Introduction
Epidermal growth factor receptor tyrosine kinase
inhibitors (EGFR-TKI) have produced dramatic anti-cancer effects in
patients with non-small cell lung cancer (NSCLC) carrying
EGFR activating mutations (1–3). The first
generation of EGFR-TKIs, including gefitinib and elrotinib,
conferred significantly prolonged progression-free survival (PFS)
in these patients. The second generation of these drugs, afatinib,
brought a remarkable prolongation of overall survival, up to 33
months, in particular in patients with exon 19 deletions (4). In spite of the effectiveness of
EGFR-TKIs, patients eventually acquire resistance. Several
treatment strategies have been evaluated in clinical trials and
practice following the onset of acquired resistance. First, agents
targeted at molecules contributing to acquired resistance have been
considered. Based on mechanisms including the secondary EGFR
mutation, T790M, MET proto-oncogene, receptor tyrosine kinase (MET)
amplification, and hepatocyte growth factor (HGF) overexpression,
second and third generation EGFR-TKIs and MET inhibitors have been
developed (5–9). Afatinib confers a potent anti-cancer
effect against lung cancer cells harboring T790M, but a phase 2b/3
randomized trial revealed that the overall response rate and PFS of
patients with lung cancer who were previously treated with EGFR-TKI
were 7% and 3.3 months, respectively, which was not satisfactory
considering the results of preclinical studies (9). Since examination of biomarkers
associated with acquired resistance to EGFR-TKI was not performed
in that trial, it was speculated that the patients included those
with cancers possessing various mechanisms of acquired resistance
to EGFR-TKI. The T790M inhibitor AZD9291 has proceeded to clinical
trials and significant anti-cancer efficacy has been demonstrated
in T790M-positive lung cancer patients, with an overall response
rate and PFS of 61% and 9.6 months, respectively (10,11). MET
inhibitors including anti-MET antibody and MET-TKI require
predictive markers for the selection of the appropriate population
according to the results of clinical trials (12–14).
The second strategy for acquired resistance is
re-challenge with EGFR-TKI. Re-challenge with gefitinib or
erlotinib, often subsequent to cytotoxic treatment following
initial EGFR-TKI, may prolong survival for patients with advanced
lung cancer who previously achieved a positive response to
EGFR-TKI. A retrospective study revealed that OS was significantly
longer in the patients who underwent gefitinib re-challenge
compared with those who did not undergo re-challenge (15). When evaluation of the efficacy of
EGFR-TKI re-challenge was limited to patients who achieved good
control of disease with the first line of EGFR-TKI, disease control
rate and PFS were 56–73% and 3.4–5.6 months, respectively (16–21). On
the other hand, patients whose disease was not be controlled with
the first line of EGFR-TKI tended to exhibit poor efficacy with
re-challenge (19,20,22–24). From
these results, the effect of first EGFR-TKI may be considered a
predictive marker of efficacy of re-challenge, but specific
molecular markers associated with mechanisms of acquired resistance
have not been identified.
Examination of biomarkers is indispensable for
accurate assessment of anti-cancer effects. However, re-biopsy is
difficult because it is an invasive procedure for elderly patients
with lung cancer and poor lung function. In addition, tumor
biological characteristics change frequently, and monitoring of
genetic alterations is required to decide treatment (25–27).
Therefore, non-invasive liquid biopsy using peripheral blood, which
it is possible to repeatedly perform, is a preferable method for
monitoring biomarkers. Our group previously developed a fully
automated T790M monitoring system using circulating plasma DNA,
mutation-biased polymerase chain reaction (PCR) and quenched probe
(MBP-QP) system (28). As the system
utilizes peripheral blood, it is possible to examine T790M
repeatedly. Our previous retrospective study using the MBP-QP
system demonstrated that T790M was detected in 53–56% of patients
who acquired resistance to EGFR-TKI (28,29). In
addition, a prospective multicenter observational study was then
performed to determine whether T790M detection using MBP-QP system
with plasma DNA was useful for monitoring acquired resistance to
EGFR-TKI, and T790M was reproducibly detected in 40% of patients
whose disease became progressive (30). HGF levels in the plasma were also
determined, and an elevation of HGF of ≥1.5-fold was observed in
38% of the population (29). A
combination of T790M detection and HGF quantification using plasma
revealed that T790M and/or elevation of HGF were detected in 69% of
that population.
The present study investigated whether detection of
these molecular markers would be useful for determining the
appropriate population for re-challenge with EGFR-TKI. In order to
administer various EGFR-TKIs appropriately, it is important to
determine what clinical characteristics and biomarkers are
predictive of treatment efficacy.
Materials and methods
Patient selection
Plasma samples were obtained from 225 patients with
lung cancer treated at Saga University Hospital (Saga, Japan)
between January 2000 and October 2013. Among these patients, 60
were treated with EGFR-TKI and 12 adenocarcinoma patients underwent
a total of 16 re-challenges with EGFR-TKI (re-challenge was
performed on the same patients 1–3 times). The clinical features of
the patients that underwent re-challenge are listed in Table I. Plasma samples were repeatedly
collected throughout the course of treatment. Clinical stage of the
cancer was determined according to criteria in the 7th edition of
the International Union Against Cancer at the times plasma samples
were obtained (31). The study
protocol was approved by the Clinical Research Ethics Committees of
Saga University (Saga, Japan). All patients provided informed
consent for blood and tissue specimen collection and genomic
testing, according to the Declaration of Helsinki.
 | Table I.Patient characteristics. |
Table I.
Patient characteristics.
|
|
|
|
|
|
| Previous EGFR-TKI
treatments | Chemotherapy
between EGFR-TKI treatments |
|
|---|
|
|
|
|
|
|
|
|
|
|
|---|
| Patient no. | Re-challenge
no. | Age | Sex | SI | EGFR activating
mutation | Effect of
therapy | PFS (days) | Frequency | Optimal effect | TKI-free interval
(days) |
|---|
| 1 | 1 | 77 | F | 0 | exon19 | PR | 301 | 1 | PD | 101 |
| 2 | 2 | 44 | F | 100 | exon19 | PR | 348 | 2 | SD | 447 |
|
| 3 |
|
|
|
| PR | 78 | 0 | NA | 3 |
| 3 | 4 | 40 | M | 200 | L858R | PR | 401 | 0 | NA | 1 |
|
| 5 |
|
|
|
| SD | 81 | 2 | SD | 618 |
| 4 | 6 | 40 | M | 600 | exon19 | PR | 322 | 2 | SD | 244 |
| 5 | 7 | 64 | M | 800 | exon19 | SD | 493 | 2 | SD | 269 |
| 6 | 8 | 55 | M | 480 | exon19 | PR | 734 | 2 | PR | 396 |
| 7 | 9 | 56 | F | 0 | exon19 | SD | 509 | 1 | SD | 290 |
|
| 10 |
|
|
|
| SD | 77 | 1 | PR | 376 |
|
| 11 |
|
|
|
| SD | 149 | 2 | SD | 337 |
| 8 | 12 | 57 | F | 0 | L858R | PR | 361 | 2 | PR | 533 |
| 9 | 13 | 78 | F | 0 | L858R | PR | 259 | 1 | SD | 185 |
| 10 | 14 | 50 | F | 50 | exon19 | PR | 377 | 3 | SD | 337 |
| 11 | 15 | 58 | M | 900 | exon19 | PR | 420 | 1 | PR | 240 |
| 12 | 16 | 58 | M | 0 | L858R | PR | 269 | 1 | SD | 266 |
DNA extraction from plasma for
detection of EGFR mutations
Peripheral blood samples were collected into tubes
containing 3.8% citric acid. Plasma was immediately separated
centrifugation at 1750 × g at 4°C for 20 min. Supernatants were
collected and stored at −80°C until assays were performed. DNA was
isolated from 200 µl patient plasma using a QIAamp DNA mini kit
(Qiagen GmbH, Hilden, Germany) according to the manufacturer's
protocol. DNA was eluted with 50 µl ultrapure water, and 4 µl was
applied for detection of EGFR mutations as described in the
next section.
Detection of EGFR mutations
Exon 19 deletions and point mutations including
L858R and T790M were detected by the wild inhibiting PCR and
quenched probe (WIP-QP) system and the MBP-QP system, respectively.
These systems were fully automated using i-densy™ IS-5320
(ARKRAY Inc., Kyoto, Japan), as described previously (28,30,32).
Briefly, WIP-QP consisted of wild inhibiting PCR (WIP) and quenched
probe (QP) systems, as described previously (32). Wild inhibitor nucleic acid (WI) is
complementary to the wild type sequence corresponding to the
deletion part. WI suppresses amplification of the wild type
sequence by binding to the wild-type template but not the mutant,
resulting in preferential amplification of the mutant sequence. The
presence of deletions in amplified sequences was determined by
monitoring the fluorescence intensity of a TAMRA-conjugated,
guanine-specific quench fluorophore probe (QProbe, J-Bio21 Center,
Tokyo, Japan), which was complementary to the wild type sequence
containing the deletion part. Fluorescence intensity was measured
at different temperatures to identify wild-type and mutant
amplicons, and quantified as previously described (32). MBP-QP for the detection of L858R and
T790M consisted of mutation-biased PCR (MBP) and quenched probe
(QP) systems, with conditions used as previously described
(28,32). For MBP, the primers for wild-type and
mutant were mixed with genomic DNA, which results in high
specificity as each primer competitively hybridizes to wild type
and mutant sequences. In addition, the length of the reverse primer
for mutant was longer than that for wild-type and the annealing
temperature was designed to be optimum to mutant primer, resulting
in higher efficiency of amplification of the mutant sequence.
Detection was performed using the QP-system as with WIP-QP, as
previously described (28,32).
Quantification of HGF plasma
levels
HGF plasma levels were measured by enzyme-linked
immunosorbent assay (Immunis HGF EIA; product code 1EH1; B-Bridge
International, Inc., Mountain View, CA, USA; limit of detection,
100 pg/ml), as described previously (30). A total of 50 µl plasma was applied to
the assay system. All samples were assayed in duplicate. Color
intensity was measured at 450 nm with a spectrophotometric plate
reader (ARVO™ MX 1420 Multilabel Counter; PerkinElmer Inc.,
Waltham, MA, USA). HGF concentrations were determined by comparison
with standard curves.
Statistical analysis
The associations between the response to EGFR-TKI
re-challenge, plasma biomarkers and clinical characteristics were
tested using the Fisher's exact test for contingency tables and the
Mann-Whitney U test for continuous data. Statistical analyses were
conducted using IBM SPSS 22 software (IBM SPSS, Armonk, NY,
USA).
Results
Patient characteristics
Table I lists the
clinical characteristics of the patients in this study.
Re-challenges were performed 16 times on 12 patients in total: 3
times on 1 patient, 2 times on 2 patients, and 1 time on 9
patients, all diagnosed with adenocarcinoma. The ages of the
patients ranged from 40–77 years (median age 57 years), there were
6 females (50%) and 5 non-smokers (42%). EGFR activating
mutation was detected in the primary tumors of all patients: 8
patients (67%) had exon 19 deletions and 4 patients (33%) harbored
the L858R mutation. Prior to the 16 re-challenges, 11 patients
achieved partial response (PR) to EGFR-TKI treatment (69%) and 5
patients exhibited stable disease (SD; 31%; Table I). All patients had EGFR-TKI
discontinued due to disease progression. Cytotoxic chemotherapy
including platinum, pemetrexed, bevacizumab, and taxans was
administered in 1–3 regimens prior to re-challenge with EGFR-TKI on
14 occasions. PR and SD were seen as the optimal response in 4 and
9 instances, respectively (Table I).
The TKI-free interval prior to EGFR-TKI re-challenge ranged from 1
to 618 days. The median EGFR-TKI free interval was 333 days in
patients whose effect of EGFR-TKI re-challenge was PR or SD and 242
days in patients whose effect was PD (Tables I and II).
 | Table II.Comparison between effect of EGFR-TKI
re-challenge and clinical parameters including biomarkers
(n=16). |
Table II.
Comparison between effect of EGFR-TKI
re-challenge and clinical parameters including biomarkers
(n=16).
|
| Effect of EGFR-TKI
re-challenge |
|
|---|
|
|
|
|
|---|
| Parameter | PR+SD, n (%) | PD, n (%) |
P-valuea |
|---|
| Effect of previous
EGFR-TKI treatment |
|
| 0.500 |
| PR | 5 (45.5) | 6 (54.5) |
|
| SD | 3 (60.0) | 2 (40.0) |
|
| PFS of previous
EGFR-TKI treatment |
|
| 0.285 |
| ≥6
months | 7 (58.3) | 5 (41.7) |
|
| <6
months | 1 (25.0) | 3 (75.0) |
|
| Effect of
chemotherapy |
|
| 0.285 |
| PR | 3 (75.0) | 1 (25.0) |
|
| SD, PA,
NA | 5 (41.7) | 7 (58.3) |
|
| TKI-free
interval |
|
| 0.248 |
| Median
(day) | 333 | 242 |
|
| EGFR
activating mutation (plasma DNA) |
|
| 0.690 |
| + | 4 (50.0) | 4 (50.0) |
|
| − | 4 (50.0) | 4 (50.0) |
|
| History of T790M
(plasma DNA) |
|
| 0.141 |
| + | 7 (63.6) | 4 (36.4) |
|
| − | 1 (20.0) | 4 (80.0) |
|
| HGF
ratiob |
|
| 0.009 |
|
<1.5 | 6 (85.7) | 1 (14.3) |
|
|
≥1.5 | 1 (12.5) | 7 (87.5) |
|
| Combination of
T790M and HGF |
|
| 0.001 |
| Neither
T790M nor HGF | 7
(100.0) | 0 (0.0) |
|
|
T790Mc and/or HGF | 1 (11.1) | 8 (88.8) |
|
Biomarker analysis with plasma
samples
Plasma samples were collected repeatedly during the
treatment, including prior to and at the time of PD to previous
EGFR-TKI treatment as well as prior to re-challenge (Table III). Prior to the previous EGFR-TKI
treatment, T790M was not detected in any patients. Following PD,
plasma DNA T790M turned to positive in four re-challenges following
the previous EGFR-TKI. Among them, T790M disappeared in three
re-challenges following treatment with cytotoxic chemotherapy
following the previous EGFR-TKI. In 1 patient, T790M appeared
following cytotoxic chemotherapy concomitant with tumor
progression. T790M was continually observed from the time of PD
following the previous EGFR-TKI to prior to re-challenge in 1
patient. EGFR activating mutations were detected with plasma
DNA in all but 2 samples that were T790M positive.
 | Table III.Biomarkers and effects of EGFR-TKI
re-challenge. |
Table III.
Biomarkers and effects of EGFR-TKI
re-challenge.
|
| T790M |
|
|
|
|---|
|
|
|
|
|
|
|---|
|
| Previous EGFR-TKI
treatment | EGFR-TKI
re-challenge |
| EGFR-TK1
re-challenge |
|---|
|
|
|
|
|
|
|---|
| Re-challenge
no. | Prior to PD | At time of PD | Prior to PD | HGF
ratiob | Effect | PFS (days) |
|---|
| 1 | − | − | −a | 1.7 | PD | 22 |
| 2 | − | − | −a | 0.4 | PR | 78 |
| 3 | − | − | −a | 3.2 | PD | 34 |
| 4 | − | − | −a | 0.6 | SD | 81 |
| 5 | − | − | − | 1.9 | PD | 16 |
| 6 | − | + | − | 2.3 | PD | 26 |
| 7 | − | − | −a | 0.6 | SD | 60 |
| 8 | − | + | − | 0.5 | PR | 95 |
| 9 | NA | − | −a | NA | SD | 77 |
| 10 | − | − | − | 0.5 | SD | 149 |
| 11 | − | + | − | 1.5 | PD | 40 |
| 12 | − | − | − | 1.7 | SD | 138 |
| 13 | − | − | −a | 1.5 | PD | 25 |
| 14 | − | − | + | 1.7 | PD | 44 |
| 15 | − | + | + | 1.4 | PD | 18 |
| 16 | − | − | −a | 0.6 | SD | 95 |
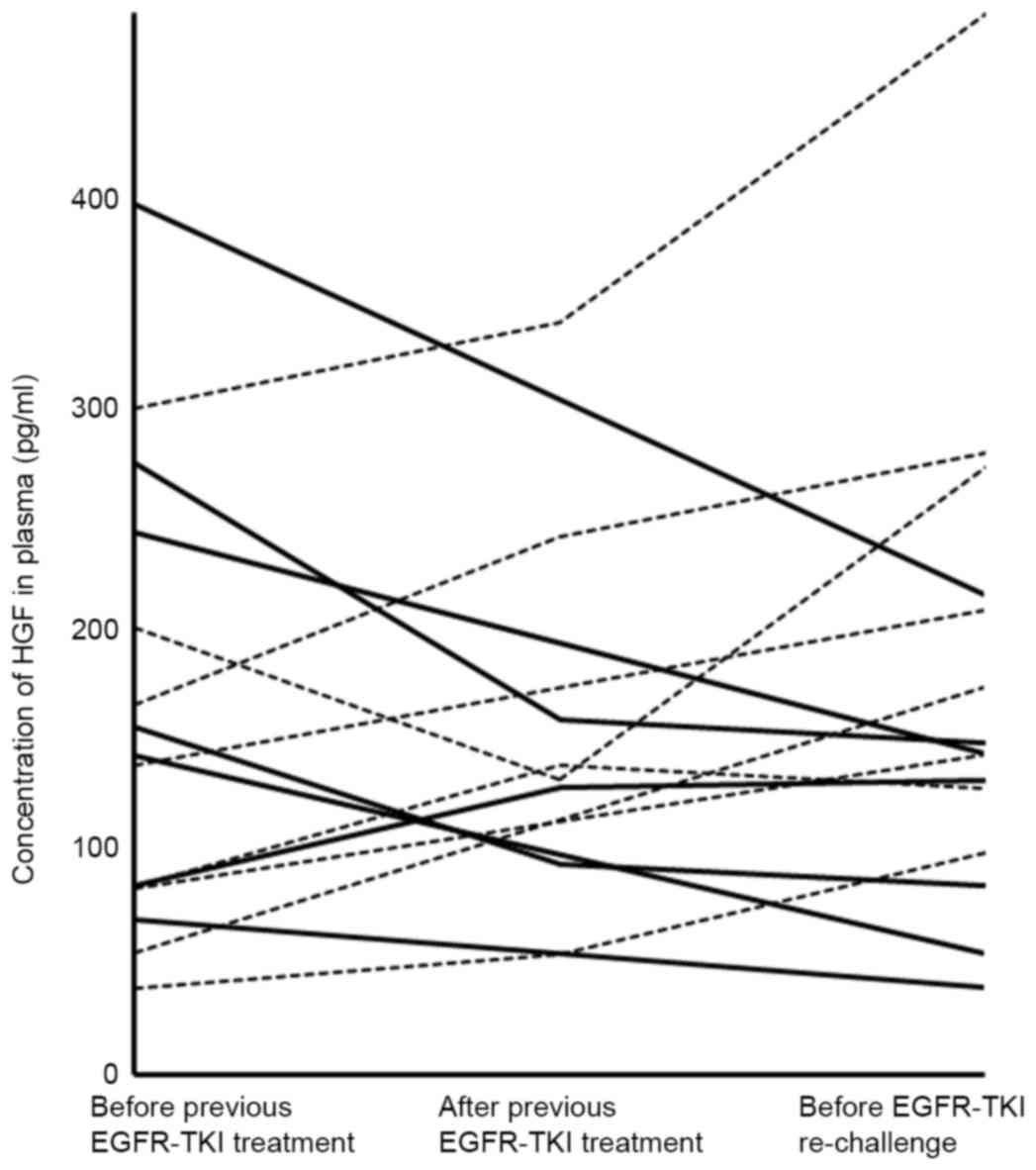
Plasma HGF levels ranged from 39–394 pg/ml prior to
previous EGFR-TKI and 39–680 pg/ml prior to re-challenge; with a
median of 144 and 147.5 pg/ml, respectively. The change of HGF
level in plasma prior to and following previous EGF-TKI and prior
to re-challenge were investigated (Fig.
1). HGF levels decreased in cases that experienced effective
re-challenge, whereas HGF levels were elevated in cases where
re-challenge was ineffective. The ratio of HGF level prior to
re-challenge to that prior to previous EGFR-TKI treatment ranged
from 0.4–3.2, and 8 patients had a ≥1.5-fold elevation of HGF
(Table III). When these ratios of
plasma HGF level were calculated in the 12 patients who remained
sensitive to EGFR-TKI treatment or who had EGFR-TKI discontinued
due to side effects, they ranged from 0.2–1.4.
Response to re-challenge with
EGFR-TKI
A total of 2 re-challenges demonstrated PR (12.5%)
and 6 remained SD (37.5%), disease control rate was 50%, and the
median PFS was 61 days (Table III).
A total of 7 out of 8 patients with PD to re-challenge demonstrated
a ≥1.5-fold elevation of the HGF ratio. Out of the 5 patients with
a history of T790M positivity in plasma DNA, 4 had PD. A total of 3
out of the 4 patients who achieved PR as the optimal response with
cytotoxic chemotherapy between EGFR-TKI treatments benefited from
EGFR-TKI re-challenge.
Clinical characteristics and plasma biomarkers
associated with the optimal response to EGFR-TKI re-challenge were
analyzed (Table II). The effect of
previous EGFR-TKI (optimal response as well as duration of
treatment), optimal response to chemotherapy, TKI-free interval and
detection of EGFR activating mutation in plasma all failed to
evidence a significant association with the effect of re-challenge.
An elevation of ≥1.5-fold in the HGF ratio was significantly
associated with poor response to EGFR-TKI re-challenge (P=0.009).
No history of T790M in the plasma and an elevation of the HGF ratio
<1.5 together were significantly associated with a positive
response to EGFR-TKI re-challenge (P=0.001).
A representative serial analysis of biomarkers for
patient 2 is depicted in Fig. 2. The
first treatment with the EGFR-TKI gefitinib demonstrated PR within
348 days of PFS. T790M and HGF elevation were not observed in
plasma collected prior to this. Two regimens of chemotherapy were
subsequently performed, and the outcomes were SD. Prior to the
first re-challenge with erlotinib, neither T790M nor elevation of
HGF ratio was observed, resulting in PR to the treatment. However,
the HGF ratio following the second re-challenge to the first
re-challenge was elevated 3.2 times. As a result, the second
re-challenge with gefitinib was ineffective.
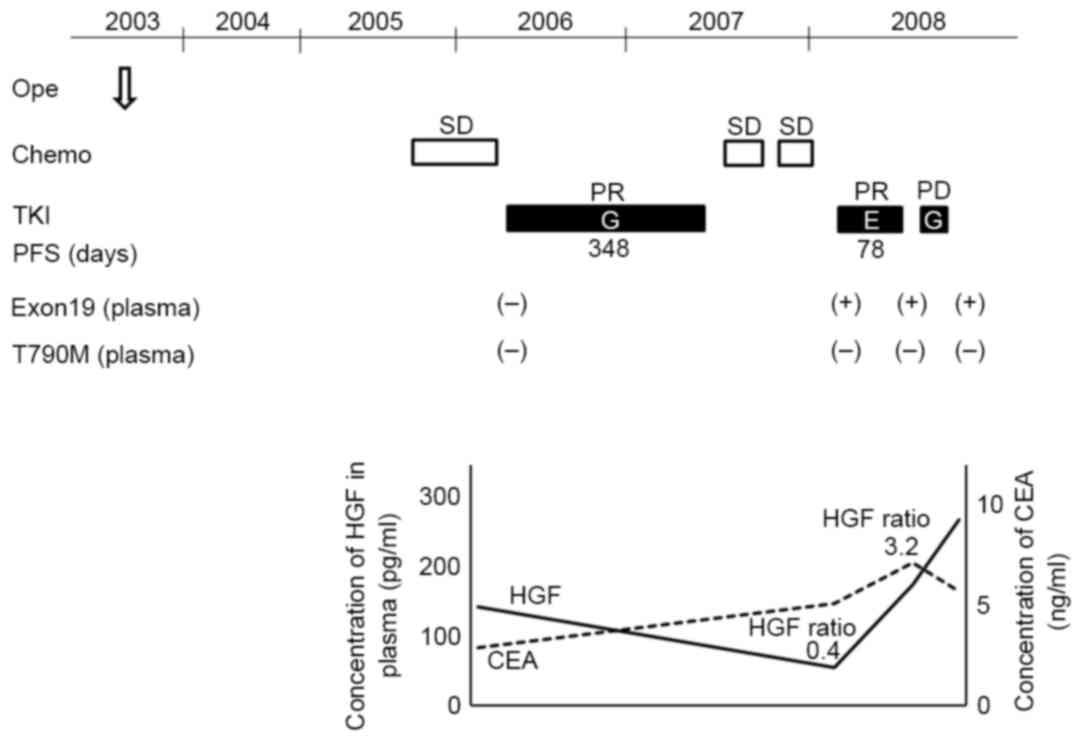 | Figure 2.Combination of exon 19 deletion with
T790M in plasma DNA, and HGF levels in plasma during treatment for
lung adenocarcinoma. Representative results of serial analysis of
the wild inhibiting PCR and quenched probe for exon 19 deletion,
mutation-biased PCR and quenched probe for T790M in plasma DNA and
ELISA for HGF from a lung adenocarcinoma patient is depicted. Plus
and minus indicate that epidermal growth factor receptor mutation
was detected or not detected, respectively. HGF, hepatocyte growth
factor; CEA, carcinoembryonic antigen; PCR, polymerase chain
reaction; ope, surgery; chemo, chemotherapy; TKI, epidermal growth
factor receptor-tyrosine kinase inhibitor; PR, partial response;
SD, stable disease; PD, progressive disease; PFS, progression free
survival; G, gefitinib; E, erlotinib. |
Discussion
As the biological characteristics of lung cancer may
alter during treatment, it is necessary to clarify the molecular
events in each individual at the time of acquired resistance to
EGFR-TKI and prior to re-challenge with EGFR-TKI for selection of
the appropriate patient population. However, analyses of biomarkers
at these times have not typically been performed due to difficulty
obtaining cancer specimens during disease progression, as the
majority of recurrences occur in distant sites including the brain,
bone, and intrapulmonary regions (33). In addition, lung cancer is
heterogeneous and the biological characteristics may vary even
among metastatic lesions within a patient. Furthermore, tumor
biological characteristics change frequently during treatment.
Therefore, a biopsy of one lesion may not reflect the mechanisms of
acquired resistance throughout the body (25,26,34–37).
To solve this problem, plasma was selected as the sample for
monitoring molecular events related with acquired resistance.
Plasma is suitable for repeated examinations to monitor acquired
resistance because collecting plasma is non-invasive, and it
appears that the molecular markers detected in plasma reflect the
main mechanism of acquired resistance of the entire body.
According to results obtained using re-biopsy,
EGFR T790M mutation and overexpression of HGF are major
mechanisms of acquired resistance to EGFR-TKI. T790M was detected
in 52–69% of cases and overexpression of HGF as assessed by
immunohistochemistry occurs in 61% of patients who acquire
resistance to EGFR-TKI (38,39). In total, 87% of patients present with
either T790M or overexpression of HGF, and 26% present with the two
combined (38,39). Therefore, T790M and HGF were selected
to be measured in the plasma, in addition to clinical parameters,
as candidate markers for predicting efficacy of re-challenge.
It has previously been reported that certain cases
achieved long-term disease control for >13 months with EGFR-TKI
re-challenge (40,41). Certain clinical characteristics,
including response or time to progression (TTP) to previous
EGFR-TKI, chemotherapy between EGFR-TKIs, and EGFR-TKI free
interval have been examined as predictive markers for the success
of EGFR-TKI re-challenge (16–21). The
association between the occurrence of disease control (PR or SD),
previous EGFR-TKI and to EGFR-TKI re-challenge has been described
frequently, but other markers are controversial. Furthermore, only
a small number of patients received biomarker analysis associated
with acquired resistance to EGFR-TKI prior to re-treatment with
EGFR-TKI.
The present study monitored EGFR activating
mutations, T790M and HGF with plasma samples from previous EGFR-TKI
treatment to re-challenge. When the transition of plasma HGF levels
was compared with the effect of re-challenge, the ratio of HGF
level was demonstrated to be useful as a predictive marker for the
efficacy of EGFR-TKI re-challenge. As the result of comparison
between acquired resistance patients and the other patients, it was
concluded that a ≥1.5-fold elevation of HGF was associated with
acquired resistance and inefficacy of EGFR-TKI re-challenge. The
relationship between plasma biomarkers and the optimal response to
EGFR-TKI re-challenge was analyzed, and the utility of using a
1.5-fold or greater elevation of HGF ratio as a negative predictive
factor was demonstrated. Neither history of T790M nor elevation of
HGF ratio were positive predictive factors. Considering the
previous results based on re-biopsy, which revealed that mechanisms
of acquired resistance including T790M and HGF overexpression may
overlap, the combined systems should be reasonable. It was not
possible to confirm concordance between plasma and re-biopsy.
However, a previous study has reported the concordance between
liquid biopsy (cell free plasma DNA and circulating tumor cell) and
concurrent re-biopsy was ~60% (42).
Considering this result, it is difficult to discuss the validity of
liquid biopsy in comparison with re-biopsy. Therefore, the
concordance between liquid biopsy and the effect of treatment is
important to establish the validity of liquid biopsy. The results
of the present study with T790M and HGF ratio using plasma are
reflective of the clinical outcomes. These results suggested that a
combination of T790M detection and HGF quantification using plasma
is useful for predicting EGFR-TKI re-challenge effectiveness.
The present study had certain imitations, being a
retrospective study with a small sample size. In addition, it was
not possible perform re-biopsy due to difficulty obtaining cancer
specimens at the point of PD or bad general condition of patients
with advanced stage cancer. Thus, it was not possible to examine
associations between T790M status and HGF expression between tissue
and liquid results. To demonstrate the usefulness of plasma T790M
and HGF monitoring, the next step will be a prospective study with
strict protocol to validate the HGF ratio in the plasma as well as
T790M with plasma DNA as predictive markers for efficacy of
re-challenge with EGFR-TKI. Eventually, more effective treatment
strategies for patients with NSCLC with EGFR activating
mutation will be developed, depending on the status of T790M and
HGF levels in the plasma. For example, second or the third
generation EGFR-TKI for detection of T790M, MET inhibitors for the
elevation of HGF levels, and EGFR-TKI re-challenge without
detection of T790M and elevation of HGF level.
Acknowledgements
The present study was supported in part by Grants-in
Aid for Cancer Research: Special Cancer Research, from the Ministry
of Education, Culture, Science, and Technology, Japan (grant no.
25870518) Mr. Toshiya Hosomi and Mr. Mitsuharu Hirai are employees
of ARKRAY, Inc., Kyoto, Japan.
References
|
1
|
Maemondo M, Inoue A, Kobayashi K, Sugawara
S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I,
et al: Gefitinib or chemotherapy for non-small-cell lung cancer
with mutated EGFR. N Engl J Med. 362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mitsudomi T, Morita S, Yatabe Y, Negoro S,
Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, et
al: Gefitinib versus cisplatin plus docetaxel in patients with
non-small-cell lung cancer harbouring mutations of the epidermal
growth factor receptor (WJTOG3405): An open label, randomised phase
3 trial. Lancet Oncol. 11:121–128. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mok TS, Wu YL, Thongprasert S, Yang CH,
Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, et
al: Gefitinib or carboplatin-paclitaxel in pulmonary
adenocarcinoma. N Engl J Med. 361:947–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang JC, Wu YL, Schuler M, Sebastian M,
Popat S, Yamamoto N, Zhou C, Hu CP, O'Byrne K, Feng J, et al:
Afatinib versus cisplatin-based chemotherapy for EGFR
mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6):
Analysis of overall survival data from two randomised, phase 3
trials. Lancet Oncol. 16:141–151. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kobayashi S, Boggon TJ, Dayaram T, Jänne
PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG and Halmos
B: EGFR mutation and resistance of non-small-cell lung cancer to
gefitinib. N Engl J Med. 352:786–792. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Engelman JA, Zejnullahu K, Mitsudomi T,
Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen
J, et al: MET amplification leads to gefitinib resistance in lung
cancer by activating ERBB3 signaling. Science. 316:1039–1043. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zakowski MF, Ladanyi M and Kris MG;
Memorial Sloan-Kettering Cancer Center Lung Cancer OncoGenome
Group, : EGFR mutations in small-cell lung cancers in patients who
have never smoked. N Engl J Med. 355:213–215. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yano S, Wang W, Li Q, Matsumoto K,
Sakurama H, Nakamura T, Ogino H, Kakiuchi S, Hanibuchi M, Nishioka
Y, et al: Hepatocyte growth factor induces gefitinib resistance of
lung adenocarcinoma with epidermal growth factor
receptor-activating mutations. Cancer Res. 68:9479–9487. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Miller VA, Hirsh V, Cadranel J, Chen YM,
Park K, Kim SW, Zhou C, Su WC, Wang M, Sun Y, et al: Afatinib
versus placebo for patients with advanced, metastatic
non-small-cell lung cancer after failure of erlotinib, gefitinib,
or both, and one or two lines of chemotherapy (LUX-Lung 1): A phase
2b/3 randomised trial. Lancet Oncol. 13:528–538. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cross DA, Ashton SE, Ghiorghiu S, Eberlein
C, Nebhan CA, Spitzler PJ, Orme JP, Finlay MR, Ward RA, Mellor MJ,
et al: AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated
resistance to EGFR inhibitors in lung cancer. Cancer Discov.
4:1046–1061. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jänne PA, Yang JC, Kim DW, Planchard D,
Ohe Y, Ramalingam SS, Ahn MJ, Kim SW, Su WC, Horn L, et al: AZD9291
in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J
Med. 372:1689–1699. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sequist LV, von Pawel J, Garmey EG,
Akerley WL, Brugger W, Ferrari D, Chen Y, Costa DB, Gerber DE,
Orlov S, et al: Randomized phase II study of erlotinib plus
tivantinib versus erlotinib plus placebo in previously treated
non-small-cell lung cancer. J Clin Oncol. 29:3307–3315. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Spigel DR, Ervin TJ, Ramlau RA, Daniel DB,
Blumenschein JH Jr, Goldschmidt GR Jr, Krzakowski MJ, Robinet G,
Godbert B, Barlesi F, et al: Randomized phase II trial of
Onartuzumab in combination with erlotinib in patients with advanced
non-small-cell lung cancer. J Clin Oncol. 31:4105–4114. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Robinson KW and Sandler AB: The role of
MET receptor tyrosine kinase in non-small cell lung cancer and
clinical development of targeted anti-MET agents. Oncologist.
18:115–122. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nishino K, Imamura F, Morita S, Mori M,
Komuta K, Kijima T, Namba Y, Kumagai T, Yamamoto S, Tachibana I, et
al: A retrospective analysis of 335 Japanese lung cancer patients
who responded to initial gefitinib treatment. Lung Cancer.
82:299–304. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Watanabe S, Tanaka J, Ota T, Kondo R,
Tanaka H, Kagamu H, Ichikawa K, Koshio J, Baba J, Miyabayashi T, et
al: Clinical responses to EGFR-tyrosine kinase inhibitor
retreatment in non-small cell lung cancer patients who benefited
from prior effective gefitinib therapy: A retrospective analysis.
BMC Cancer. 11:12011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Oh IJ, Ban HJ, Kim KS and Kim YC:
Retreatment of gefitinib in patients with non-small-cell lung
cancer who previously controlled to gefitinib: A single-arm,
open-label, phase II study. Lung Cancer. 77:121–127. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yokouchi H, Yamazaki K, Kinoshita I,
Konishi J, Asahina H, Sukoh N, Harada M, Akie K, Ogura S, Ishida T,
et al: Clinical benefit of readministration of gefitinib for
initial gefitinib-responders with non-small cell lung cancer. BMC
Cancer. 7:512007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wong AS, Soong R, Seah SB, Lim SW, Chuah
KL, Nga ME, Chin TM and Soo RA: Evidence for disease control with
erlotinib after gefitinib failure in typical gefitinib-sensitive
Asian patients with non-small cell lung cancer. J Thorac Oncol.
3:400–404. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wong MK, Lo AI, Lam B, Lam WK, Ip MS and
Ho JC: Erlotinib as salvage treatment after failure to first-line
gefitinib in non-small cell lung cancer. Cancer Chemother
Pharmacol. 65:1023–1028. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vasile E, Tibaldi C, Chella A and Falcone
A: Erlotinib after failure of gefitinib in patients with advanced
non-small cell lung cancer previously responding to gefitinib. J
Thorac Oncol. 3:912–914. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Costa DB, Nguyen KS, Cho BC, Sequist LV,
Jackman DM, Riely GJ, Yeap BY, Halmos B, Kim JH, Jänne PA, et al:
Effects of erlotinib in EGFR mutated non-small cell lung cancers
with resistance to gefitinib. Clin Cancer Res. 14:7060–7067. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cho BC, Im CK, Park MS, Kim SK, Chang J,
Park JP, Choi HJ, Kim YJ, Shin SJ, Sohn JH, et al: Phase II study
of erlotinib in advanced non-small-cell lung cancer after failure
of gefitinib. J Clin Oncol. 25:2528–2533. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee DH, Kim SW, Suh C, Yoon DH, Yi EJ and
Lee JS: Phase II study of erlotinib as a salvage treatment for
non-small-cell lung cancer patients after failure of gefitinib
treatment. Ann Oncol. 19:2039–2042. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sequist LV, Waltman BA, Dias-Santagata D,
Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger
S, Cosper AK, et al: Genotypic and histological evolution of lung
cancers acquiring resistance to EGFR inhibitors. Sci Transl Med.
3:75ra262011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kuiper JL, Heideman DA, Thunnissen E, Paul
MA, van Wijk AW, Postmus PE and Smit EF: Incidence of T790M
mutation in (sequential) rebiopsies in EGFR-mutated NSCLC-patients.
Lung Cancer. 85:19–24. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hata A, Katakami N, Yoshioka H, Kaji R,
Masago K, Fujita S, Imai Y, Nishiyama A, Ishida T, Nishimura Y and
Yatabe Y: Spatiotemporal T790M heterogeneity in individual patients
with EGFR-mutant non-small-cell lung cancer after acquired
resistance to EGFR-TKI. J Thorac Oncol. 10:1553–1559. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nakamura T, Sueoka-Aragane N, Iwanaga K,
Sato A, Komiya K, Abe T, Ureshino N, Hayashi S, Hosomi T, Hirai M,
et al: A noninvasive system for monitoring resistance to epidermal
growth factor receptor tyrosine kinase inhibitors with plasma DNA.
J Thorac Oncol. 6:1639–1648. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Umeguchi H, Sueoka-Aragane N, Kobayashi N,
Nakamura T, Sato A, Takeda Y, Hayashi S, Sueoka E and Kimura S:
Usefulness of plasma HGF level for monitoring acquired resistance
to EGFR tyrosine kinase inhibitors in non-small cell lung cancer.
Oncol Rep. 33:391–396. 2015.PubMed/NCBI
|
|
30
|
Sueoka-Aragane N, Katakami N, Satouchi M,
Yokota S, Aoe K, Iwanaga K, Otsuka K, Morita S, Kimura S, Negoro S,
et al: Monitoring EGFR T790M with plasma DNA from lung cancer
patients in a prospective observational study. Cancer Sci.
107:162–167. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Goldstraw P, Crowley J, Chansky K, Giroux
DJ, Groome PA, Rami-Porta R, Postmus PE, Rusch V and Sobin L;
International Association for the Study of Lung Cancer
International Staging Committee, : Participating Institutions: The
IASLC lung cancer staging project: Proposals for the revision of
the TNM stage groupings in the forthcoming (seventh) edition of the
TNM classification of malignant tumours. J Thorac Oncol. 2:706–714.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nakamura T, Sueoka-Aragane N, Iwanaga K,
Sato A, Komiya K, Kobayashi N, Hayashi S, Hosomi T, Hirai M, Sueoka
E and Kimura S: Application of a highly sensitive detection system
for epidermal growth factor receptor mutations in plasma DNA. J
Thorac Oncol. 7:1369–1381. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bach PB, Silvestri GA, Hanger M and Jett
JR: American College of Chest Physicians: Screening for lung
cancer: ACCP evidence-based clinical practice guidelines (2nd
edition). Chest. 132 Suppl 3:69S–77S. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Suda K, Murakami I, Katayama T, Tomizawa
K, Osada H, Sekido Y, Maehara Y, Yatabe Y and Mitsudomi T:
Reciprocal and complementary role of MET amplification and EGFR
T790M mutation in acquired resistance to kinase inhibitors in lung
cancer. Clin Cancer Res. 16:5489–5498. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Marusyk A and Polyak K: Tumor
heterogeneity: Causes and consequences. Biochim Biophys Acta.
1805:105–117. 2010.PubMed/NCBI
|
|
36
|
Gerlinger M, Rowan AJ, Horswell S, Larkin
J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A,
Tarpey P, et al: Intratumor heterogeneity and branched evolution
revealed by multiregion sequencing. N Engl J Med. 366:883–892.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Swanton C: Intratumor heterogeneity:
Evolution through space and time. Cancer Res. 72:4875–4882. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yano S, Yamada T, Takeuchi S, Tachibana K,
Minami Y, Yatabe Y, Mitsudomi T, Tanaka H, Kimura T, Kudoh S, et
al: Hepatocyte growth factor expression in EGFR mutant lung cancer
with intrinsic and acquired resistance to tyrosine kinase
inhibitors in a Japanese cohort. J Thorac Oncol. 6:2011–2017. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yu HA, Arcila ME, Rekhtman N, Sima CS,
Zakowski MF, Pao W, Kris MG, Miller VA, Ladanyi M and Riely GJ:
Analysis of tumor specimens at the time of acquired resistance to
EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers.
Clin Cancer Res. 19:2240–2247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chang JW, Chou CL, Huang SF, Wang HM,
Hsieh JJ, Hsu T and Cheung YC: Erlotinib response of EGFR-mutant
gefitinib-resistant non-small-cell lung cancer. Lung Cancer.
58:414–417. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gridelli C, Maione P, Galetta D,
Colantuoni G, Del Gaizo F, Ferrara C, Guerriero C, Nicolella D and
Rossi A: Three cases of long-lasting tumor control with erlotinib
after progression with gefitinib in advanced non-small cell lung
cancer. J Thorac Oncol. 2:758–761. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sundaresan TK, Sequist LV, Heymach JV,
Riely GJ, Jänne PA, Koch WH, Sullivan JP, Fox DB, Maher R,
Muzikansky A, et al: Detection of T790M, the acquired resistance
EGFR mutation, by tumor biopsy versus noninvasive blood-based
analyses. Clin cancer res. 22:1103–1110. 2016. View Article : Google Scholar : PubMed/NCBI
|