Proc Natl Acad Sci USA.
99:6812–6815. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Azuma N, Yamaguchi Y, Handa H, Hayakawa M,
Kanai A and Yamada M: Missense mutation in the alternative splice
region of the PAX6 gene in eye anomalies. Am J Hum Genet.
65:656–663. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chauhan BK, Reed NA, Zhang W, Duncan MK,
Kilimann MW and Cvekl A: Identification of genes downstream of Pax6
in the mouse lens using cDNA microarrays. J Biol Chem.
277:11539–11548. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chauhan BK, Yang Y, Cveklová K and Cvekl
A: Functional interactions between alternatively spliced forms of
Pax6 in crystallin gene regulation and in haploinsufficiency.
Nucleic Acids Res. 32:1696–1709. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pinson J, Mason JO, Simpson TI and Price
DJ: Regulation of the Pax6: Pax6(5a) mRNA ratio in the developing
mammalian brain. BMC Dev Biol. 5:132005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang W, Cveklova K, Oppermann B, Kantorow
M and Cvekl A: Quantitation of PAX6 and PAX6(5a) transcript levels
in adult human lens, cornea, and monkey retina. Mol Vis. 7:1–5.
2001.PubMed/NCBI
|
|
40
|
Sand LG, Scotlandi K, Berghuis D,
Snaar-Jagalska BE, Picci P, Schmidt T, Szuhai K and Hogendoorn PC:
CXCL14, CXCR7 expression and CXCR4 splice variant ratio associate
with survival and metastases in Ewing sarcoma patients. Eur J
Cancer. 51:2624–2633. 2015. View Article : Google Scholar : PubMed/NCBI
|