Introduction
Despite increasing knowledge about lung cancer and
its treatment modalities over the last few decades, this disease
continues to be responsible for the largest number of mortalities
in males and females worldwide (1).
Lung cancer was estimated to account for 27% of all
cancer-associated mortalities in 2014 (2). There are two major histological types of
lung cancer: Small cell lung cancer (SCLC) and non-SCLC (NSCLC)
(3). Standard treatments for NSCLC
include combinations of surgery, chemotherapy and radiotherapy
(4). Although most advanced-stage
patients receive chemotherapy and achieve clinical responses to a
certain extent, the majority eventually experience relapse
(5,6).
The 5-year survival rate of patients with NSCLC only marginally
improved from 15.9 in 2008 to 18.0% in 2014 (2,7,8). Therefore, novel therapies are required
to ensure improved management of NSCLC.
Immunotherapy is a novel therapeutic strategy that
is currently being evaluated for the treatment of NSCLC (4). Until recently, studies of vaccine
treatment for patients with lung cancer have not yielded very
positive results either due to non-specific immune system
activation or due to toxicity (9–13). The
lung, which has a high level of environmental exposure, has long
been considered a poorly immunogenic tumor site (14). Therefore, an effective vaccine against
NSCLC should contain antigens that are specific to the tumor cells
and have the ability to generate immunogenicity following
administration (15). Previously,
cDNA microarray and laser microdissection were used to identify
three genes for which transcripts were observed at high levels in
cells from lung cancer, esophageal cancer, testis and placenta, but
not in normal cells: Cell division-associated 1 (CDCA1) (16,17),
lymphocyte antigen 6 complex locus K (LY6K) (17,18) and
insulin-like growth factor-II mRNA-binding protein 3 (IMP-3)
(17,19) Accordingly, a vaccine comprising three
human leukocyte antigen (HLA)-A24-restricted epitope peptides
derived from these genes was developed for NSCLC and esophageal
cancer (20). A Phase I clinical
study of a combination of three peptides, including LY6K and IMP-3,
in patients with advanced esophageal squamous cell carcinoma
demonstrated that the vaccine was well tolerated and that strong
T-cell responses to these specific antigens were induced following
vaccination (21). Previously, a
Phase II clinical study of a combination of three peptides (LY6K,
IMP-3 and CDCA1) in patients with advanced head and neck squamous
cell carcinoma also reported peptide-specific cytotoxic T
lymphocyte (CTL) responses in the majority of the
HLA-A2402-positive patients (22). In
the present study, a Phase II clinical trial with exploratory
investigations was conducted using a vaccine comprising these three
peptides in patients with advanced NSCLC who were refractory to
standard therapies (15).
One important predictive biomarker of vaccine
therapy efficacy is the ability to induce an immunogenic response
against specific cancer cells (23).
Traditionally, enzyme-linked immune-spot and HLA-multimer assays
have been used to measure CTL responses, since a high level of CTL
infiltration into a tumor was reported to be associated with a good
response to treatment (24). However,
these assays require the ex vivo expansion of peripheral
blood lymphocytes (PBLs) via stimulation and are not practical in
clinical settings (25). In addition,
lung tumor biopsy to examine intratumoral CTL infiltration is
invasive (26). Therefore, it may be
preferable to identify biomarkers from easily available human
materials, including blood, using the predictive power that has
been demonstrated to associate well with overall survival (OS) rate
(27). The expansion and activation
of certain T-cell populations, including cytotoxic CD8+
T cells, has been reported to be beneficial for the recognition and
elimination of cancer cells (28,29). By
contrast, the expansion of regulatory T cells may be harmful, as
these T cells protect cancer cells by suppressing tumor-specific
CD8+ cytotoxic T cells (30–33).
Therefore, it is important to quantitatively characterize the
T-cell receptor (TCR) repertoires of patients with cancer prior to
and following immunotherapy, including cancer vaccine treatment, to
improve the understanding of the molecular mechanism underlying
treatment efficacy.
In total ~95% of T cells express TCR, which is a
hetero-dimer of the TCRα and TCRβ chains, a signature of each T
lymphocyte (34). To date, the TCRα
gene on chromosome 14 has been reported to comprise a total of 70
variable (V) exons, 61 joining (J) exons and 1 constant (C) exon,
whereas the TCRβ gene on chromosome 7 comprises 60 V exons, 15 J
exons and 2 C exons (35). In
addition, TCRβ contains two diversity (D) exons; accordingly, the
TCR genes undergo somatic V(D)J recombination, resulting in a
significant increase in TCR diversity (36–38). This
V(D)J segment rearrangement results in the highly variable
complementary-determining region (CDR3), thus allowing the
recognition of any possible antigens presented by HLA molecules
(39). It has been estimated that
~1018 different TCRs are generated in humans (25).
Advances in next-generation sequencing technology
have made it possible to sequence millions of TCR cDNAs, and thus
characterize patient TCR repertoires in a single experiment
(40–42). In the present study, it was
hypothesized that the expansion and activation of a large number of
T-cell populations, particularly the cytotoxic T-cell population,
may be used as predictive biomarkers in response to vaccine
treatment to assess the outcomes of patients with NSCLC who had
received cancer vaccine therapy. In the present study, T-cell
repertoires and certain immune-associated molecules in patients
with advanced NSCLC with an HLA-A*2402 who received the cancer
vaccine treatment were characterized using cDNA-sequencing
technology and a gene expression assay.
Materials and methods
Vaccines and patients
A total of 53 patients with advanced NSCLC resistant
to standard therapies were enrolled between 21 May 2012 and 4 April
2013 in a Phase II open-label multicenter non-randomized clinical
cancer vaccination trial conducted in an exploratory setting.
Patients were vaccinated with a mixture of 1 mg each of three
HLA-A24-restricted epitope peptides derived from CDCA1, LY6K and
IMP-3 mixed with incomplete Freund's adjuvant (Montanide ISA 51;
SEPPIC, Puteaux, France) (trial no. NCT01592617). The clinical
characteristics and treatment information for all 53 patients
enrolled in the clinical trial are summarized in Table I. Patients received weekly
subcutaneous injections of the peptides into the axillary region
until disease progression was observed or the patient declined
continued vaccine treatment. Written informed consent was obtained
from all individuals enrolled in the trial. The trials were carried
out in accordance with The Declaration of Helsinki on
experimentation on human subjects, under the approval of the
institutional ethics committees of the individual institutes. TCRβ
sequencing and gene expression analysis were performed for blood
samples from 24/35 HLA-A*2402-positive patients obtained.
 | Table I.Background of patients. |
Table I.
Background of patients.
|
Characteristics | Total, n | HLA-A*2402(+),
n | HLA-A*2402(−),
n |
|---|
| Total | 53 | 35 | 18 |
| Median
age ± SD, years | 64.0±7.7 | 64.1±7.5 | 63.9±8.2 |
| Sex |
|
|
|
|
Female | 21 | 16 | 5 |
|
Male | 32 | 19 | 13 |
| Location of primary
lesion |
|
|
|
|
Pulmonary hilum | 5 | 5 | 0 |
| Lung
field | 43 | 28 | 15 |
| Pleural
effusion | 1 | 0 | 1 |
|
Missing | 4 | 2 | 2 |
| Histological
type |
|
|
|
|
Adenocarcinoma | 43 | 28 | 15 |
|
Squamous cell carcinoma | 9 | 7 | 2 |
|
Pleomorphic carcinoma | 1 | 0 | 1 |
| T factor |
|
|
|
| TX | 19 | 9 | 10 |
| T0 | 2 | 1 | 1 |
|
T1a | 3 | 3 | 0 |
|
T1b | 1 | 1 | 0 |
|
T2a | 4 | 4 | 0 |
|
T2b | 1 | 0 | 1 |
| T3 | 7 | 6 | 1 |
| T4 | 16 | 11 | 5 |
| N factor |
|
|
|
| NX | 4 | 2 | 2 |
| N0 | 18 | 11 | 7 |
| N1 | 2 | 2 | 0 |
| N2 | 18 | 14 | 4 |
| N3 | 11 | 6 | 5 |
| M factor |
|
|
|
| MX | 0 | 0 | 0 |
| M0 | 9 | 8 | 1 |
|
M1a | 20 | 13 | 7 |
|
M1b | 17 | 9 | 8 |
|
M1a+1b | 7 | 5 | 2 |
| ECOG performance
status score |
|
|
|
| 0 | 38 | 25 | 13 |
| 1 | 11 | 8 | 3 |
| 2 | 4 | 2 | 2 |
| Smoking status |
|
|
|
| Current
or former smoker | 30 | 20 | 10 |
| Never
smoked | 23 | 15 | 8 |
| EGFR mutation
status |
|
|
|
|
Positive | 13 | 6 | 7 |
| Not
detected | 36 | 27 | 9 |
| Not
reported | 4 | 2 | 2 |
Peripheral blood mononuclear cell
collection
Blood samples were collected from patients prior to
and following 8 weeks of vaccine treatment. Blood was drawn into BD
Vacutainer® CPT™ cell preparation tubes containing
sodium citrate (CPT; BD Biosciences, Franklin Lakes, NJ, USA).
Samples were immediately centrifuged at 400 × g for 20 min at room
temperature. Peripheral blood mononuclear cells (PBMCs) were
collected from the second layer of the column and washed with PBS.
Cell numbers were determined using a hemocytometer, and cell
viability was assessed via trypan blue staining. Following
treatment with trypan blue, cells inside the four large corner
squares were counted at ×100 magnification under the light
microscope (CKX41; Olympus Corporation, Tokyo, Japan). A total of
~8×106 viable cells/sample were used for RNA
isolation.
RNA isolation and polymerase chain
reaction (PCR) amplification
An RNeasy mini kit (Qiagen, Inc., Valencia, CA, USA)
was used to isolate total RNA from PBMCs. In total 10 µl DNase I
(from the RNeasy mini kit) treatment was applied to remove
undesirable genomic DNA contamination. A SMART cDNA library
construction kit (Clontech Laboratories, Inc., Mountain View, CA,
USA) was used to synthesize cDNA with a common adaptor (SMART IV
oligonucleotide) at the 5′-ends. PCRs were performed to amplify
TCRβ cDNAs. All possible TCRβ combinations were captured using a
common forward primer
(5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTATCAACGCAGAGTGGCCAT-3′)
complementary to the SMART IV adaptor and a reverse primer specific
for the constant region
(5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGDVHDVTCTGATGGCTCAAACACAGC-3′).
The PCR protocol was as follows: 94°C for 3 min; followed by 30
cycles of 94°C for 30 sec, 65°C for 30 sec and 72°C for 1 min. Size
selection (between 300 and 950 base pairs) was conducted using
Pippin Prep (Sage Science, Beverly, MA, USA) to collect products
known to be of the expected size (43). This experiment was performed once with
the patient blood sample.
Library preparation and
sequencing
A Nextera XT DNA library kit (Illumina, Inc., San
Diego, CA, USA) was used to add adapter sequences onto template DNA
to generate multiplexed sequencing libraries, allowing the
sequencing and distinction of multiple samples in a single
experiment. The PCR protocol was as follows: 95°C for 3 min; 8
cycles of 95°C for 30 sec, 55°C for 30 sec and 72°C for 30 sec; and
a hold step at 72°C for 5 min. Multiple dual index-encoded samples
were combined in a single sequencing library. This library was
loaded onto the MiSeq Reagent kit (version 3; Illumina, Inc.) and
sequenced using an Illumina MiSeq Desktop Sequencer (Illumina,
Inc.).
TCR sequence analysis
FASTQ files containing sequencing reads were
generated using the MiSeq sequencer and mapped to the reference
sequences derived from IMGT/GENE-DB (www.imgt.org)
with the Bowtie2 alignment program (version 2.1.0; http://bowtie-bio.sourceforge.net/bowtie2/index.shtml)
(44). To determine CDR3 in TCRβ, a
conserved cysteine residue encoded in the 3′ end of the V segment
and a conserved phenylalanine residue encoded in the 5′ end of the
J segment, which signal CDR3, were identified as described
previously (25). Amino acid
sequences were determined using the nucleotide sequences between
the conserved TCR V cysteine residue and TCR J phenylalanine
residue.
Gene expression assay
First-strand cDNA products as aforementioned were
used in a gene expression assay to analyze the expression of CD4,
CD8 and granzyme A (GZMA) transcripts. A PCR TaqMan gene expression
assay was performed on an ABI ViiA™ 7 system (Applied Biosystems;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. All CD4, CD8 and GZMA transcript expression levels were
normalized to the transcript expression of the housekeeping gene
GAPDH.
Statistical analysis
OS rates were analyzed using the Kaplan-Meier
estimator method, and survival was measured in days between the
first vaccination and mortality. Progression-free survival (PFS)
was measured in days from the first vaccination to the date of
first documented disease progression or the date of mortality from
any cause. The statistical significance of the survival period was
analyzed using the Harrington-Fleming test.
The diversity indexes (DIs) of CDR3 sequences were
calculated using inverse Simpson's index formula as previously
reported (25). The DI reflects the
total number and also the evenness of the identified
clonotypes.
The Mann-Whitney test was used to compare the DIs
between the long-survival and short-survival groups. A paired
Student's t-test was performed to compare the DI prior to
and following vaccine treatment within each group. These
statistical tests were conducted using Prism software (version 6.0;
GraphPad, Inc., La Jolla, CA, USA). The median was used as a
threshold point to divide into two groups: High GZMA/CD8 and low
GZMA/CD8 or high CD8/CD4 and low CD8/CD4. A log-rank test was
performed using R software (version 3.2.0; The R Project for
Statistical Computing, Vienna, Austria) to compare the percentage
survival in these two groups. Data are presented as the mean ±
standard error. P<0.05 was considered to indicate a
statistically significant difference.
Results
Association between the TCRβ DI and
survival status of patients with NSCLC treated with cancer peptide
vaccines
Patients with NSCLC (35 HLA-A*2402-positive and 18
others) who had previously failed standard therapies were
recruited, as summarized in Table I.
The median OS time of these patients was 400 days, whereas the PFS
was 57 days. No significant difference in the clinical outcomes
between the HLA-A*2402-positive and -negative groups was
identified. None of the patients exhibited a complete response
according to the Response Evaluation Criteria in Solid Tumors
(45). The most common adverse events
of any grades observed during the present study are described in
Table II.
 | Table II.Treatment-associated adverse
effects. |
Table II.
Treatment-associated adverse
effects.
|
| HLA-A*2402(+)
(n=35) | HLA-A*2402(−)
(n=18) | Total (n=53) |
|---|
|
|
|
|
|
|---|
| Adverse events | Cases (incidence
rate), n (%) | Events, n | Cases (incidence
rate), n (%) | Events, n | Cases (incidence
rate), n (%) | Events, n |
|---|
| Total | 34 (97.1) | 159 | 16 (88.9) | 56 | 51 (94.3) | 215 |
| Infectious
disease | 13 (37.1) | 20 | 3 (16.7) | 4 | 16 (30.2) | 24 |
|
Nasopharyngitis | 6 (17.1) | 7 | 1 (5.6) | 1 | 7 (13.2) | 8 |
| Herpes
zoster | 2 (5.7) | 2 | 0 (0.0) | 0 | 2 (3.8) | 2 |
|
Periodontitis | 1 (2.9) | 1 | 1 (5.6) | 1 | 2 (3.8) | 2 |
|
Bronchitis | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Cellulitis | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Influenza | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Esophageal candidiasis | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Paronychia | 0 (0.0) | 0 | 1 (5.6) | 1 | 1 (1.9) | 1 |
|
Pneumonitis | 0 (0.0) | 0 | 1 (5.6) | 1 | 1 (1.9) | 1 |
|
Pulpitis | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Rhinitis | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
| Foot
tinea pedis | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Tonsillitis | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
| Urinary
tract infection | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
| Device
related infection | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
| Malignant
neoplasm | 9 (25.7) | 9 | 3 (16.7) | 4 | 12 (22.6) | 13 |
|
Malignant pleuritis | 2 (5.7) | 2 | 2 (11.1) | 2 | 4 (7.5) | 4 |
| Central
nervous system metastasis | 2 (5.7) | 2 | 0 (0.0) | 0 | 2 (3.8) | 2 |
|
NSCLC | 2 (5.7) | 2 | 0 (0.0) | 0 | 2 (3.8) | 2 |
|
Malignant ascites | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Esophageal cancer | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
| Tumor
infiltration of bone marrow | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Meningeal metastasis | 0 (0.0) | 0 | 1 (5.6) | 1 | 1 (1.9) | 1 |
| Brain
tumor | 0 (0.0) | 0 | 1 (5.6) | 1 | 1 (1.9) | 1 |
| Metabolic
disease | 2 (5.7) | 2 | 0 (0.0) | 0 | 2 (3.8) | 2 |
|
Hyperuricemia | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Hyperlipidemia | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
| Mental disease | 1 (2.9) | 1 | 1 (5.6) | 1 | 2 (3.8) | 2 |
|
Insomnia | 1 (2.9) | 1 | 1 (5.6) | 1 | 2 (3.8) | 2 |
| Nervous system
disorder | 6 (17.1) | 8 | 1 (5.6) | 1 | 7 (13.2) | 9 |
|
Headache | 3 (8.6) | 3 | 0 (0.0) | 0 | 3 (5.7) | 3 |
| Brain
compression | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Convulsion | 1 (2.9) | 2 | 0 (0.0) | 0 | 1 (1.9) | 2 |
|
Dizziness | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Hypoesthesia | 0 (0.0) | 0 | 1 (5.6) | 1 | 1 (1.9) | 1 |
|
Drowsiness | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
| Eye disease | 1 (2.9) | 1 | 1 (5.6) | 1 | 2 (3.8) | 2 |
|
Cataract | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
| Dry
eye | 0 (0.0) | 0 | 1 (5.6) | 1 | 1 (1.9) | 1 |
| Ear disease and
labyrinthine disturbance | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
| Rotary
vertigo | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
| Heart disease | 2 (5.7) | 4 | 0 (0.0) | 0 | 2 (3.8) | 4 |
|
Arrhythmia | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Pericardial effusion
collection | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Angina | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Supraventricular
tachycardia | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
| Angiopathy | 0 (0.0) | 0 | 1 (5.6) | 1 | 1 (1.9) | 1 |
|
Hypertension | 0 (0.0) | 0 | 1 (5.6) | 1 | 1 (1.9) | 1 |
| Respiratory
disease | 15 (42.9) | 18 | 4 (22.2) | 7 | 19 (35.8) | 25 |
| Upper
respiratory infection | 11 (31.4) | 13 | 2 (11.1) | 4 | 13 (24.5) | 17 |
|
Allergic rhinitis | 1 (2.9) | 1 | 1 (5.6) | 1 | 2 (3.8) | 2 |
|
Atelectasis | 0 (0.0) | 0 | 1 (5.6) | 1 | 1 (1.9) | 1 |
| Voice
disturbance | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Dyspnea | 0 (0.0) | 0 | 1 (5.6) | 1 | 1 (1.9) | 1 |
|
Pleuritis | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Sneezing | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Hypertrophic rhinitis | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
| Gastrointestinal
injury | 13 (37.1) | 15 | 5 (27.8) | 7 | 18 (34.0) | 22 |
|
Constipation | 5 (14.3) | 5 | 2 (11.1) | 2 | 7 (13.2) | 7 |
|
Diarrhea | 3 (8.6) | 3 | 0 (0.0) | 0 | 3 (5.7) | 3 |
|
Vomiting | 1 (2.9) | 1 | 2 (11.1) | 3 | 3 (5.7) | 4 |
|
Nausea | 2 (5.7) | 2 | 0 (0.0) | 0 | 2 (3.8) | 2 |
|
Abdominal discomfort | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Enteritis | 0 (0.0) | 0 | 1 (5.6) | 1 | 1 (1.9) | 1 |
|
Gastritis | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Hemorrhoid | 0 (0.0) | 0 | 1 (5.6) | 1 | 1 (1.9) | 1 |
|
Esophageal stenosis | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Stomatitis | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
| Biliary system
disorders | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Cholecystitis | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
| Skin disease | 7 (20.0) | 9 | 1 (5.6) | 1 | 8 (15.1) | 10 |
|
Rash | 2 (5.7) | 2 | 0 (0.0) | 0 | 2 (3.8) | 2 |
|
Alopecia | 0 (0.0) | 0 | 1 (5.6) | 1 | 1 (1.9) | 1 |
| Xerosis
cutis | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Asteatotic eczema | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Erythema | 1 (2.9) | 3 | 0 (0.0) | 0 | 1 (1.9) | 3 |
| Skeletal muscle or
soft tissue disorder | 5 (14.3) | 5 | 1 (5.6) | 2 | 6 (11.3) | 7 |
| Back
pain | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Fasciitis | 0 (0.0) | 0 | 1 (5.6) | 2 | 1 (1.9) | 2 |
|
Melalgia | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Periarthritis | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Spondylosis deformans | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Synovial cyst | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
| Kidney or urinary
disorder | 0 (0.0) | 0 | 1 (5.6) | 1 | 1 (1.9) | 1 |
|
Strangury | 0 (0.0) | 0 | 1 (5.6) | 1 | 1 (1.9) | 1 |
| Body or injection
site disorder | 34 (97.1) | 51 | 14 (77.8) | 22 | 48 (90.6) | 73 |
|
Injection site reaction | 33 (94.3) | 41 | 13 (72.2) | 20 | 46 (86.8) | 61 |
| High
fever | 2 (5.7) | 5 | 1 (5.6) | 1 | 3 (5.7) | 6 |
| Disease
progression | 2 (5.7) | 2 | 1 (5.6) | 1 | 3 (5.7) | 3 |
| General
fatigue | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Injection site bleeding | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Pain | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
| Laboratory
examination | 7 (20.0) | 9 | 2 (11.1) | 3 | 9 (17.0) | 12 |
|
Elevated GTPγ | 2 (5.7) | 2 | 1 (5.6) | 1 | 3 (5.7) | 3 |
|
Lymphocytopenia | 2 (5.7) | 2 | 0 (0.0) | 0 | 2 (3.8) | 2 |
|
Elevated hepatic
transaminase | 0 (0.0) | 0 | 1 (5.6) | 1 | 1 (1.9) | 1 |
|
Elevated CPK | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Elevated serum potassium
level | 0 (0.0) | 0 | 1 (5.6) | 1 | 1 (1.9) | 1 |
|
Elevated hepatic
transaminase | 0 (0.0) | 0 | 1 (5.6) | 1 | 1 (1.9) | 1 |
|
Increases in urine glucose
levels | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Abnormal liver function
test | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Albuminuria | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
| Weight
loss | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
| Injury, toxicosis
and procedural complication | 5 (14.3) | 5 | 1 (5.6) | 1 | 6 (11.3) | 6 |
|
Fractured sacrum | 0 (0.0) | 0 | 1 (5.6) | 1 | 1 (1.9) | 1 |
|
Lacerated wound | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Ligament sprain | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Ecchymoma | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Bruise | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
|
Radiation dermatitis | 1 (2.9) | 1 | 0 (0.0) | 0 | 1 (1.9) | 1 |
Blood samples from 24 of 35 HLA-A*2402-positive
patients were obtained, 14 of which achieved stable disease and/or
remained alive >12 months after enrollment, and possible
predictive immune biomarkers, including TCRβ analysis, were
examined. The TCRβ DIs were compared between long-term survivors
(lived for >12 months after enrollment) and short-term survivors
(died within 12 months of enrollment) by calculating an inverse
Simpson's DI (1/D) of the TCRβ repertoire, as described in the
Materials and methods section. The present data revealed that prior
to vaccine treatment, patients in the long-term survival group
exhibited significantly lower TCRβ DIs compared with short-term
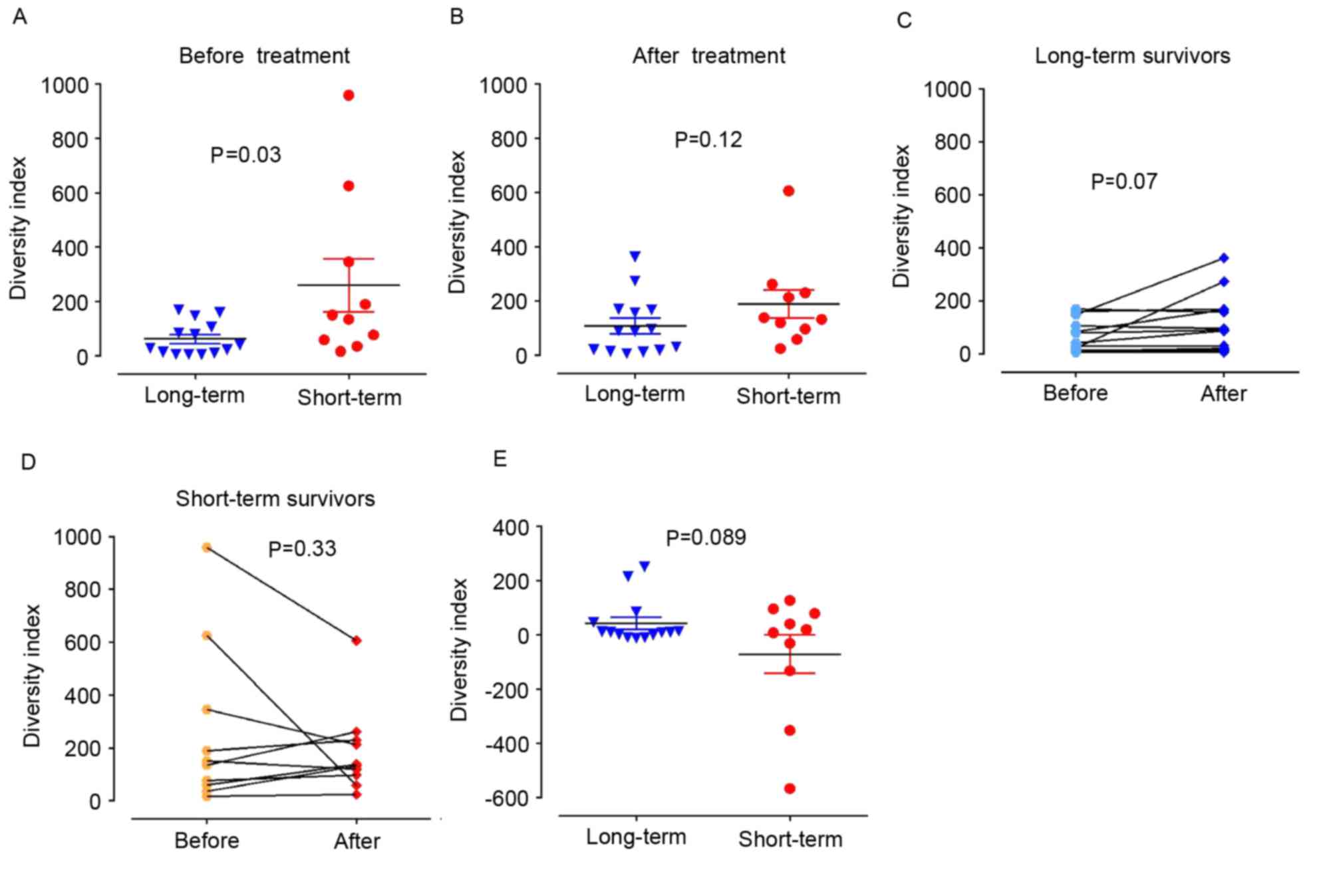
survivors (63±61 vs. 259±307; P=0.03; Fig. 1A). However, the difference between the
two groups was not significant in blood samples following 8 weeks
of treatment (107±108 vs. 188±165; P=0.12; Fig. 1B) although a trend in the increase in
DI was observed in the long-term survival group (mean of
differences, 44±7; P=0.07; Fig. 1C).
By contrast, no significant change in DI between samples prior to
and following the treatment was observed in the short-term survival
group (Fig. 1D). The difference in
the changes in DI in the long-term and short-term groups is
presented in Fig. 1E.
Association between the immunogenic
gene expression and survival status of patients with NSCLC treated
with cancer peptide vaccines
To assess alterations in T-cell populations in
response to treatment, TaqMan assays were performed to analyze the
relative expression of CD8, CD4 and GZMA prior to the vaccine
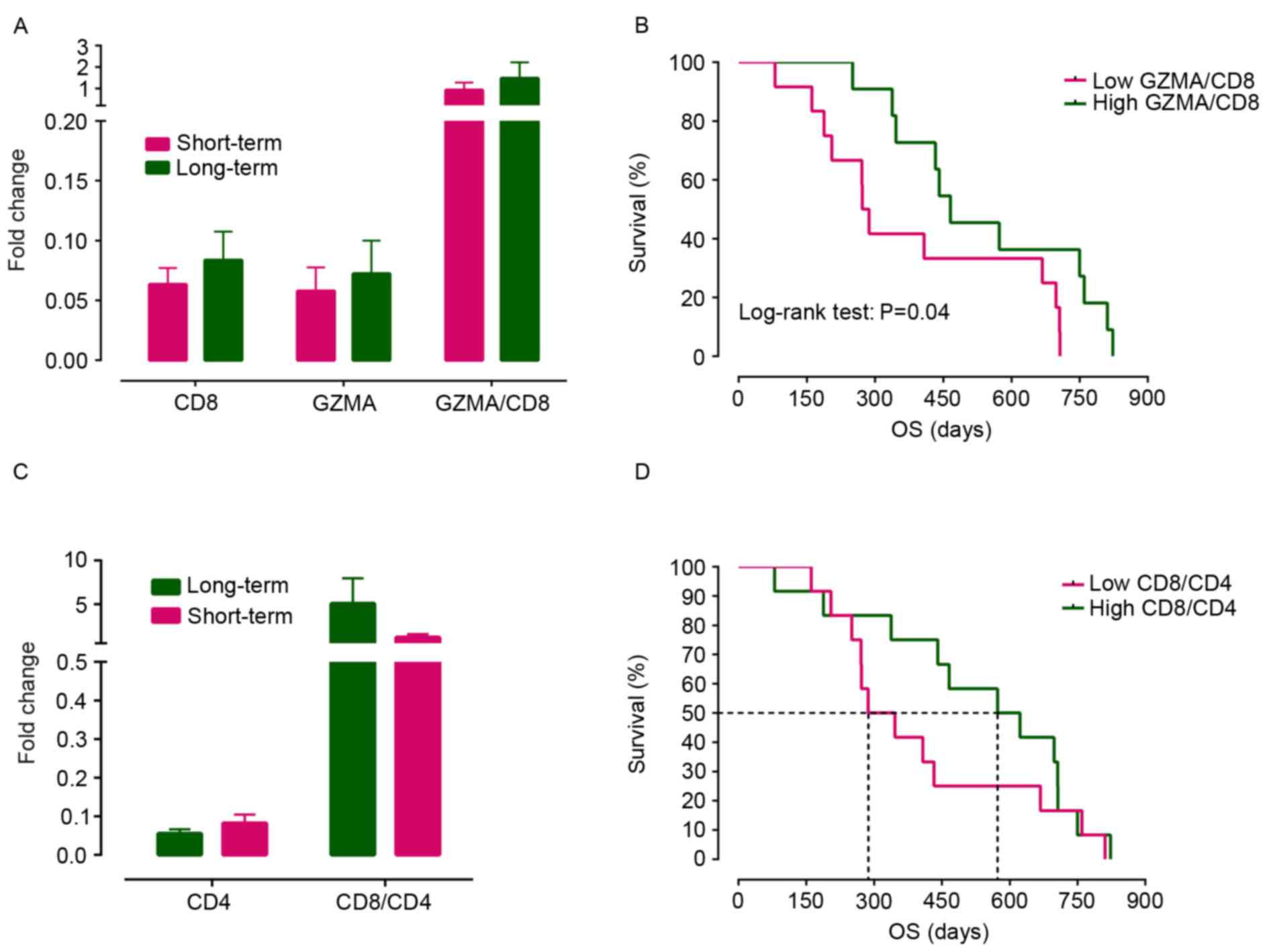
treatment. As presented in Fig. 2A,
no significant differences were identified for CD8 and GZMA
expression levels and the GZMA/CD8 ratio between the long-term and
short-term survival groups. However, when the patients were
separated into two groups by a median value of the GZMA/CD8 ratio,
which may reflect the level of activated cytotoxic T cells, the
survival curves were significantly different, favoring the group
with a high GZMA/CD8 ratio (log-rank test, P=0.04; Fig. 2B). In addition, a tendency was
observed for the CD8/CD4 ratio to be increased in the long-term
survival group compared with the short-term survival group (this
difference was not statistically significant, Fig. 2C). Notably, the median overall
survival time in the low CD8/CD4 group was only 316 days compared
with 598 days in the high CD8/CD4 group (Fig. 2D).
Discussion
Although numerous attempts have been made to develop
an effective peptide cancer vaccine therapy to treat NSCLC, the
results have not been positive (9,12,13). The present study aimed to screen
possible predictive biomarkers that were able to distinguish
responders from non-responders in patients with HLA-A24-positive
NSCLC who received a cancer peptide vaccine treatment of a mixture
of three peptides. Certain long-term survivors were observed among
these patients who had previously failed standard therapies.
Injection site reaction was the most common type of adverse event
possibly associated with treatment. One patient in whom a coronary
artery stent had been placed for angina succumbed to myocardial
infarction as confirmed by autopsy, indicating a non-deniable
causal association between treatment and this adverse event.
The immunogenic properties of this vaccine were
evaluated through an analysis of the TCRβ repertoire in blood
samples prior to and following 8 weeks of treatment. The main
results included the following: i) A lower DI prior to vaccine
treatment appeared to be beneficial, as it was associated with an
improved survival status; and ii) a higher ratio of activated
cytotoxic T cell at baseline, as indicated by GZMA/CD8 prior to
treatment, appeared to be a favorable clinical outcome. A previous
study demonstrated that cancer vaccination may cause an increase in
circulating tumor antigen-specific T cells (46). However, the profiles of these T cells
have not been well characterized with regard to their association
with treatment outcomes. The results of the present study
demonstrated that patients with a lower baseline DI tended to have
a longer survival time. Notably, patients enrolled in the trial had
advanced NSCLC refractory to standard chemotherapies. Prior
treatment with cytotoxic chemotherapies may lead to generation of
cancer-specific antigens that possibly generate immunological
effects, although intensive chemotherapy may also kill these CTL
clones (47). It was speculated that
the residual immunogenic effects of prior chemotherapies led to the
polyclonal expansion of T-cell clones and that this expansion,
reflected as a lower DI of the TCRβ repertoire, contributed to the
positive effect of the vaccine. Therefore, the pretreatment TCRβ DI
may be used as a predictive marker of the ability of a patient to
generate an immune response against either chemotherapeutic or
vaccine treatment. Peptide vaccination was reported to boost a
preexisting dominant clonotypic response (48). Therefore, a suggestion was made to
administer vaccine therapy concomitantly with chemotherapy to
generate the most effective immunological effects (49). A trend towards an increased TCRβ DI
was observed in long-term survivors but not in short-term
survivors, supporting earlier evidence that TCRβ repertoire
diversification due to chemotherapy and vaccination is beneficial
in the prevention of immune-resistant mutant cancer cells, since
more cancer-specific T-cell clones; particularly cytotoxic T cells
are generated to target cancer cells (49). This phenomenon may be explained as a
secondary immune response to vaccine treatment. As the immune
system (either cytotoxic T cells or macrophages) eliminated greater
numbers of cancer cells, phagocytosis of these cells may result in
the presentation of cancer-specific antigens by antigen-presenting
cells. The results of the present study were consistent with those
of a study by Fang et al (25), in which patients with NSCLC were
demonstrated to benefit from chemotherapy prior to peptide
vaccination due to an increase in the TCR repertoire diversity.
Therefore, the TCRβ DI prior to treatment and the increase in the
DI of PBLs following treatment may be used to monitor the responses
of patients with NSCLC to the peptide vaccine.
A TaqMan gene expression assay was performed to
quantify the relative presence of these two populations in the
patient samples in the present study. GZMA has been proposed as a
biomarker of activated cytolytic T lymphocytes (50). A high level of GZMA expression
indicates a high level of inflammatory cells in allograft,
autoimmune diabetes and chronic Chagas' myocardial lesions
(51,52). Therefore, GZMA/CD8 ratios were
calculated to study the activated subset of cytotoxic CD8 cells.
Although no difference was observed in the expression of CD8 or
GZMA between the long-term and short-term survivor groups, the
results indicated that an increased level of GZMA/CD8 ratio was
beneficial for the survival status. Although the difference was not
statistically significant, the relative ratio of
CD8+/CD4+ was increased prior to treatment in
the long-term survival group. A limitation of the present study was
that the limited number of available T lymphocytes did not allow
for T-cell sorting prior to sequencing. Therefore, future studies
are required to improve the characterization of the specific TCR
repertoires of different T-cell populations (CD8+,
CD4+CD25+ and
CD4+CD25−), and thus elucidate the specific
immunogenic effect of the three-peptide cocktail vaccine.
In conclusion, TCRβ DI and the immunogenic markers,
including GZMA, may serve as predictive biomarkers for successful
cancer vaccine treatment. Limitations of the present study included
the limited number of patients and the lack of placebo control arm.
Future prospective studies, in which these markers are used to
predict the outcomes of vaccine treatment, are warranted.
Acknowledgements
The present study was partly supported by a research
grant from the Japanese Ministry of Health Labor and Welfare.
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small-cell lung cancer
|
|
TCR
|
T-cell receptor
|
|
HLA
|
human leukocyte antigen
|
|
CDR3
|
complementary-determining region 3
|
|
PBMC
|
peripheral blood mononuclear cell
|
|
PBL
|
peripheral blood lymphocyte
|
|
DI
|
diversity index
|
References
|
1
|
Global status report on noncommunicable
diseases 2014. World Health Organization; Geneva: pp. 11–14.
2014
|
|
2
|
Cancer Facts & Figures 2014. American
Cancer Society; Atlanta, GA: pp. 14–15. 2014
|
|
3
|
Travis WD, Brambilla E, Nicholson AG,
Yatabe Y, Austin JH, Beasley MB, Chirieac LR, Dacic S, Duhig E,
Flieder DB, et al: The 2015 World Health Organization
Classification of Lung Tumors: Impact of Genetic, Clinical and
Radiologic Advances Since the 2004 Classification. J Thorac Oncol.
10:1243–1260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Santana-Davila R and Martins R: Treatment
of stage IIIA non-small-cell lung cancer: A concise review for the
practicing oncologist. J Oncol Pract. 12:601–606. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Katayama R, Shaw AT, Khan TM,
Mino-Kenudson M, Solomon BJ, Halmos B, Jessop NA, Wain JC, Yeo AT,
Benes C, et al: Mechanisms of acquired crizotinib resistance in
ALK-rearranged lung Cancers. Sci Transl Med. 4:120ra172012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kobayashi S, Boggon TJ, Dayaram T, Jänne
PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG and Halmos
B: EGFR mutation and resistance of non-small-cell lung cancer to
gefitinib. N Engl J Med. 352:786–792. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ettinger DS, Akerley W, Borghaei H, Chang
AC, Cheney RT, Chirieac LR, D'Amico TA, Demmy TL, Govindan R,
Grannis FW Jr, et al: Non-small cell lung cancer, version 2.2013. J
Natl Compr Canc Netw. 11:645–653. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: Epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:pp.
584–594. 2008; View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Butts C, Socinski MA, Mitchell PL,
Thatcher N, Havel L, Krzakowski M, Nawrocki S, Ciuleanu TE, Bosquée
L, Trigo JM, et al: Tecemotide (L-BLP25) versus placebo after
chemoradiotherapy for stage III non-small-cell lung cancer (START):
A randomized, double-blind, phase 3 trial. Lancet Oncol. 15:59–68.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dessureault S, Noyes D, Lee D, Dunn M,
Janssen W, Cantor A, Sotomayor E, Messina J and Antonia SJ: A
phase-I trial using a universal GM-CSF-producing and
CD40L-expressing bystander cell line (GM.CD40L) in the formulation
of autologous tumor cell-based vaccines for cancer patients with
stage IV disease. Ann Surg Oncol. 14:869–884. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nemunaitis J, Nemunaitis M, Senzer N,
Snitz P, Bedell C, Kumar P, Pappen B, Maples PB, Shawler D and
Fakhrai H: Phase II trial of Belagenpumatucel-L, a TGF-beta2
antisense gene modified allogeneic tumor vaccine in advanced non
small cell lung cancer (NSCLC) patients. Cancer Gene Ther.
16:620–624. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Palmer M, Parker J, Modi S, Butts C,
Smylie M, Meikle A, Kehoe M, MacLean G and Longenecker M: Phase I
study of the BLP25 (MUC1 peptide) liposomal vaccine for active
specific immunotherapy in stage IIIB/IV non-small-cell lung cancer.
Clin Lung Cancer. 3:49–58. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Quoix E, Ramlau R, Westeel V, Papai Z,
Madroszyk A, Riviere A, Koralewski P, Breton JL, Stoelben E, Braun
D, et al: Therapeutic vaccination with TG4010 and first-line
chemotherapy in advanced non-small-cell lung cancer: A controlled
phase 2B trial. Lancet Oncol. 12:1125–1133. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang N, Zhong WZ and Wu YL: Special issue
on personalized therapy in lung cancer. Transl Lung Cancer Res.
3:358–359. 2014.PubMed/NCBI
|
|
15
|
Daigo Y, Takano A, Teramoto K, Chung S and
Nakamura Y: A systematic approach to the development of novel
therapeutics for lung cancer using genomic analyses. Clin Pharmacol
Ther. 94:218–223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hayama S, Daigo Y, Kato T, Ishikawa N,
Yamabuki T, Miyamoto M, Ito T, Tsuchiya E, Kondo S and Nakamura Y:
Activation of CDCA1-KNTC2, members of centromere protein complex,
involved in pulmonary carcinogenesis. Cancer Res. 66:10339–10348.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kikuchi T, Daigo Y, Katagiri T, Tsunoda T,
Okada K, Kakiuchi S, Zembutsu H, Furukawa Y, Kawamura M, Kobayashi
K, et al: Expression profiles of non-small cell lung cancers on
cDNA microarrays: Identification of genes for prediction of
lymph-node metastasis and sensitivity to anti-cancer drugs.
Oncogene. 22:2192–2205. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ishikawa N, Takano A, Yasui W, Inai K,
Nishimura H, Ito H, Miyagi Y, Nakayama H, Fujita M, Hosokawa M, et
al: Cancer-testis antigen lymphocyte antigen 6 complex locus K is a
serologic biomarker and a therapeutic target for lung and
esophageal carcinomas. Cancer Res. 67:11601–11611. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tomita Y, Harao M, Senju S, Imai K, Hirata
S, Irie A, Inoue M, Hayashida Y, Yoshimoto K, Shiraishi K, et al:
Peptides derived from human insulin-like growth factor-II mRNA
binding protein 3 can induce human leukocyte antigen-A2-restricted
cytotoxic T lymphocytes reactive to cancer cells. Cancer Sci.
102:71–78. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Suda T, Tsunoda T, Daigo Y, Nakamura Y and
Tahara H: Identification of human leukocyte antigen-A24-restricted
epitope peptides derived from gene products upregulated in lung and
esophageal cancers as novel targets for immunotherapy. Cancer Sci.
98:1803–1808. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kono K, Mizukami Y, Daigo Y, Takano A,
Masuda K, Yoshida K, Tsunoda T, Kawaguchi Y, Nakamura Y and Fujii
H: Vaccination with multiple peptides derived from novel
cancer-testis antigens can induce specific T-cell responses and
clinical responses in advanced esophageal cancer. Cancer Sci.
100:1502–1509. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yoshitake Y, Fukuma D, Yuno A, Hirayama M,
Nakayama H, Tanaka T, Nagata M, Takamune Y, Kawahara K, Nakagawa Y,
et al: Phase II clinical trial of multiple peptide vaccination for
advanced head and neck cancer patients revealed induction of immune
responses and improved OS. Clin Cancer Res. 21:312–321. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Harrop R: Cancer vaccines: Identification
of biomarkers predictive of clinical efficacy. Hum Vaccin
Immunother. 9:800–804. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Grivennikov SI, Greten FR and Karin M:
Immunity, inflammation, and cancer. Cell. 140:883–899. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fang H, Yamaguchi R, Liu X, Daigo Y, Yew
PY, Tanikawa C, Matsuda K, Imoto S, Miyano S and Nakamura Y:
Quantitative T cell repertoire analysis by deep cDNA sequencing of
T cell receptor α and β chains using next-generation sequencing
(NGS). Oncoimmunology. 3:e9684672015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wong AK and Walkey AJ: Open lung biopsy
among critically Ill, mechanically ventilated patients: A
Metaanalysis. Ann Am Thorac Soc. 12:1226–1230. 2015.PubMed/NCBI
|
|
27
|
Mehta S, Shelling A, Muthukaruppan A,
Lasham A, Blenkiron C, Laking G and Print C: Predictive and
prognostic molecular markers for cancer medicine. Ther Adv Med
Oncol. 2:125–148. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang G, Zhao H, Wu J, Li J, Xiang Y, Wang
G, Wu L and Jiao S: Adoptive immunotherapy for non-small cell lung
cancer by NK and cytotoxic T lymphocytes mixed effector cells:
Retrospective clinical observation. Int Immunopharmacology.
21:396–405. 2014. View Article : Google Scholar
|
|
29
|
Santegoets SJ, Turksma AW, Suhoski MM,
Stam AG, Albelda SM, Hooijberg E, Scheper RJ, van den Eertwegh AJ,
Gerritsen WR, Powell DJ Jr, et al: IL-21 promotes the expansion of
CD27+ CD28+ tumor infiltrating lymphocytes with high cytotoxic
potential and low collateral expansion of regulatory T cells. J
Transl Med. 11:372013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Khazaie K and von Boehmer H: The impact of
CD4+CD25+ Treg on tumor specific CD8+ T cell cytotoxicity and
cancer. Semin Cancer Biol. 16:124–136. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Erfani N, Mehrabadi SM, Ghayumi MA,
Haghshenas MR, Mojtahedi Z, Ghaderi A and Amani D: Increase of
regulatory T cells in metastatic stage and CTLA-4 over expression
in lymphocytes of patients with non-small cell lung cancer (NSCLC).
Lung Cancer. 77:306–311. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Phillips JD, Knab LM, Blatner NR, Haghi L,
DeCamp MM, Meyerson SL, Heiferman MJ, Heiferman JR, Gounari F,
Bentrem DJ and Khazaie K: Preferential expansion of
pro-inflammatory Tregs in human non-small cell lung cancer. Cancer
Immunol Immunother. 64:1185–1191. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gounaris E, Blatner NR, Dennis K,
Magnusson F, Gurish MF, Strom TB, Beckhove P, Gounari F and Khazaie
K: T-regulatory cells shift from a protective anti-inflammatory to
a cancer-promoting proinflammatory phenotype in polyposis. Cancer
Res. 69:5490–5497. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Glusman G, Rowen L, Lee I, Boysen C, Roach
JC, Smit AF, Wang K, Koop BF and Hood L: Comparative genomics of
the human and mouse T cell receptor loci. Immunity. 15:337–349.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Litman GW, Rast JP and Fugmann SD: The
origins of vertebrate adaptive immunity. Nat Rev Immunol.
10:543–553. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Folch G and Lefranc MP: The human T cell
receptor beta variable (TRBV) genes. Exp Clin Immunogenet.
17:42–54. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Haynes MR and Wu GE: Gene discovery at the
human T-cell receptor alpha/delta locus. Immunogenetics.
59:109–121. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Scaviner D and Lefranc MP: The human T
cell receptor alpha variable (TRAV) genes. Exp Clin Immunogenet.
17:83–96. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Robinson MW, Hughes J, Wilkie GS, Swann R,
Barclay ST, Mills PR, Patel AH, Thomson EC and McLauchlan J:
Tracking TCRβ sequence clonotype expansions during antiviral
therapy using high-throughput sequencing of the hypervariable
region. Front Immunol. 7:1312016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Britanova OV, Putintseva EV, Shugay M,
Merzlyak EM, Turchaninova MA, Staroverov DB, Bolotin DA, Lukyanov
S, Bogdanova EA, Mamedov IZ, et al: Age-related decrease in TCR
repertoire diversity measured with deep and normalized sequence
profiling. J Immunol. 192:2689–2698. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li Z, Liu G, Tong Y, Zhang M, Xu Y, Qin L,
Wang Z, Chen X and He J: Comprehensive analysis of the T-cell
receptor beta chain gene in rhesus monkey by high throughput
sequencing. Sci Rep. 5:100922015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li D, Gao G, Li Z, Sun W, Li X, Chen N,
Sun J and Yang Y: Profiling the T-cell receptor repertoire of
patient with pleural tuberculosis by high-throughput sequencing.
Immunol Lett. 162:170–180. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yew PY, Alachkar H, Yamaguchi R, Kiyotani
K, Fang H, Yap KL, Liu HT, Wickrema A, Artz A, van Besien K, et al:
Quantitative characterization of T-cell repertoire in allogeneic
hematopoietic stem cell transplant recipients. Bone Marrow
Transplant. 50:1227–1234. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Giudicelli V, Chaume D and Lefranc MP:
IMGT/GENE-DB: A comprehensive database for human and mouse
immunoglobulin and T cell receptor genes. Nucleic Acids Res.
33(Database Issue): D256–D261. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Spiotto MT and Schreiber H: Rapid
destruction of the tumor microenvironment by CTLs recognizing
cancer-specific antigens cross-presented by stromal cells. Cancer
Immun. 5:82005.PubMed/NCBI
|
|
47
|
Bonmassar E, Bonmassar A, Vadlamudi S and
Goldin A: Immunological alteration of leukemic cells in vivo after
treatment with an antitumor drug. Proc Natl Acad Sci USA. 66:pp.
1089–1095. 1970; View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Speiser DE, Baumgaertner P, Barbey C,
Rubio-Godoy V, Moulin A, Corthesy P, Devevre E, Dietrich PY,
Rimoldi D, Liénard D, et al: A novel approach to characterize
clonality and differentiation of human melanoma-specific T cell
responses: Spontaneous priming and efficient boosting by
vaccination. J Immunol. 177:1338–1348. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Palermo B, Del Bello D, Sottini A, Serana
F, Ghidini C, Gualtieri N, Ferraresi V, Catricalà C, Belardelli F,
Proietti E, et al: Dacarbazine treatment before peptide vaccination
enlarges T-cell repertoire diversity of melan-a-specific,
tumor-reactive CTL in melanoma patients. Cancer Res. 70:7084–7092.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cornberg M, Chen AT, Wilkinson LA, Brehm
MA, Kim SK, Calcagno C, Ghersi D, Puzone R, Celada F, Welsh RM and
Selin LK: Narrowed TCR repertoire and viral escape as a consequence
of heterologous immunity. J Clin Invest. 116:1443–1456. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Reis DD, Jones EM, Tostes S Jr, Lopes ER,
Gazzinelli G, Colley DG and McCurley TL: Characterization of
inflammatory infiltrates in chronic chagasic myocardial lesions:
Presence of tumor necrosis factor-alpha+ cells and dominance of
granzyme A+, CD8+ lymphocytes. Am J Trop Med Hyg. 48:637–644. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen RH, Ivens KW, Alpert S, Billingham
ME, Fathman CG, Flavin TF, Shizuru JA, Starnes VA, Weissman IL and
Griffiths GM: The use of granzyme A as a marker of heart transplant
rejection in cyclosporine or anti-CD4 monoclonal antibody-treated
rats. Transplantation. 55:146–153. 1993. View Article : Google Scholar : PubMed/NCBI
|