Introduction
Gefitinib is widely used for patients with lung
adenocarcinoma in Asia harboring sensitizing epidermal growth
factor receptor (EGFR) mutations, and usually results in a high
response rate and prolonged progression-free survival (PFS)
(1–3).
However, almost every patient ultimately develops drug resistance
following a median duration of ~12 months (1–3). A
secondary T790M point mutation is the underlying mechanism in ~60%
of patients whose disease progresses beyond the first generation of
EGFR tyrosine kinase inhibitors (TKIs) (4–6). Novel
TKIs have been developed that specifically target the T790M
mutation, including AZD9291 (7) and
CO-1686 (8). Therefore, creating a
reliable and cost-effective approach for the real-time monitoring
of EGFR mutations is of great importance.
Collection of serial re-biopsy tissues in routine
clinical practice is not always feasible, and thus, circulating
cell-free DNA (cfDNA) extracted from patient plasma has been
proposed as a promising alternative (9–11).
Numerous cutting-edge platforms have been developed (12,13), and
droplet digital polymerase chain reaction (ddPCR) is one of the
most accurate and robust methods for absolute nucleic acid
quantification (13). Previous
studies using these platforms have demonstrated a sensitivity and
specificity >90% for detecting EGFR mutations in cfDNA (14–17).
Notably, early detection of T790M mutation up to 16 weeks prior to
radiographic progression was shown in a proof-of-concept study that
collected a fraction of representative plasma samples in each
patient (15). However, this result
requires confirmation in larger studies with consecutive samples
from a more homogeneous patient population with a predefined and
unified time interval. In addition, the growth rate of tumor burden
represented by tumor size or tumor volume was found to be
associated with patient survival (18). However, the prognostic significance of
parameters derived from cfDNA-based EGFR mutation quantification,
including the growth rate of EGFR mutations, has not been
investigated.
In order to explore the association between the
dynamics of EGFR mutations in cfDNA and disease evolution, 20
patients with EGFR-mutant advanced lung adenocarcinoma receiving
first-line gefitinib therapy were prospectively enrolled in the
present study. Clinically relevant activating and resistant EGFR
mutations in monthly collected samples were quantified using ddPCR
for a duration of up to 12 months. In addition, the prognostic
significance of pretreatment T790M mutation status, as well as the
growth rate of EGFR mutations in cfDNA, were examined.
Materials and methods
Patient population
Patients with metastatic lung adenocarcinoma
receiving first-line gefitinib therapy and meeting the following
criteria were prospectively enrolled. Firstly, patients must harbor
one of the two most common sensitizing EGFR mutations, exon 19
E746-A750 deletion (19del) or L858R mutation (L858R), and not the
T790M mutation, in their pretreatment tumor tissues. Detection of
EGFR mutations in tumor tissues was performed using the
Amplification Refractory Mutation System (ARMS), which was approved
by the Chinese Food and Drug Administration for in vitro
diagnostic use, covering the 29 most common EGFR mutations in lung
cancer (17). Patients with 19del
were categorized as 19del patients, while patients with the L858R
mutation were categorized as L858R patients. Secondly, patients
must have adequate baseline plasma samples and baseline tumor
information, including primary and metastatic locations, size,
histology and radiological images. Finally, patients must have a
minimum duration of gefitinib therapy of 3 months without
radiological progression (to rule out patients with primary
resistance as much as possible), and consent to monthly follow-ups
in the clinics of the Department of Medical Oncology of Peking
Union Medical College Hospital (PUMCH; Beijing, China).
Patients with measurable or immeasurable disease
were included. The present study was approved by the Institutional
Review Boards of PUMCH. All patients provided written informed
consent.
Sample processing and quantification
of EGFR mutations
For each eligible patient, baseline blood samples
were retrospectively gathered from the biobank of PUMCH and
subsequent samples were collected during monthly follow-ups. The
present study also collected five blood samples from five healthy
donors (three males and two females, with a median age of
33-years). Each blood sample was spun into plasma by centrifugation
for 10 min at 1,200 × g and the plasma supernatant was further
cleared by centrifugation for 10 min at 3,000 × g, all at ≤4°C for
2 h, and cfDNA was extracted using the QIAamp DNA blood mini kit
(Qiagen GmbH, Hilden, Germany). L858R and T790M mutations were
evaluated in samples from L858R patients, while 19del and T790M
mutations were evaluated in samples from 19del patients. All three
mutations were evaluated in the samples from the 5 healthy donors.
EGFR mutations were detected using the PrimePCR™
ddPCR™ Mutation Detection Assay kits (Bio-Rad
Laboratories, Inc. Hercules, CA, USA; catalog nos., 1863103,
1863104 and 1863105) and the QX100™ AutoDG™
Droplet Digital™ PCR system (Bio-Rad Laboratories,
Inc.), according to the manufacturer's protocol. All assays were
performed in triplicate and the results were reported as copies of
mutant allele per ml of plasma, as described in a previous study
(15).
Clinical data collection
Standard clinicopathological data were collected
from each patient, including demographics, primary and metastatic
tumor characteristics, tumor tissue genotyping results, bimonthly
radiological images, treatment and response, most recent follow-up
date and progression status. PFS was defined as the time interval
between the beginning of gefitinib therapy and disease progression
or mortality. Treatment response and disease progression were
defined according to Response Evaluation Criteria in Solid Tumors
1.1 (RECIST 1.1). Taking the lowest EGFR mutation concentration
recorded since the treatment started as reference, molecular
progressive disease was defined as ≥20% increase of sensitizing
EGFR mutations and/or T790M mutations, which is similar to
RECIST.
Growth rate of tumor burden was calculated in each
patient with measurable disease. In line with a previous study
(18), it was defined as the increase
of the target tumor lesion diameter during the last two follow-ups,
prior to documentation of disease progression. For the 2 patients
that did not experience disease progression, the last two
follow-ups prior to the end of the present study were used. Growth
rate of EGFR mutation concentration was calculated for each patient
in the same manner and the geometric mean was calculated when
growth rates of sensitizing mutation and T790M mutation could be
calculated.
Statistical analysis
All analyses were performed using SPSS version 12.0
(SPSS, Inc., Chicago, IL, USA). All the data are presented as the
mean ± standard deviation. Comparisons of proportions were
performed by χ2 tests. Survival curves for PFS were
created by the Kaplan-Meier method. Log-rank tests were used to
compare the survival curves between different subgroups. Pearson's
test was used to determine the significance of linear correlations
between different parameters. Two-tailed tests were used and
P<0.05 was considered to indicate a statistically significant
difference.
Results
Characteristics of patients
A total of 20 patients were enrolled in the present
study (Table I), including 16 (80%)
female and 4 (20%) male individuals. The average age of the
patients was 58.9±10.2 years, with no significant difference being
observed between male and female patients (56.2 vs. 59.5 years).
Bone (particularly vertebrae), the contralateral lobe, brain, liver
and malignant pleural dissemination were the most common sites of
metastases. In terms of EGFR mutations, 12 patients (60%) harbored
the L858R mutation and 8 patients (40%) harbored the 19 deletion.
The average duration of gefitinib therapy at the time of enrollment
was 7.4 months (range, 4–38 months). Regarding the optimal
treatment response, 1 patient reached complete response, 10
patients reached partial response and the remaining 9 patients had
stable disease. By the end of the study, 18 patients had disease
progression (7 19del patients and 11 L858R patients), with a median
PFS of 12.0 months (95% CI, 10.3–13.7 months).
 | Table I.Characteristics and clinical data of
patients. |
Table I.
Characteristics and clinical data of
patients.
| No. | Age/sex | Sensitizing
mutation | Primary foci | Metastases | Optimal response | Progression
status | PFS | T790M status |
|---|
| 1 | 57/M | 19del | Superior lobe of
left lung | Liver, brain and
bone | PR | Yes | 9 | No |
| 2 | 58/F | L858R | Superior lobe of
left lung | Bones | PR | No | 8 | No |
| 3 | 43/F | 19del | Right Lung | Left lung | CR | No | 16 | Yes |
| 4 | 60/F | 19del | Superior lobe of
left lung | Right lung | SD | Yes | 13 | Yes |
| 5 | 57/F | L858R | Inferior lobe of
right lung | Vertebrae | PR | Yes | 13 | No |
| 6 | 70/F | L858R | Middle lobe of
right lung | Brain | SD | Yes | 29 | Yes |
| 7 | 70/F | L858R | Inferior lobe of
right lung | Left lung | PR | Yes | 12 | No |
| 8 | 62/F | L858R | Superior lobe of
right lung | Left lung | PR | Yes | 15 | No |
| 9 | 74/F | 19del | Left lung | Bilateral MPE | PR | Yes | 47 | Yes |
| 10 | 64/F | 19del | Left lung | Right lung | SD | Yes | 11 | No |
| 11 | 72/F | L858R | Superior lobe of
left lung | Vertebrae | SD | Yes | 24 | Yes |
| 12 | 50/F | 19del | Inferior lobe of
left lung | Right lung,
bones | PR | Yes | 12 | Yes |
| 13 | 37/F | L858R | Inferior lobe of
left lung | Bilateral MPE,
bones | SD | Yes | 15 | Yes |
| 14 | 53/F | L858R | Superior lobe of
left lung | Vertebrae | PR | Yes | 10 | Yes |
| 15 | 53/F | L858R | Superior lobe of
left lung | Vertebrae | SD | Yes | 11 | Yes |
| 16 | 63/F | 19del | Both lungs | Cervical LN | PR | Yes | 6 | No |
| 17 | 67/F | L858R | Left lung | Cervical LN | PR | Yes | 30 | Yes |
| 18 | 56/M | L858R | Inferior lobe of
right lung | Bones | SD | Yes | 8 | Yes |
| 19 | 44/M | 19del | Inferior lobe of
right lung | Bones | SD | Yes | 10 | No |
| 20 | 68/M | L858R | Inferior lobe of
left lung | Bones | SD | Yes | 12 | No |
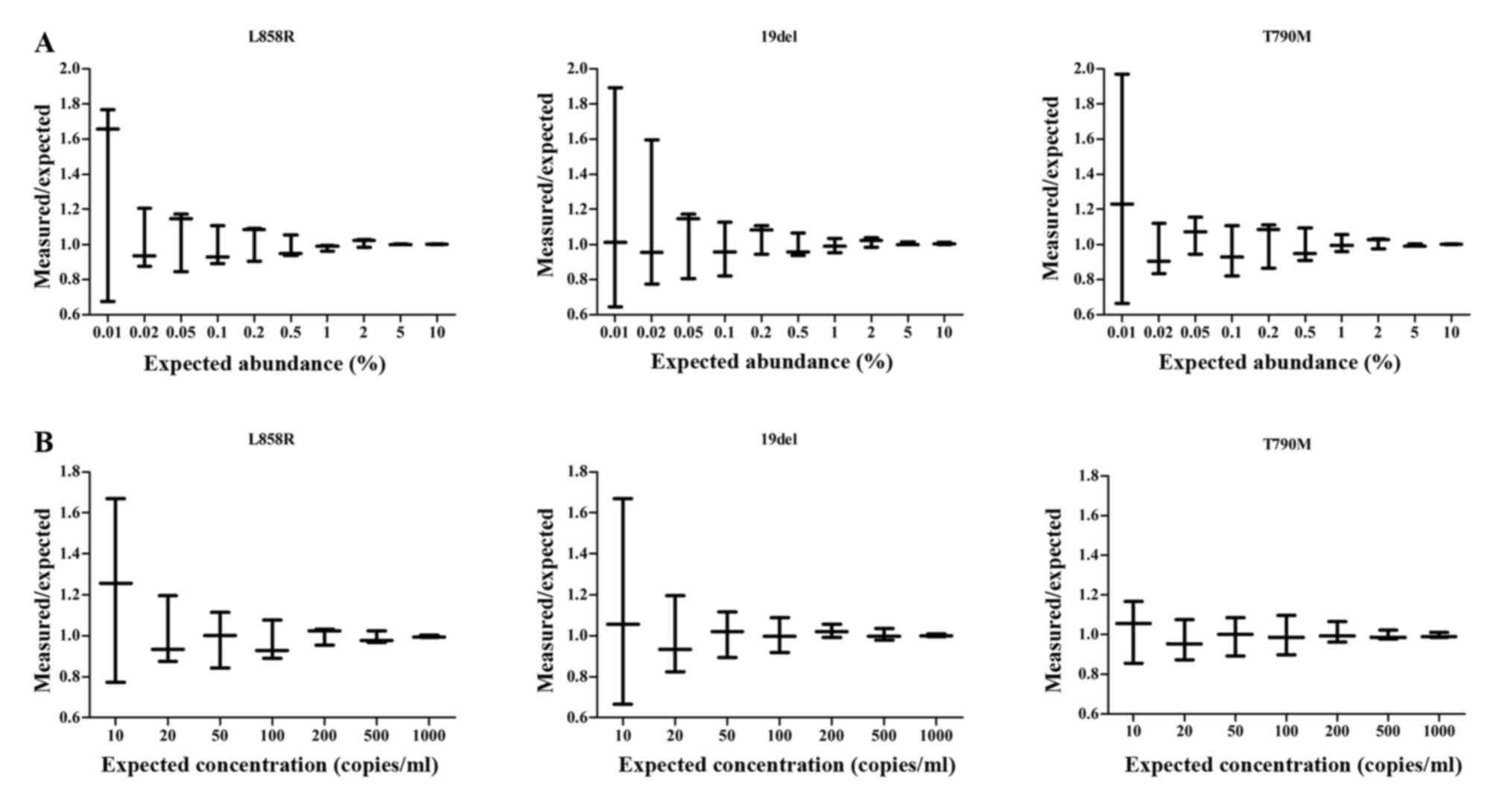
Validation of ddPCR assays
To test the analytic sensitivity and specificity of
the three ddPCR assays, mutant DNA was serially diluted from
NCI-H1650 cells (harboring 19 deletion) and NCIH1975 cells
(harboring L858R and T790M mutations) in human reference genomic
DNA (catalog no., G1471; Promega Corporation, Madison, WI, USA) in
decreasing ratios (1:10 to 1:10,000). The assays were able to
detect mutation abundance as low as 0.05%, with a relative error of
<20%. Similarly, by serially decreasing the amount of mutant DNA
added, a detection limit of 20 copies/ml of mutant DNA was
demonstrated (Fig. 1). With a
mutation abundance of >0.05% and a mutation concentration of
>20 copies/ml, the assays demonstrated linear quantification
across a dynamic range spanning 4 orders of magnitude (data not
shown).
To determine the reference range of the assays, EGFR
mutations were examined in the samples from the 5 healthy donors.
Low levels of EGFR mutations were detected, with a peak value of 7
copies/ml, 13 copies/ml and 5 copies/ml for the L858R mutation,
19del and T790M mutation, respectively. As these detection limits
were <20 copies/ml, 20 copies/ml was selected as the threshold
for a positive result.
EGFR mutations in cfDNA may be
quantified for the majority of patients
Each patient enrolled in the present study was
followed up monthly for a duration of 4–12 months. Together with
the 20 baseline samples, 106 serial plasma samples were collected
from 12 L858R patients, and 62 plasma samples were collected from 8
19del patients.
In the baseline samples, corresponding sensitizing
mutations were successfully detected in 19 patients (with the
exception of patient 7) with an average concentration of 614.8
copies/ml (range, 457.1–1272.7 copies/ml) for L858R patients and
668.9 copies/ml (range, 543.7–874.2 copies/ml) for 19del patients.
Notably, T790M mutations were also detected in 3 patients (patient
15, 18 and 19), with a concentration of 70.21 copies/ml, 98.10
copies/ml and 54.37 copies/ml, respectively.
At the time of disease progression, corresponding
sensitizing mutations were detected in all patients, with an
average concentration of 572.9 copies/ml (range, 192.1–1057.9
copies/ml) for L858R patients and 629.8 copies/ml (range,
213.4–965.3copies/ml) for 19del patients. T790M mutations were
detected in 10 patients (7 L858R patients and 3 19del patients)
with an average concentration of 473.69 copies/ml (range,
106.4–793.8 copies/ml). In addition, EGFR sensitizing mutations and
T790M mutations were also detected in patient 2 and patient 3,
whose disease did not reach progression by the end of the present
study.
Dynamics of EGFR mutations are
associated with treatment response
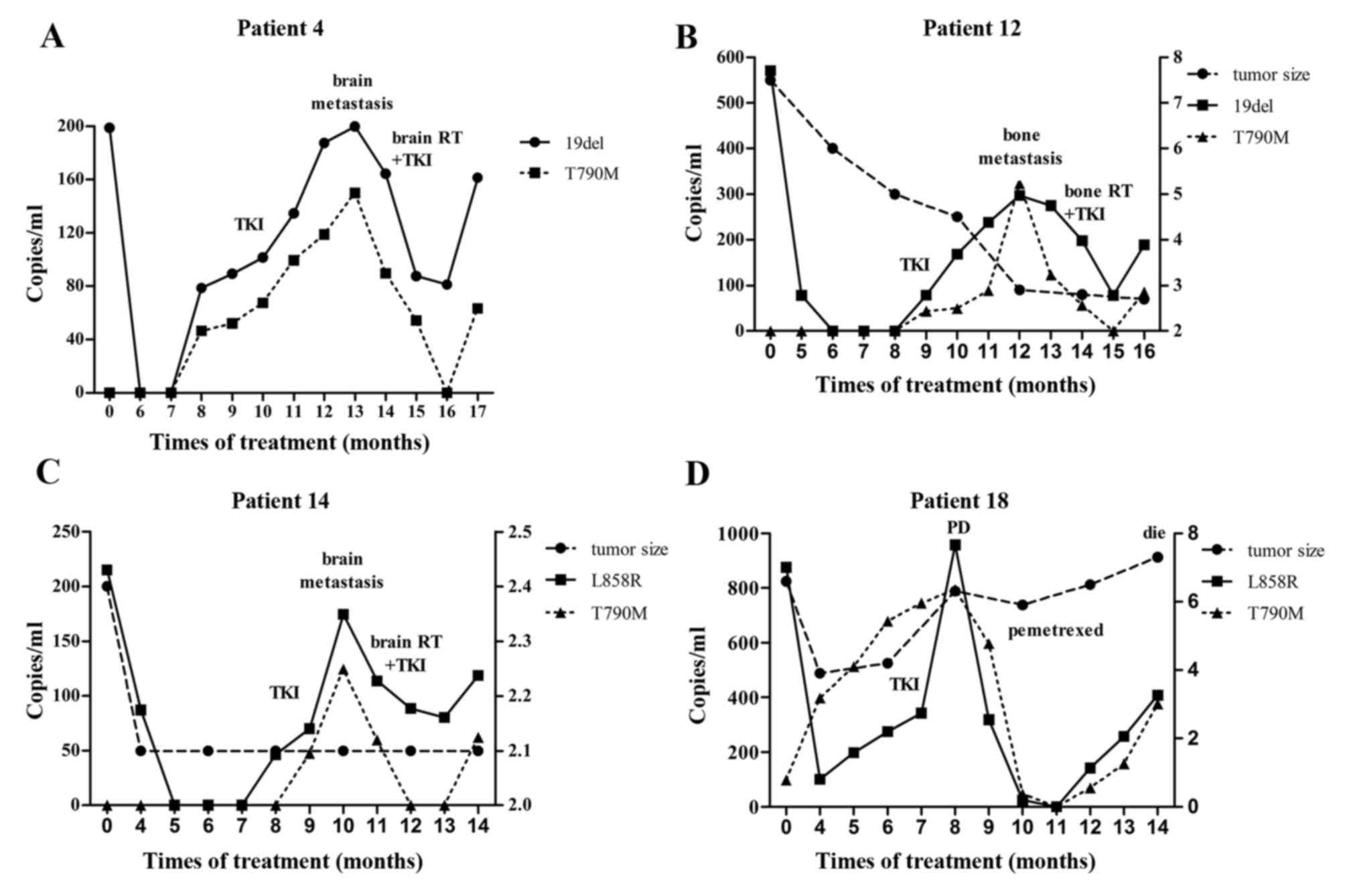
The serial changes of the EGFR mutation
concentrations (L858R or 19del and/or T790M) were generally
associated with treatment response observed in images or reflected
from clinical manifestations, during or beyond first-line gefitinib
therapy (Fig. 2).
Notably, compared with tumor burden (as measured by
the sum of diameters of target tumor lesions), the dynamics of EGFR
mutations were found to be more helpful in disease monitoring.
First, for patients with immeasurable disease (patient 3, 4, 5, 9
and 17), follow-up parameters were otherwise limited. Second, this
may also be the case for patients with measurable disease, but
presenting exceptional phenomena. The present cohort included
patients who had shrinking primary tumors and immeasurable
metastatic lesions, including patients with progressive bone
metastases confirmed by bone scintigraphy (patient 12; Fig. 1), or accumulating malignant pleural
effusion. There were also patients with stable primary disease but
sudden metastases, including an emergent brain metastasis that was
absent 2 months earlier at the last follow-up, but presented as a
considerable tumor mass at the most recent cranial magnetic
resonance imaging (patient 14; Fig.
1). For all these patients, the dynamics of EGFR mutations were
associated with disease evolution.
Molecular progressive disease may
predict drug resistance
Generally, the concentration of sensitizing EGFR
mutation dropped upon initiation of gefitinib treatment. In certain
patients, the value dropped to zero. Later, this concentration
gradually increased during continuous gefitinib treatment. With the
exception of the 3 patients with pretreatment T790M mutation, the
occurrence of T790M mutation usually accompanied an increase of
sensitizing mutation, and the concentration of T790M mutation also
continued increasing (Fig. 3).
Consequently, molecular progressive disease may be tracked at a
median time interval of 4 months (range, 0–8 months) prior to
objective progression.
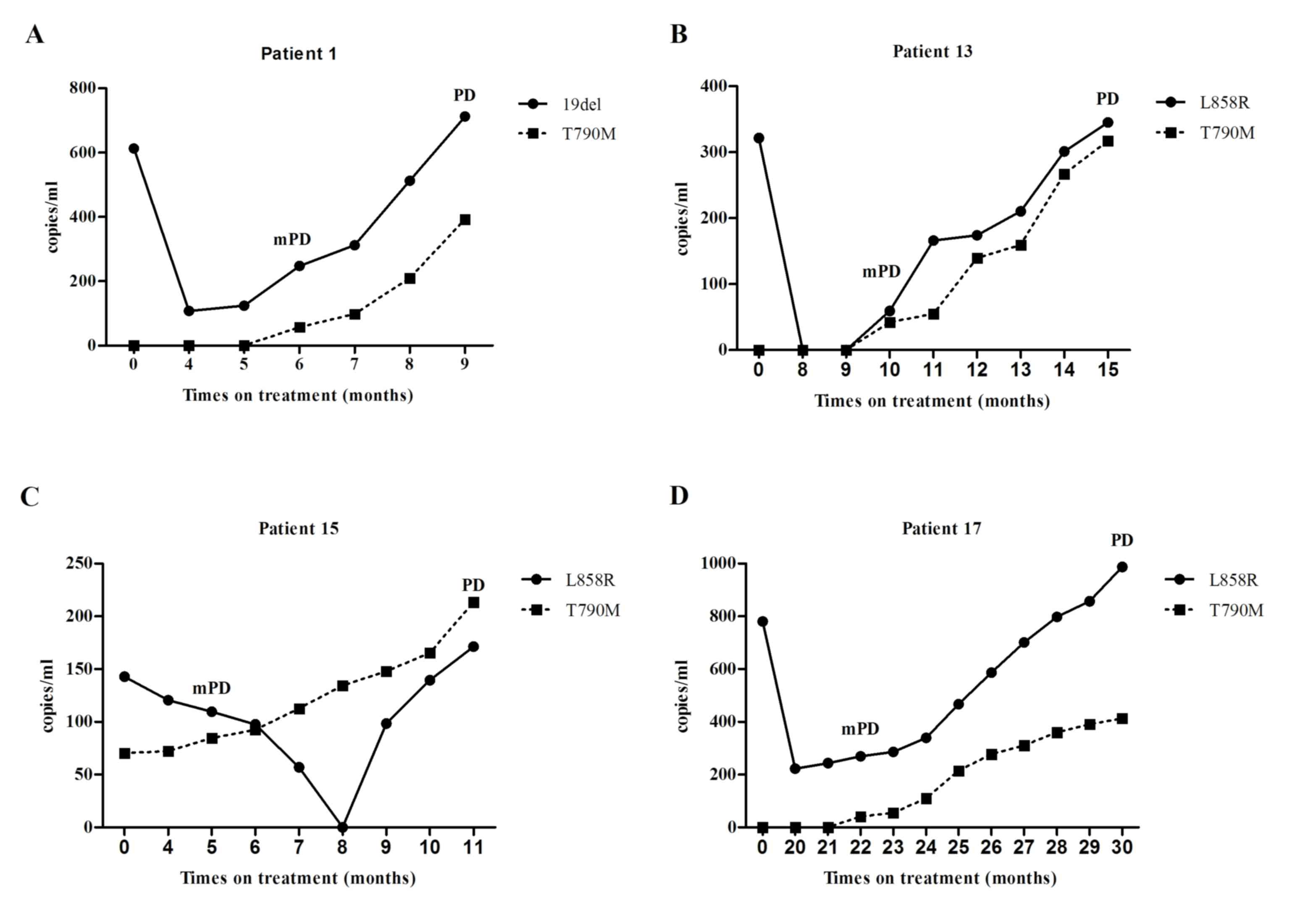 | Figure 3.Molecular progression disease, defined
as a ≥20% increase of epidermal growth factor receptor mutation
concentration compared with the lowest concentration recorded
during treatment, usually emerged several months earlier than the
documentation of objective progression disease defined by Response
Evaluation Criteria in Solid Tumors 1.1. (A) Patient 1 was
diagnosed with plasma-positive EGFR 19del mutation and
plasma-negative EGFR T790M mutation. On the 6th month of gefitinib
treatment, EGFR T790M mutation was detected (demonstrating mPD),
accompanied by the elevation of EGFR 19del mutation concentration
reaching mPD (the lowest concentration of EGFR 19del mutation was
recorded on the 4th month). Development of a new bone metastasis on
the 9th month, marked PD. (B) For patient 13, the concentration of
plasma EGFR L858R mutation reduced upon the initiation of gefitinib
treatment and it was undetectable on the 8th month. However, 2
months later, the activating EGFR mutation (L858R) was re-detected,
along with the emergence of EGFR T790M mutation, marking the
development of mPD. On the 15th month, a significant
re-accumulation of malignant pleural effusion demonstrated PD. (C)
Patient 15 was diagnosed with plasma-positive pretreatment EGFR
T790M mutation (pretreatment EGFR T790M mutation was undetectable
in tumor tissues, using Amplification Refractory Mutation System;
and there was inadequate tissue sample for further analysis using
droplet digital polymerase chain reaction). During continuous
gefitinib treatment, the concentration of EGFR T790M mutation
increased, reaching mPD on the 5th month. During the same time, the
concentration of the sensitizing EGFR mutation (L858R) was still
decreasing. On the 11th month, PD was documented when multiple
lesions in the left lung progressed. (D) Elevation of the
activating EGFR mutation (L858R) occurred prior to the emergence of
the resistant EGFR mutation (T790M) in patient 17, and mPD was
demonstrated on the 22th month of gefitinib treatment. A new lymph
node metastasis was noted on the 30th month, marking PD. mPD,
molecular progression disease; PD, progression disease. |
In addition, the dynamics of EGFR mutation may also
perform an important role in predicting disease progression under
second-line therapy. Specifically, in 5 T790M-positive patients,
sensitizing EGFR mutation and T790 M mutation concentration dropped
markedly upon the initiation of second-line treatment (local
radiation + gefitinib, gefitinib + chemotherapy or changing to
chemotherapy), but elevated or reoccurred prior to the second
objective progression (Fig. 2).
Prognostic significance of EGFR
mutations in cfDNA
In survival analysis, patient PFS was stratified
according to age, sex, activating EGFR mutation type (19del vs.
L858R), best treatment response to gefitinib (complete response,
partial response vs. stable disease), pretreatment tumor size,
pretreatment T790M mutation status (pre-T790M+ vs.
pre-T790M-), T790M mutation status at the time of disease
progression (T790M+ vs. T790M−), growth rate of tumor burden and
growth rate of EGFR mutation concentrations. Results showed that
sex (P=0.005), pretreatment T790M mutation status (P=0.006), T790M
mutation status at the time of disease progression (P=0.043) and
growth rate of EGFR mutation concentration (P=0.023), were
significantly associated with patient survival (Fig. 4).
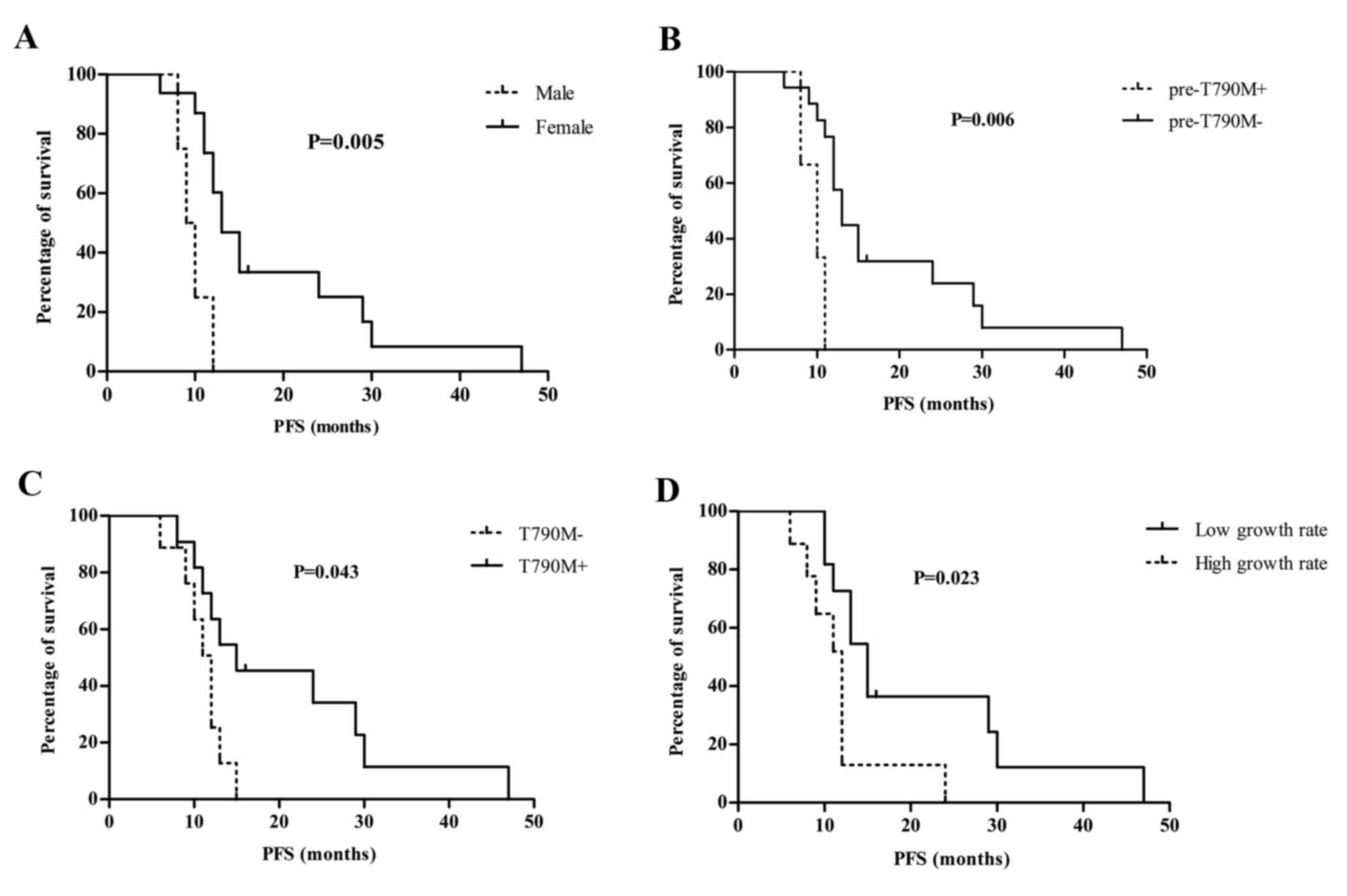 | Figure 4.Sex, T790M mutation status, growth
rate of EGFR mutations and survival. (A) Male patients had a
significantly shorter PFS compared with female counterparts (9.5
vs. 13.0 months, P=0.005; HR, 16.62; 95% CI, 2.308–119.7). (B)
Presence of pretreatment T790M mutation negatively affected patient
median PFS (10.0 vs. 13.0 months, P=0.006; HR, 29.38; 95% CI,
2.627–328.7). (C) Absence of T790M mutation at the time of disease
progression negatively affected patient median PFS (12 vs. 15
months, P=0.043; HR, 3.785; 95% CI, 1.134–12.62). (D) Patients with
a low growth rate (monthly growth rate <50%) of EGFR mutation
concentrations in cell-free DNA had a superior median PFS (15.0 vs.
12.0 months, P=0.023; HR, 0.251; 95% CI, 0.075–0.833) compared with
those having a high growth rate (monthly growth rate >50%). PFS,
progression-free survival; HR, hazard ratio; CI, confidence
interval; EGFR, epidermal growth factor receptor. |
When examining the prognostic significance of T790M
mutation, the results were conflicting. Presence of pretreatment
T790M mutation negatively affected patient median PFS [10.0 vs.
13.0 months; hazard ration (HR) =29.38; 95% confidence interval
(CI), 2.627–328.7], while patients with negative T790M mutation at
the time of disease progression had an inferior median PFS (12
months vs. 15 months; HR=3.785; 95% CI, 1.134–12.62).
In addition, growth rate of EGFR mutation, but not
growth rate of tumor burden, was associated with patient survival.
First, growth rates of tumor burden were calculated in 15 patients
with measurable disease and the median growth rate was 11% (range,
0–26%). No significant difference of median PFS stratified by the
growth rate of tumor burden was found, no matter which cut-off was
selected, including 11% (the median growth in the present study) or
20% (proposed in a previous study) (18). The growth rates of EGFR mutation were
calculated in all 20 patients and the median growth rate was 50%
(range, 0–101%). Using 50% as a cut-off value, patients with a
growth rate <50% had a superior median PFS (15.0 months vs. 12.0
months; HR=0.251; 95% CI, 0.075–0.833) compared with those with a
growth rate >50%.
Discussion
Drug resistance to first- and second-generation EGFR
TKIs is a major concern for patients with lung cancer harboring
activated EGFR mutations. With the emergence of third-generation
EGFR TKIs specifically targeting T790M mutation, the development of
noninvasive tools for cancer genotyping and disease monitoring is
required (7–8). In the present study, EGFR mutations were
successfully quantified in cfDNA from 168 monthly collected
samples, and their effectiveness in disease monitoring and
prognosis characterization was demonstrated. To the best of our
knowledge, this is the first prospective cohort that consecutively
quantified EGFR mutations in cfDNA for patients with metastatic
EGFR-mutant lung adenocarcinoma receiving gefitinib treatment in
China.
In line with previous studies (14–17,19,20),
the present study confirmed the feasibility and accuracy of
quantifying EGFR mutations in cfDNA and found it to be a promising
approach for early prediction of drug resistance. Corresponding
sensitizing EGFR mutations were correctly detected in 19 (95.0%) of
the 20 patients in the baseline samples, highlighting the
ultra-sensitivity of ddPCR (13,15,17,19–21).
At the time of disease progression, T790M mutations were detected
in 10 out of 18 (55.6%) patients, the frequency of which was
comparable to former studies using re-biopsy tissue samples
(6,22,23). In
addition, molecular progressive disease occurred up to 8 months
prior to objective progression, indicating the approach of drug
resistance. With the emergence of AZD9291 and CO1686, future
clinical trials are warranted to explore the best timing and
schedule of these novel EGFR-TKIs in cfDNA-based T790M-positive
patients.
Pretreatment T790M mutation or de novo T790M
mutation has been observed in tumor tissues using ultra-sensitive
detection platforms, with an incidence up to 79% (24–26). In
the present study, patients with negative T790M mutations in their
pretreatment tissues, confirmed by ARMS, were enrolled. However,
pretreatment T790M mutation in cfDNA was detected in 3 (15%) cases.
The different sensitivity between ARMS and ddPCR may explain this
disparity. The sensitivity of detecting EGFR mutations using ARMS
is ~1% (27), and thus, mutations
with frequencies <1% may end up with false-negative results. For
surgically resected tumor samples, pretreatment T790M mutation
commonly presented at a low ratio (<0.1%) (26). To determine the incidence and
abundance of de novo T790M in advanced patients with lung
cancer, quantifying pretreatment T790M mutation in tissue samples
and cfDNA using ultra-sensitive detection methods, including ddPCR,
is recommended.
In addition, the presence of T790M mutation in cfDNA
prior to the initiation of EGFR TKIs as a second-line therapy has
been significantly associated with a shorter PFS (28). Similarly, positive pretreatment T790M
mutation, in the present study, was found to negatively affect
patient PFS under first-line gefitinib therapy. These observations
raise questions regarding the best regimen for this subgroup of
patients. Whether third-generation EGFR TKIs, including AZD9291,
should be administered in the first-line or used until the
abundance of T790M mutation reaches a certain threshold, is now an
open question. Additional well-designed randomized controlled
trials are required to answer these questions.
In the current study, although the presence of
pretreatment T790M mutation negatively affected patient PFS,
presence of T790M mutation at the time of disease progression had
the opposite effect. There are multiple mechanisms underlying drug
resistance to EGFR TKIs, and numerous studies have identified that
presence of T790M mutation at time of disease progression
positively affects patient survival, regardless of whether T790M
mutation is detected in re-biopsy tissues or in cfDNA, or whether
the prognosis is presented as median PFS, median overall survival
or 5-year survival rate (21–24). One of the explanations may come from
the in vitro study that demonstrated EGFR-mutated
T790M-positive cells have a slower proliferation rate compared with
T790M-negative cells (29).
Circulating tumor DNA in the plasma may be passively
accumulated or actively released and may reflect the systemic tumor
burden and the overall tumor activity (30,31). The
dynamics of EGFR mutation concentrations were associated with
treatment response in almost every patient in the present study,
highlighting the potential role of cfDNA-based parameters in
disease monitoring. Notably, the dynamics of EGFR mutations were
consistent with disease evolution, even in patients with
immeasurable disease and patients with exceptional phenomena. In
these patients, estimating tumor burden using bimonthly
radiographic examinations may be misleading, and thus quantifying
EGFR mutations in cfDNA may be a valuable alternative or at least
adjuvant.
In addition to disease monitoring, parameters
derived from cfDNA-based mutation quantification may also have
prognostic significance. It has been well recognized that tumor
burden expressed as the number of metastatic sites, uptake of
18F-fluorodeoxyglucose measured by positron emission
tomography, or tumor volume analyzed using special software
associated well with patient survival (32–35).
However, the prognostic significance of genetic mutation
concentrations in cfDNA is largely unknown (21,34). In
the present study, it was discovered that a growth rate of EGFR
mutations >50% negatively affected patient median PFS, and this
observation requires validation in larger cohorts.
Acknowledgements
The authors thank Paul Horak from the Johns Hopkins
University School of Medicine and Professor Zhang Li from Sun
Yat-Sen University Cancer Center for reading the manuscript. The
present study was supported by Wu Jieping Medical Foundation (grant
no. 320.6750.14281).
References
|
1
|
Mok TS, Wu YL, Thongprasert S, Yang CH,
Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, et
al: Gefitinib or carboplatin-paclitaxel in pulmonary
adenocarcinoma. N Engl J Med. 361:947–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Maemondo M, Inoue A, Kobayashi K, Sugawara
S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I,
et al: Gefitinib or chemotherapy for non-small-cell lung cancer
with mutated EGFR. N Engl J Med. 362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mitsudomi T, Morita S, Yatabe Y, Negoro S,
Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, et
al: Gefitinib versus cisplatin plus docetaxel in patients with
non-small-cell lung cancer harbouring mutations of the epidermal
growth factor receptor (WJTOG3405): An open label, randomised phase
3 trial. Lancet Oncol. 11:121–128. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kobayashi S, Boggon TJ, Dayaram T, Jänne
PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG and Halmos
B: EGFR mutation and resistance of non-small-cell lung cancer to
gefitinib. N Engl J Med. 352:786–792. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maheswaran S, Sequist LV, Nagrath S, Ulkus
L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ,
Bell DW, et al: Detection of mutations in EGFR in circulating
lung-cancer cells. N Engl J Med. 359:366–377. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yu HA, Arcila ME, Rekhtman N, Sima CS,
Zakowski MF, Pao W, Kris MG, Miller VA, Ladanyi M and Riely GJ:
Analysis of tumor specimens at the time of acquired resistance to
EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers.
Clin Cancer Res. 19:2240–2247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jänne PA, Yang JC, Kim DW, Planchard D,
Ohe Y, Ramalingam SS, Ahn MJ, Kim SW, Su WC, Horn L, et al: AZD9291
in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J
Med. 372:1689–1699. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sequist LV, Soria JC, Goldman JW, Wakelee
HA, Gadgeel SM, Varga A, Papadimitrakopoulou V, Solomon BJ, Oxnard
GR, Dziadziuszko R, et al: Rociletinib in EGFR-mutated
non-small-cell lung cancer. N Engl J Med. 372:1700–1709. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Murtaza M, Dawson SJ, Tsui DW, Gale D,
Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong AS,
et al: Non-invasive analysis of acquired resistance to cancer
therapy by sequencing of plasma DNA. Nature. 497:108–112. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Del Re M, Vasile E, Falcone A, Danesi R
and Petrini I: Molecular analysis of cell-free circulating DNA for
the diagnosis of somatic mutations associated with resistance to
tyrosine kinase inhibitors in non-small-cell lung cancer. Expert
Rev Mol Diagn. 14:453–468. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tseng JS, Yang TY, Tsai CR, Chen KC, Hsu
KH, Tsai MH, Yu SL, Su KY, Chen JJ and Chang GC: Dynamic plasma
EGFR mutation status as a predictor of EGFR-TKI efficacy in
patients with EGFR-mutant lung adenocarcinoma. J Thorac Oncol.
10:603–610. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ottesen EA, Hong JW, Quake SR and
Leadbetter JR: Microfluidic digital PCR enables multigene analysis
of individual environmental bacteria. Science. 314:1464–1467. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hindson CM, Chevillet JR, Briggs HA,
Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL and Tewari M:
Absolute quantification by droplet digital PCR versus analog
real-time PCR. Nat Methods. 10:1003–1005. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yung TK, Chan KC, Mok TS, Tong J, To KF
and Lo YM: Single-molecule detection of epidermal growth factor
receptor mutations in plasma by microfluidics digital PCR in
non-small cell lung cancer patients. Clin Cancer Res. 15:2076–2084.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Oxnard GR, Paweletz CP, Kuang Y, Mach SL,
O'Connell A, Messineo MM, Luke JJ, Butaney M, Kirschmeier P,
Jackman DM and Jänne PA: Noninvasive detection of response and
resistance in EGFR-mutant lung cancer using quantitative
next-generation genotyping of cell-free plasma DNA. Clin Cancer
Res. 20:1698–1705. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Watanabe M, Kawaguchi T, Isa S, Ando M,
Tamiya A, Kubo A, Saka H, Takeo S, Adachi H, Tagawa T, et al:
Ultra-sensitive detection of the pretreatment EGFR T790M mutation
in non-small cell lung cancer patients with an EGFR-activating
mutation using droplet digital PCR. Clin Cancer Res. 21:3552–3560.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu G, Ye X, Dong Z, Lu YC, Sun Y, Liu Y,
McCormack R, Gu Y and Liu X: Highly sensitive droplet digital PCR
method for detection of EGFR activating mutations in plasma
cell-free DNA from patients with advanced non-small cell lung
cancer. J Mol Diagn. 17:265–272. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cha YK, Lee HY, Ahn MJ, Choi YL, Lee JH,
Park K and Lee KS: Survival outcome assessed according to tumor
burden and progression patterns in patients with epidermal growth
factor receptor mutant lung adenocarcinoma undergoing epidermal
growth factor receptor tyrosine kinase inhibitor therapy. Clin Lung
Cancer. 16:228–236. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sorensen BS, Wu L, Wei W, Tsai J, Weber B,
Nexo E and Meldgaard P: Monitoring of epidermal growth factor
receptor tyrosine kinase inhibitor-sensitizing and resistance
mutations in the plasma DNA of patients with advanced non-small
cell lung cancer during treatment with erlotinib. Cancer.
120:3896–3901. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ishii H, Azuma K, Sakai K, Kawahara A,
Yamada K, Tokito T, Okamoto I, Nishio K and Hoshino T: Digital PCR
analysis of plasma cell-free DNA for non-invasive detection of drug
resistance mechanisms in EGFR mutant NSCLC: Correlation with paired
tumor samples. Oncotarget. 6:30850–30858. 2015.PubMed/NCBI
|
|
21
|
Wang Z, Chen R, Wang S, Zhong J, Wu M,
Zhao J, Duan J, Zhuo M, An T, Wang Y, et al: Quantification and
dynamic monitoring of EGFR T790M in plasma cell-free DNA by digital
PCR for prognosis of EGFR-TKI treatment in advanced NSCLC. PLoS
One. 9:e1107802014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kuiper JL, Heideman DA, Thunnissen E, Paul
MA, van Wijk AW, Postmus PE and Smit EF: Incidence of T790M
mutation in (sequential) rebiopsies in EGFR-mutated NSCLC-patients.
Lung Cancer. 85:19–24. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hata A, Katakami N, Yoshioka H, Takeshita
J, Tanaka K, Nanjo S, Fujita S, Kaji R, Imai Y, Monden K, et al:
Rebiopsy of non-small cell lung cancer patients with acquired
resistance to epidermal growth factor receptor-tyrosine kinase
inhibitor: Comparison between T790M mutation-positive and
mutation-negative populations. Cancer. 119:4325–4332. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Su KY, Chen HY, Li KC, Kuo ML, Yang JC,
Chan WK, Ho BC, Chang GC, Shih JY, Yu SL and Yang PC: Pretreatment
epidermal growth factor receptor (EGFR) T790M mutation predicts
shorter EGFR tyrosine kinase inhibitor response duration in
patients with non-small-cell lung cancer. J Clin Oncol. 30:433–440.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fujita Y, Suda K, Kimura H, Matsumoto K,
Arao T, Nagai T, Saijo N, Yatabe Y, Mitsudomi T and Nishio K:
Highly sensitive detection of EGFR T790M mutation using colony
hybridization predicts favorable prognosis of patients with lung
cancer harboring activating EGFR mutation. J Thorac Oncol.
7:1640–1644. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Watanabe M, Kawaguchi T, Isa SI, Ando M,
Tamiya A, Kubo A, Saka H, Takeo S, Adachi H, Tagawa T, et al:
Ultra-sensitive detection of the pretreatment EGFR T790M mutation
in non-small-cell lung cancer patients with an EGFR-activating
mutation using droplet digital PCR. Clin Cancer Res. 21:3552–3560.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou Q, Zhang XC, Chen ZH, Yin XL, Yang
JJ, Xu CR, Yan HH, Chen HJ, Su J, Zhong WZ, et al: Relative
abundance of EGFR mutations predicts benefit from gefitinib
treatment for advanced non-small-cell lung cancer. J Clin Oncol.
29:3316–3321. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zheng D, Ye X, Zhang MZ, Sun Y, Wang JY,
Ni J, Zhang HP, Zhang L, Luo J, Zhang J, et al: Plasma EGFR T790M
ctDNA status is associated with clinical outcome in advanced NSCLC
patients with acquired EGFR-TKI resistance. Sci Rep. 6:209132016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chmielecki J, Foo J, Oxnard GR, Hutchinson
K, Ohashi K, Somwar R, Wang L, Amato KR, Arcila M, Sos ML, et al:
Optimization of dosing for EGFR-mutant non-small cell lung cancer
with evolutionary cancer modeling. Sci Transl Med. 3:90ra59. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Crowley E, Di Nicolantonio F, Loupakis F
and Bardelli A: Liquid biopsy: Monitoring cancer-genetics in the
blood. Nat Rev Clin Oncol. 10:472–484. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Haber DA and Velculescu VE: Blood-based
analyses of cancer: Circulating tumor cells and circulating tumor
DNA. Cancer Discov. 4:650–661. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Park JH, Kim TM, Keam B, Jeon YK, Lee SH,
Kim DW, Chung DH, Kim YT, Kim YW and Heo DS: Tumor burden is
predictive of survival in patients with non-small-cell lung cancer
and with activating epidermal growth factor receptor mutations who
receive gefitinib. Clin Lung Cancer. 14:383–389. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zander T, Scheffler M, Nogova L, Kobe C,
Engel-Riedel W, Hellmich M, Papachristou I, Toepelt K, Draube A,
Heukamp L, et al: Early prediction of nonprogression in advanced
non-small-cell lung cancer treated with erlotinib by using
[(18)F]fluorodeoxyglucose and [(18)F]fluorothymidine positron
emission tomography. J Clin Oncol. 29:1701–1708. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mok T, Wu YL, Lee JS, Yu CJ, Sriuranpong
V, Sandoval-Tan J, Ladrera G, Thongprasert S, Srimuninnimit V, Liao
M, et al: Detection and dynamic changes of EGFR mutations from
circulating tumor DNA as a predictor of survival outcomes in NSCLC
patients treated with first-line intercalated erlotinib and
chemotherapy. Clin Cancer Res. 21:3196–3203. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nishino M, Dahlberg SE, Cardarella S,
Jackman DM, Rabin MS, Ramaiya NH, Hatabu H, Jänne PA and Johnson
BE: Volumetric tumor growth in advanced non-small cell lung cancer
patients with EGFR mutations during EGFR-tyrosine kinase inhibitor
therapy: Developing criteria to continue therapy beyond RECIST
progression. Cancer. 119:3761–3768. 2013. View Article : Google Scholar : PubMed/NCBI
|