Introduction
Osteosarcoma, a highly malignant and aggressive
tumor generated from mesenchymal tissue that produces osteoid and
bone, occurs more frequently in the early years of life (1). Osteosarcoma develops rapidly and is
usually associated with high mortality due to its progressive
pulmonary metastasis, which eventually leads to respiratory failure
(1). Despite the development of the
multidisciplinary treatment for osteosarcoma, the prognosis of
osteosarcoma patients remains poor (1).
Tumor-initiating cells (TICs), also known as cancer
stem cells (CSCs), comprise a unique subpopulation of cells within
a tumor. Relative to the remainder of the tumor bulk, CSCs are
often more tumorigenic and chemoresistant or radioresistant in
comparison (2). Evidence has
indicated that novel therapeutics targeting CSCs, which are
critical for tumorigenicity and tumor progression, could
significantly improve the clinical outcome of cancer treatment
(2). Therefore, it is of paramount
importance to uncover the existence of CSCs and to elucidate the
pathogenesis of osteosarcoma.
Octamer-binding transcription factor 4 (Oct-4), a
member of the POU-domain transcription factors family, is a key
reprogramming factor that balances the pluripotent and
differentiated states in stem cell and cancer development (3). Notably, Oct-4 is crucial role for
regulating the self-renewal of stem cells, and its expression
dowregulates substantially during cell differentiation (4). Nanog homeobox (Nanog), another
transcription regulator, is a key factor for maintaining
pluripotency during embryonic development (5). The messenger RNA (mRNA) expression of
Nanog is abundant in pluripotent cell lines, including embryonic
stem, embryonic germ and embryonic carcinoma cells, compared with
in adult tissues (5). Similar to
Oct-4, Nanog expression is also downregulated when pluripotent
cells undergo differentiation (5).
Collectively, these findings indicate the possibility that Oct-4
and Nanog may also be important in stem-like cells derived from
osteosarcoma.
Dihydrofolate reductase (DHFR), an enzyme that
reduces dihydrofolic acid to tetrahydrofolic acid, is important for
the synthesis of nucleic acid precursors, which are used for cell
proliferation and growth (6). DHFR,
the target of methotrexate (MTX) in osteosarcoma chemotherapy
(6), is demonstrably important for
the development of chemoresistance to MTX in human osteosarcoma
cells (7,8). The reemergence of chemotherapy-resistant
CSCs has been shown to contribute to cancer relapse following
conventional therapeutic treatment (9). However, the functional role of DHFR in
chemoresistance in CSCs has not been determined.
To further elucidate the roles of Oct-4 and Nanog in
the tumorigenesis of osteosarcoma, the present study aimed to
compare and distinguish the phenotypic differences between
osteosarcoma TICs (OS-TICs) and parental osteosarcoma cells, as
well as isolate the OS-TICs using sphere formation assays and
defined serum-depleted media containing basic fibroblast growth
factor (bFGF) and epidermal growth factor (EGF). In addition, the
present study sought to evaluate the expression of several
stem-like markers, including Oct-4, Nestin, Nanog, cluster of
differenatiation (CD)117 and CD133, and the anticancer drug
transporters, MDR1 and ATP binding cassette subfamily G member 2
(ABCG2). The capabilities of OS-TICs for migration, invasion and
tumorigenicity were simultaneously assessed in vitro and
in vivo. The findings of the present study may help to
elucidate the mechanisms underlying the chemoresistance in
osteosarcoma patients.
Materials and methods
Isolation of OS-TICs from osteosarcoma
cells by tumor sphere-forming assay
The human osteosarcoma cell lines (U2OS and MG-63)
were purchased from the American Type Culture Collection (Manassas,
VA, USA). Originally, U2OS and MG-63 cells were grown in Dulbecco's
modified Eagle's medium (DMEM) (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(HyClone; GE Healthcare Life Sciences, Logan, UT, USA). U2OS and
MG-63 cells were cultured as sphere-forming OS-TICs, as described
previously (10). For expanding
OS-TICs, primary tumor spheres were dissociated into single cell
suspension using HyQTase solution (HyClone; GE Healthcare Life
Sciences) at 37°C for 5 min, and seeded at a density of
104 live cells/10-mm into ultralow attachment 6-well
plates (Corning Life Sciences, Tewksbury, MA, USA), with defined
serum-depleted DMEM/F-12 (Gibco; Thermo Fisher Scientific, Inc.)
containing 10 ng/ml bFGF (Invitrogen; Thermo Fisher Scientific,
Inc.) and 10 ng/ml EGF (Invitrogen; Thermo Fisher Scientific,
Inc.).
RNA extraction, RNA quantification and
quantitative reverse transcription-polymerase chain reaction
(qRT-PCR)
Total RNA was extracted from cells using RNeasy Mini
Kits (Qiagen, Inc., Valencia, CA, USA), according to the
manufacturer's instructions. RNA concentration was measured using a
NanoVue Plus spectrophotometer (GE Healthcare Life Sciences). RNA
was then reverse transcribed to complementary DNA using the
SuperScript III First-Strand Synthesis System (Invitrogen; Thermo
Fisher Scientific, Inc.). The TaqMan mRNA Assay (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was used to determine
the mRNA expression levels on the StepOnePlus Fast Real-Time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
cycling conditions were as follows: 50°C for 2 min, 95°C for 10
min, and 40 cycles of 95°C for 10 sec and 60°C for 1 min.
Endogenous control gene, GAPDH, was used to normalize the relative
gene expression level. Primer sequences listed in Table I were synthesized and purchased from
Mission Biotech (Taipei, Taiwan). Fold-changes were determined
using the 2−ΔΔCq method (11).
 | Table I.The sequences of the primers for
quantitative reverse transcription-quantitiative polymerase chain
reaction. |
Table I.
The sequences of the primers for
quantitative reverse transcription-quantitiative polymerase chain
reaction.
| Gene (accession
no.) | Primer sequence, 5′
to 3′ | Product size, base
pairs | Temperature,
°C |
|---|
| Oct-4 | F:
GTGGAGAGCAACTCCGATG | 86 | 60 |
| (NM_002701) | R:
TGCTCCAGCTTCTCCTTCTC |
|
|
| Nanog | F:
ATTCAGGACAGCCCTGATTCTTC | 76 | 60 |
| (NM_024865) | R:
TTTTTGCGACACTCTTCTCTGC |
|
|
| GAPDH | F:
CATCATCCCTGCCTCTACTG | 180 | 60 |
| (NM_002046) | R:
GCCTGCTTCACCACCTTC |
|
|
Western blot analysis
Parental osteosarcoma or OS-TICs were lysed using
NP-40 lysis buffer (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany),
and the protein concentration was determined using bicinchoninic
acid protein assay reagent (Thermo Fisher Scientific Inc.). Samples
(25 µg of total protein) were boiled at 95°C for 5 min and
separated by 10% SDS-PAGE. The proteins were wet-transferred to
polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA,
USA). After blocking with 5% milk for 30 min, the membranes were
washed three times with 0.1% Tween-20 in Tris buffer (TBS-T) for 5
min each. Subsequently, the primary antibodies were diluted in 0.1%
TBS-T and incubated with the membrane at room temperature for 1 h.
Upon washing three times for 5 min each with 0.1% TBS-T, the
membrane was incubated for 30 min at room temperature with
horseradish peroxidase-conjugated anti-rabbit (1:3,000; catalog no.
7074; Cell Signaling Technology, Inc., Danvers, MA, USA) or
anti-mouse immunoglobulin G polyclonal antibodies (1:3,000; catalog
no. 7076; Cell Signaling Technology, Inc.) were used as secondary
antibodies diluted with 0.1% TBS-T. Following three washes with
0.1% TBS-T for 5 min each, the signals were developed using an
Enhanced Chemiluminescence Plus substrate (PerkinElmer, Inc.,
Waltham, MA, USA) and captured using a LAS-1000plus Luminescent
Image Analyzer (GE Healthcare Life Sciences). The following primary
antibodies were used: Anti-Oct-4 antibody (1:1,000; #2750; Cell
Signaling Technology, Inc.), anti-Nanog antibody (1:500; #3850;
Cell Signaling Technology, Inc.), anti-Nestin antibody (1:1,000;
MAB5326; EMD Millipore), anti-GAPDH antibody (1:2,000; GTX627408;
GeneTex, Inc., Irvine, CA, USA), anti-DHFR antibody (1:1,000;
ab49881; Abcam, Cambridge, UK) and anti-MDR1 antibody (1:1,000;
#13978; Cell Signaling Technology, Inc.).
Immunofluorescent staining
Parental and OS-TICs cells were plated on glass
coverslips and then fixed with 4% buffered formalin in PBS. Cells
were permeabilized with 0.1% Triton X-100/PBS, washed with PBS, and
incubated with a mouse anti-Nestin monoclonal antibody (1:100;
MAB5326; EMD Millipore), an antibody against Oct-4 (1:50; #2750;
Cell Signaling Technology, Inc.), a rabbit polyclonal antibody
against Nanog (1:50; #3850; Cell Signaling Technology, Inc.) or a
rabbit anti-MDR1 monoclonal antibody (1:100; #13342; Cell Signaling
Technology, Inc.). Upon washing with 0.1% Tween-20 in PBS, the
cells were then incubated with fluorescent probe-conjugated
secondary antibodies (1:1,000; #Z25002; Molecular Probes; Thermo
Fisher Scientific, Inc.) at room temperature for 1 h. Cells were
further counterstained with 0.1 µg/ml DAPI. The images were
captured using the Leica Application Suite X with Leica DMi8 (Leica
Microsystems GmbH, Wetzlar, Germany).
Flow cytometry analysis
Parental and OS-TICs cells were stained with
fluorescein isothiocyanate-labeled anti-CD133 (1:100; 130-105-225;
Miltenyi Biotech, Inc., Auburn, CA, USA), anti-CD117 (1:100;
313232; BioLegend, Inc., San Diego, CA, USA) or anti-ABCG2 (1:10;
130-104-957; Miltenyi Biotech, Inc.) antibodies, according to the
manufacturer's instructions. All antibodies were diluted in 0.1%
bovine serum albumin (A8531; Sigma-Aldrich; Merck KGaA). The
results were measured using the FACSCalibur flow cytometer (BD
Biosciences, Franklin Lakes, NJ, USA) and analyzed by FlowJo 7.6
software (FlowJo, LLC, Ashland, OR, USA).
Transwell chamber assay for
migration/invasion analysis
Cell migration and invasion assays were performed
using 5×104 parental osteosarcoma or derived OS-TICs in
24-well plate Transwell chambers (pore size, 8 µm; Millicell; EMD
Millipore), as described previously (10). Cell suspensions were seeded into the
upper compartment of the Transwell chamber at a cell density of
1×105 cells per 100 µl in serum-free medium. The lower
chamber was filled with medium containing 10% serum. After 24 h of
incubation, the medium was removed, and the filter membrane was
fixed with methanol for 1 h. Subsequently, the remaining cells in
the filter membrane facing the lower chamber were stained with
crystal violet (Sigma-Aldrich; Merck KGaA). The migrated and
invasive cancer cells were then visualized and counted from five
different visual fields at ×100 magnification under an inverted
microscope.
Xenografts animal experiment
All the NOD/SCID mice (18–22 g; National Laboratory
Animal Center, Zhunan, Taiwan) practices in the present study were
approved and performed in accordance with the Institutional Animal
Care and Use Committee of Chung Shan Medical University (Taichung,
Taiwan). All mice were housed with a regular 12-h light/dark cycle,
and ad libitum access to water and a standard rodent chow
diet (Laboratory Rodent Diet 5001; LabDiet, St. Louis, MO, USA),
and were kept in a pathogen-free environment at the Laboratory
Animal Unit of Chung Shan Medical University (temperature, 22°C;
humidity, 30–70%; n=5 mice/cage), according to the requirements of
the Institutional Animal Care and Use Committee of Chung Shan
Medical University, Taichung, Taiwan. Single-cell suspensions
containing serial dilutions of parental and OS-TICs in 100 µl
serum-free medium (Table II) were
mixed with 100 µl Matrigel (BD Biosciences) and subcutaneously
injected into 6-week-old, male NOD/SCID mice. Each cell injected
group consisted of 3 mice, all of which were male. A total of 24
mice are used for the experiment. At 6 weeks after injection, the
mice were sacrificed by CO2 inhalation. Tumor volume
(TV) was calculated using the following formula: TV
(mm3) = (length × width2)/2.
 | Table II.In vivo tumorigenicity of
parental U2OS and derived OS-TICs was examined in NOD/SCID mice by
xenotransplantation analysis. |
Table II.
In vivo tumorigenicity of
parental U2OS and derived OS-TICs was examined in NOD/SCID mice by
xenotransplantation analysis.
|
| No. of cells for
injection |
|---|
|
|
|
|---|
| Cells |
1×104 |
5×104 |
1×105 |
2×105 |
|---|
| U2OS | 0/3 | 0/3 | 1/3 | 2/3 |
| OS-TICs | 3/3 | 3/3 | 3/3 | 3/3 |
MTT assay
The viability of parental and OS-TICs cells treated
with increasing concentrations of MTX or doxorubicin was measured
by MTT assay (Sigma-Aldrich; Merck KGaA), according to the
manufacturer's instructions. Cells were plated in 24-well plates
(5×104 cells/well) in different concentrations of
doxorubicin or MTX and cultured at 37°C for 24 h. The concentration
of doxorubicin was initiated at 0 µM and increased at 50 µM
increments. The concentration of MTX was also started at 0 µM, but
was increased at 25 µM increments. The attached cells were
incubated with 0.5 mg/ml of MTT at 37°C for 4 h. The blue formazan
crystals of viable cells were dissolved in dimethyl sulfoxide
(DMSO) and then evaluated spectrophotometrically at 570 nm. The
DMSO-treated group was set as 100%, and data were presented as a
percentage of the DMSO control. Cell survival was measured using
Infinite M200 PRO (Tecan Group Ltd., Männedorf, Switzerland) and
analyzed with Magellan 7.1 software (Tecan Group Ltd.).
Statistical analysis
Data are presented as mean ± standard error of the
mean. Statistical analyses were performed using the unpaired
Student's t-test in SPSS 16 (SPSS, Inc., Chicago, IL, USA).
Kaplan-Meier survival analysis was used to analyze animal survival
data. P<0.05 was considered to indicate a statistically
significant difference.
Results
Characterization of progenitor/stem
cell properties in isolated OS-TICs
To explore the existence of stem-like cells/TICs in
osteosarcoma, the osteosarcoma U2OS and MG-63 cell lines were
incubated with defined serum-free medium with bFGF and EGF cultured
in low-attachment 6-well plates. After 10 days incubation, the
cancer cells gradually detached from the culture dishes, aggregated
and became spheres-forming OS-TICs (Fig.
1A). To further characterize the stem-like properties of the
enriched OS-TICs, flow cytometry was used to analyze the expression
profiles of stem-like cell surface markers in parental U2OS cells
and OS-TICs. As shown in Fig. 1B, the
majority of isolated OS-TICs were positively stained for CD133 and
CD117, two well-known cell surface markers of CSCs. Similarly, the
upregulation of ABCG2 was also observed to be increased in OS-TICs
compared with parental U2OS cells (Fig.
1B). In total, ~90% of OS-TICs derived from U2OS cells were
stained positive for CD133, CD117 and ABCG2.
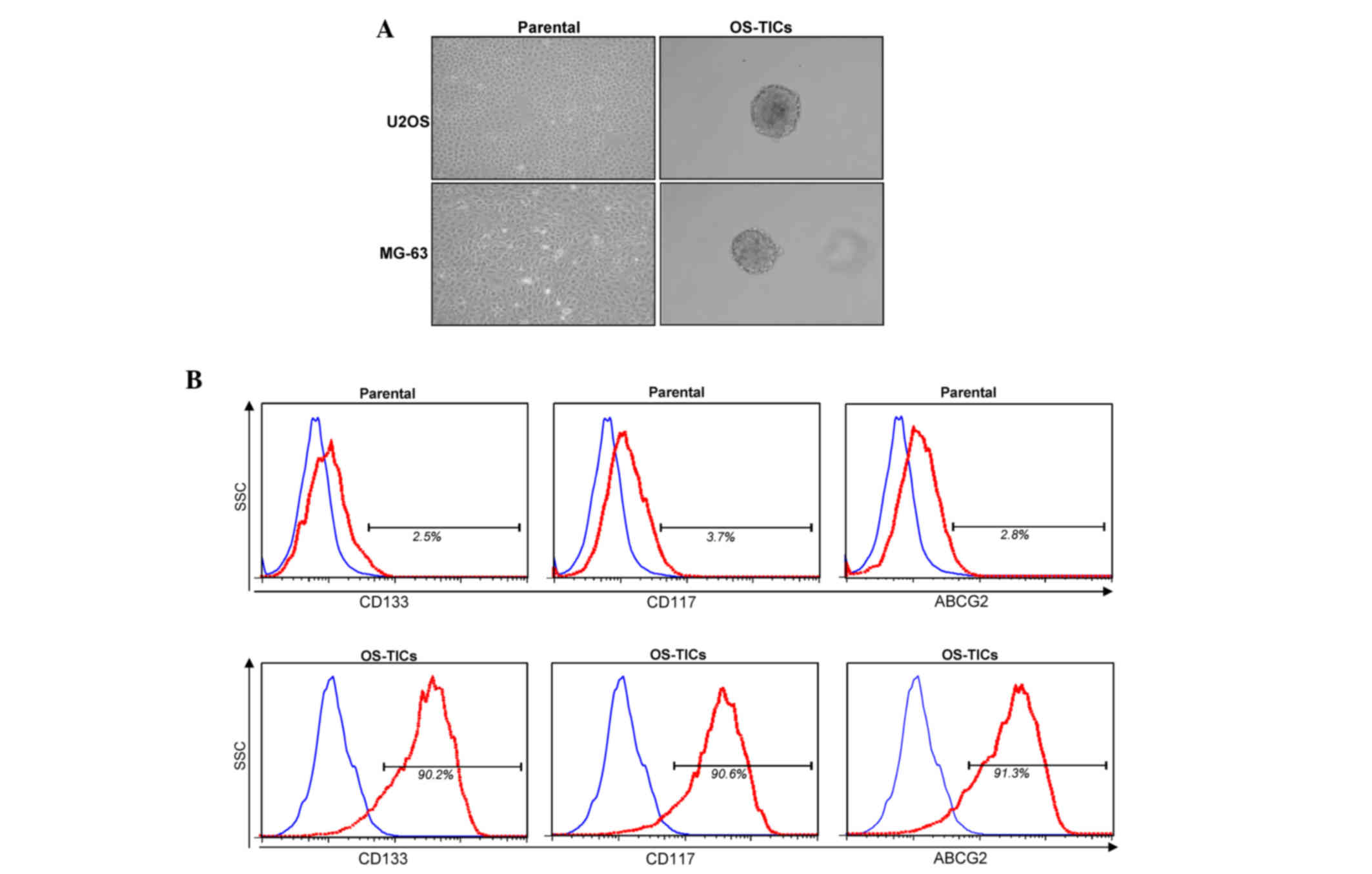 | Figure 1.Isolation and characterization of
OS-TICs. (A) Osteosarcoma cancer cell lines were used and cultured
in serum-depleted Dulbecco's modified Eagle's medium/F-12 medium
containing basic fibroblast growth factor and epidermal growth
factor into low-attachment 6-well plates. After 10 days of culture,
cancer cells gradually detached from culture dishes, aggregated and
formed tumor spheres. (B) Expression profiles of progenitor/stem
cell-specific surface markers, including CD133, CD117 and ABCG2, in
parental U2OS cells or derived OS-TICs, analyzed by flow cytometry.
Single-cell suspension from parental cells or derived OS-TICs was
either stained with control immunoglobulin G antibody or
experimental antibodies, including anti-CD133 (left), anti-CD117
(middle) and anti-ABCG2 (right). OS-TICs, osteosarcoma
tumor-initiating cells; CD, cluster of differentiation; ABCG2, ATP
binding cassette subfamily G member 2. |
Elevated expression of progenitor/stem
cell genes in OS-TICs
An MTT assay was then used to evaluate the
proliferation rate of parental U2OS cells and OS-TICs. Notably,
OS-TICs exhibited an increased proliferation capability compared
with parental cells (Fig. 2A). The
expression of stem cell-specific genes, Oct-4, Nanog, and Nestin,
was examined transcriptionally and translationally. The total RNA
of parental U2OS cells or OS-TICs was extracted and qRT-PCR was
performed. The results confirmed the high expression of Oct-4,
Nanog and Nestin transcripts in the enriched OS-TICs (Fig. 2B). Similar to the observations of
qRT-PCR, the results of the western blot analysis also showed that
the protein levels of Oct-4, Nanog, and Nestin were also
upregulated in enriched OS-TICs compared with in parental U2OS
cells (Fig. 2C). Furthermore,
immunofluorescent staining validated the high levels of Oct-4
(Fig. 2D; top panel), Nestin
(Fig. 2D; middle panel), and Nanog
(Fig. 2D; bottom panel) in the
intracellular compartments in enriched OS-TICs compared with in
parental U2OS cells.
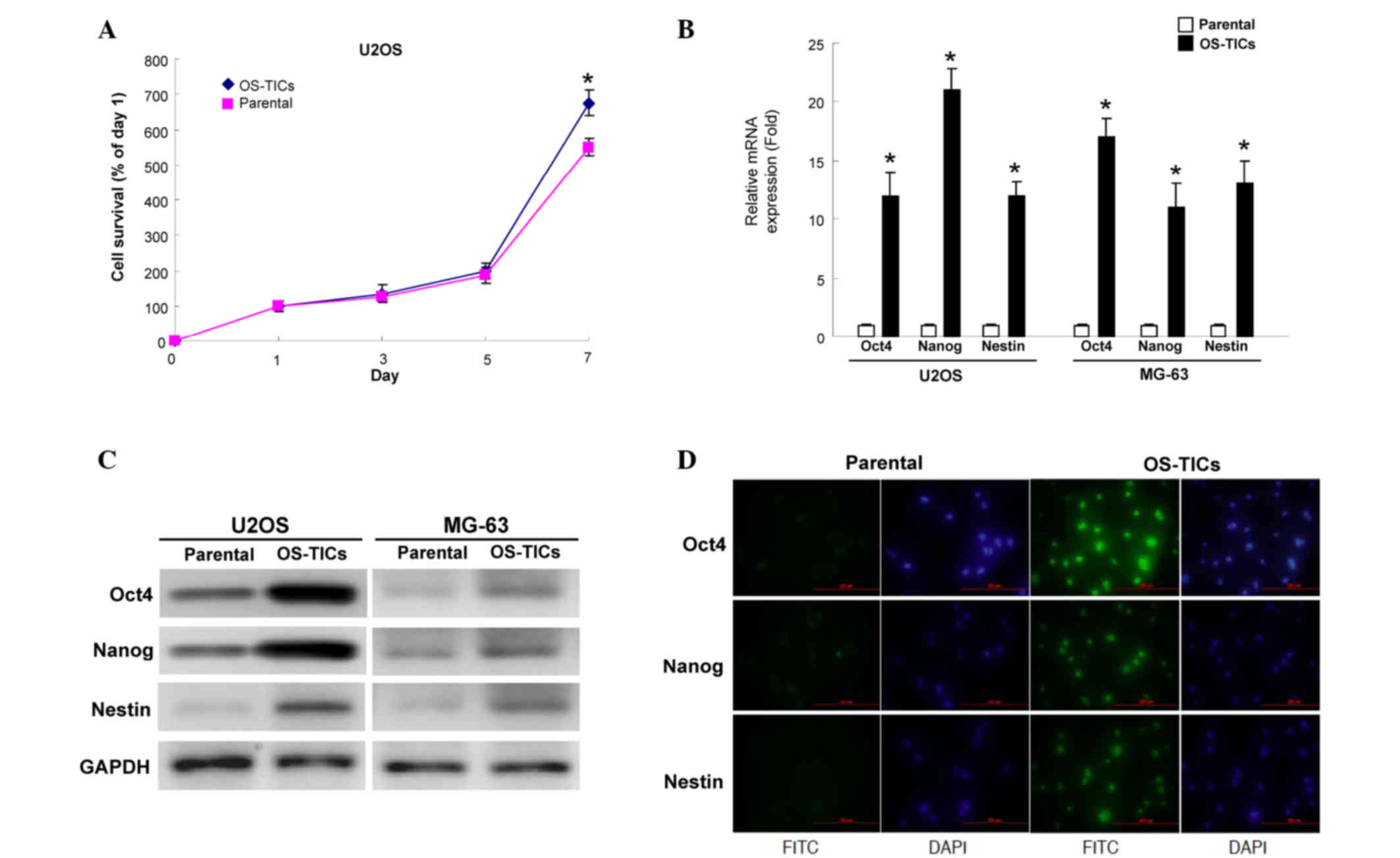 | Figure 2.Detection of the expression levels of
stemness markers in OS-TICs and parental osteosarcoma cells. (A)
Proliferation rate of parental and OS-TICs was determined by
3-(4,5-dimethyl thiazol)-2,5-diphenyltetrazolium bromide assay. (B)
Quantitative reverse transcription-polymerase chain reaction
analysis revealed the transcription amounts of Nanog, Oct-4 and
Nestin in OS-TICs and parental osteosarcoma cells. (C) Total
proteins were prepared from parental osteosarcoma or OS-TICs and
analyzed by immunoblotting with anti-Oct-4, -Nanog, -Nestin
or-GAPDH antibodies as indicated. The amount of GAPDH protein of
various crude cell extracts was referred as loading control. (D) By
immunofluorescence analysis, parental and enriched OS-TICs from
U2OS cells were stained with anti-Oct-4 (top panel), anti-Nestin
(middle panel) or anti-Nanog, respectively. *P<0.05. OS-TICs,
osteosarcoma-tumor-initiating cells; Oct-4, octamer-binding
transcription factor 4; Nanog, Nanog homeobox; FITC, fluorescein
isothiocyanate. |
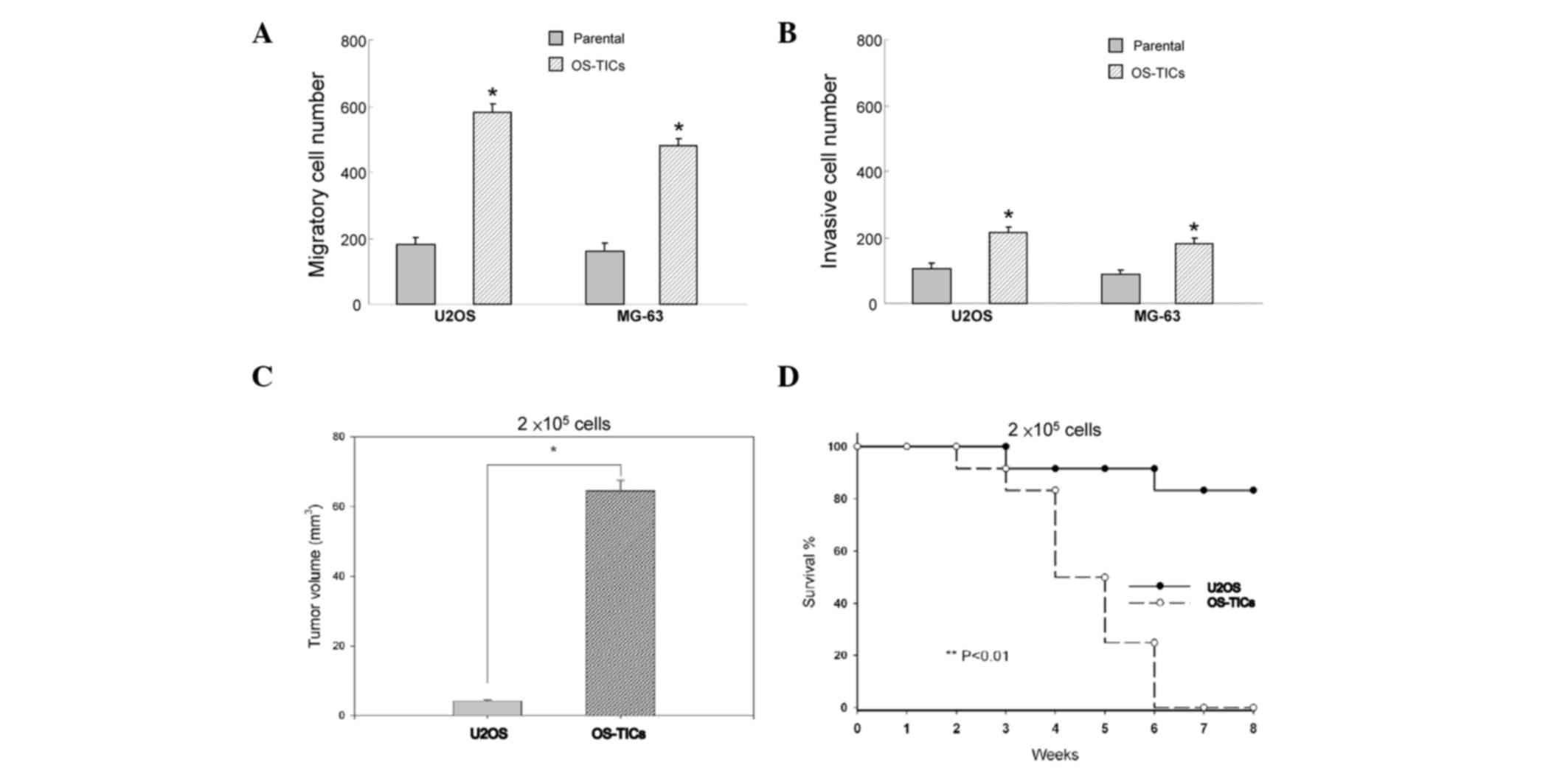
Enhanced tumorigenic potential of
OS-TICs in vitro and in vivo
To evaluate whether these enriched OS-TICs possess
enhanced tumorigenicity, an in vitro Matrigel assay combined
Transwell invasion/migration assay was performed. The results
indicated that enriched OS-TICs have increased invasion and
migration capabilities compared with the parental U2OS cells
(Fig. 3A and B; P<0.05). To
further validate the enhanced tumor-initiating abilities of OS-TICs
in vivo, the parental cells and OS-TICs derived from U2OS
cells were injected into the subcutaneous space of NOD/SCID mice
for xenograft tumorigenicity assay. As shown in Table II, only one recipient of U2OS
parental cells at 1×105 cells/mice resulted in a tumor.
However, all of three recipients of U2OS OS-TICs developed
tumor-like formation when only 1×104 cells were
injected. After 6 weeks of xenotransplantation, the tumor volume
was significantly increased in OS-TIC-transplanted mice compared
with recipients transplanted with parental U2OS cells at the same
injected cell numbers (2×105 cells) (Fig. 3C; P<0.05). Furthermore, survival
curve analysis indicated that the mean survival rate of
OS-TIC-transplanted recipients was significantly lower compared
with the parental U2OS cells (Fig.
3D; P<0.01).
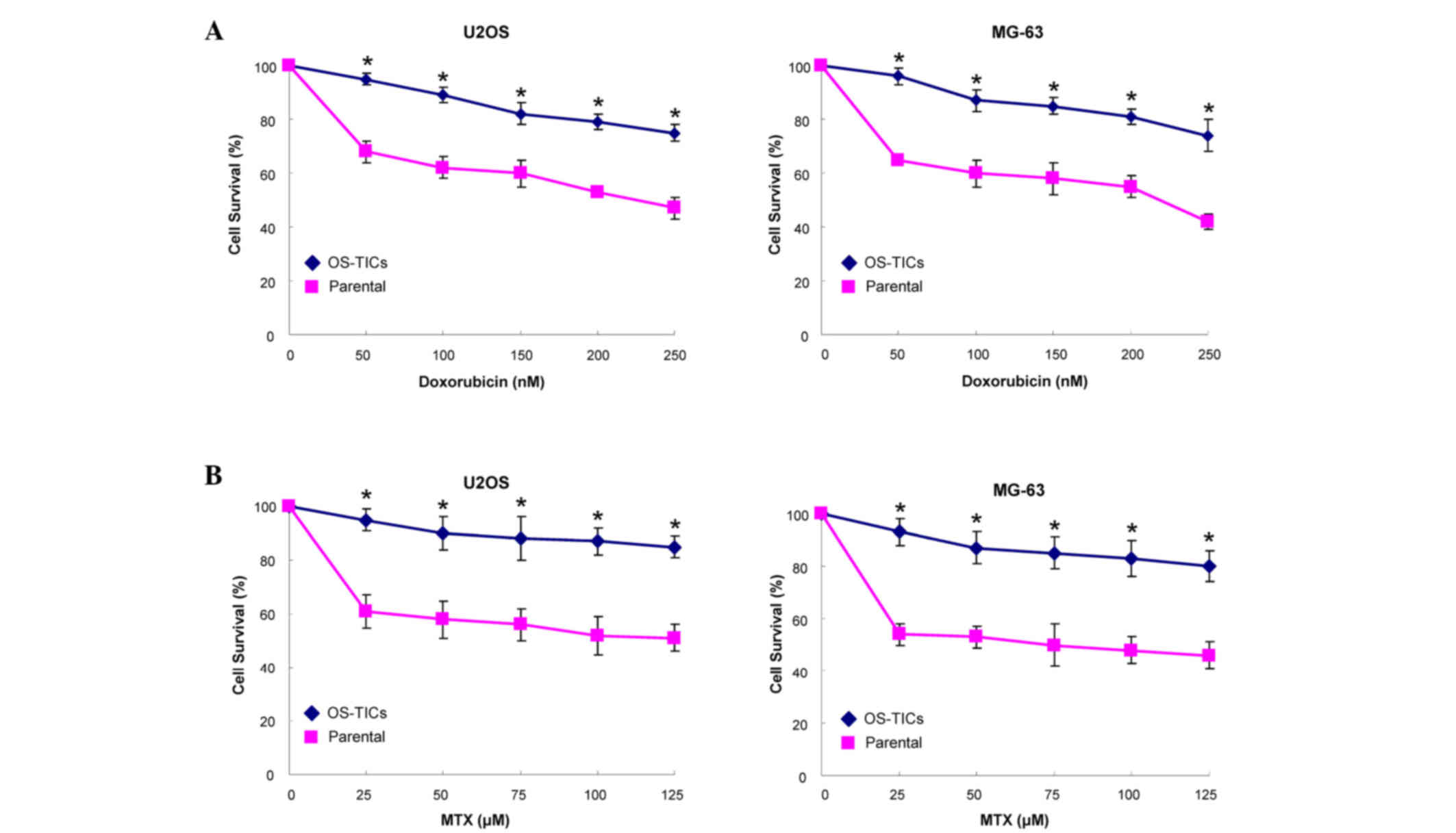
Enhanced resistance of OS-TICs to
chemotherapy
Resistance to chemotherapy treatment is the major
clinical criterion to characterize CSCs (12). Accordingly, an MTT assay was used to
monitor the sensitivity of OS-TICs to the treatment of MTX and
doxorubicin. Notably, OS-TICs were more chemoresistant to
doxorubicin (Fig. 4A) and MTX
(Fig. 4B) in dose-dependent
concentrations compared with parental U2OS cells (P<0.05).
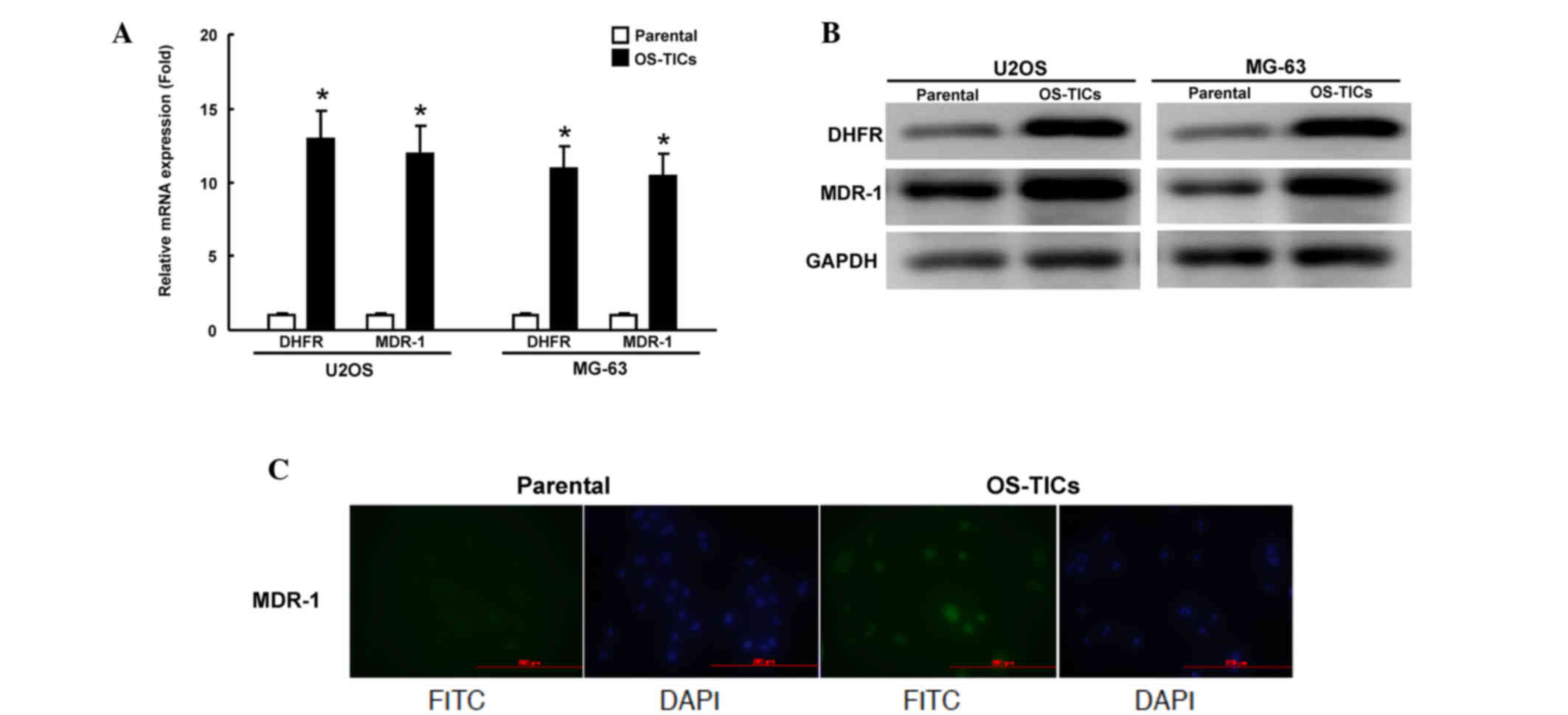
Elevated expression of DHFR and MDR1
genes in OS-TICs
Since chemoresistance to doxorubicin and MTX is
enhanced in enriched OS-TICs, the present study sought to
investigate the mechanisms underlying this regulation of
chemoresistance. DHFR, a target of MTX, was previously shown to
regulate the acquisition of chemoresistance to MTX in human
osteosarcoma cells (7,8). MDR1, a member of ATP-binding cassette
(ABC) transporter superfamily, is critical for regulating the
resistance to multiple chemotherapeutic drugs (13). The expression of DHFR and MDR1 mRNA
was examined transcriptionally by qRT-PCR. The total RNA of
parental U2OS cells or OS-TICs was extracted. The amount of DHFR
and MDR1 transcripts of enriched OS-TICs was significantly
increased compared with parental U2OS cells (Fig. 5A). In accordance with the results of
the qRT-PCR, the western blot analysis also showed that DHFR and
MDR1 were also upregulated at the protein level in enriched OS-TICs
(Fig. 5B). Furthermore,
immunofluorescent staining showed that MDR1 expression was
dramatically increased in OS-TICs derived from U2OS cells, compared
with those in parental U2OS cells (Fig.
5C).
Discussion
Osteosarcomas are highly aggressive tumors, which
most commonly occur in children and young adults (13). Previous studies have demonstrated the
existence of a highly tumorigenic cell subset, commonly known as
CSCs, within the tumor bulk (14),
and the existence of CSCs within osteosarcoma has also been
documented in several studies (15–20).
CD117-positive osteosarcoma cells show highly metastatic,
tumorigenic capacity and resistance to chemotherapy (17). CD133-positive osteosarcoma cells also
possess CSC features, including a high proliferation rate, enhanced
abilities for sphere cluster formation, clonogenic efficiency and
in vivo tumorigenicity (18,20). In
addition, the embryonic stem cell-specific transcriptional factor,
sex determining region Y-box 2, also maintains self-renewal and
tumorigenic properties in osteosarcoma CSCs (15). In the present study, OS-TICs were
found to express ABCG2, a membrane-associated protein that is
usually associated with side population phenotype and ATP-dependent
exclusion of cellular toxic agents (21). Given that the expression of ABC
transporters, including MDR1 and ABCG2, may be important for
multidrug resistance to chemotherapeutic agents (22), the expression of ABCG2 can be
considered as an additional biomarker for the identification of
OS-TICs.
Oct-4 and Nanog were previously suggested as two of
the four major factors that render the reprogramming capability of
adult cells into germ-line-competent induced pluripotent stem cells
(23,24). Nanog blocks differentiation
functionally, and the clinical results showed that the elevated
expression of Nanog has been associated with retinoblastoma,
prostate cancer, embryonal carcinoma, metastatic germ cell tumor
and ovarian cancer (25–29). The expression of Oct-4 and Nanog has
also been shown in human oral cancer stem-like cells, suggesting
that its expression may be implicated in self-renewal and
tumorigenesis (10). In the present
study, a subpopulation of CSCs from OS-TICs were successfully
isolated and enriched using tumor sphere formation assays (Fig. 1A). Notably, the enriched OS-TICs
exhibited CSC-like features. For example, the results of
immunofluorescent staining and flow cytometry analysis revealed
that enriched OS-TICs were stained positive for several stem cell
markers, including Oct-4, Nanog, CD133 and CD117, as well as the
ABC transporters, MDR1 and ABCG2. Consistent with these findings,
another study proposed that aberrant expression of Oct-4 may
contribute to the neoplastic process in cells (3). Overall, these findings indicated that
the abnormal expression of Oct-4 or Nanog in stem cells may be
critical for regulating tumorigenicity (30,31). The
chemoresistant properties of CSCs have been associated with the
expression of Oct-4 and Nanog (32–36). In
one study, the overexpression of Nanog promoted cisplatin
resistance and decreased the proportion of apoptotic cells in
esophageal cancer cells (36).
Another study found that the ectopic overexpression of Oct-4 and
Nanog enhanced the mRNA levels of ABCB1, resulting in an increased
tolerance of lung adenocarcinoma cells to cisplatin treatment
(32). Further studies on the
regulatory networks between Oct-4 or Nanog and chemoresistance may
be required to update the current knowledge for the future
development of therapies against osteosarcoma.
Doxorubicin is widely used as a chemotherapeutic
agent in the treatment of malignant cancers, including osteosarcoma
(37,38). The cytotoxic effects mediated by
doxorubicin on malignant cells have been shown to involve: i) DNA
base-pair intercalation; ii) the interaction of drug molecules with
topoisomerase II to induce the formation of DNA-cleavable
complexes; and iii) the interaction of drug molecules with electron
transport chain, which may result in the generation of superoxide
anion radicals in the cells (39).
The mechanisms involved in the chemoresistance of tumor cells to
doxorubicin have been shown to include: i) The overexpression of
membrane-associated efflux pump, P-glycoprotein, mediating
multidrug resistance; ii) the altered expression of topoisomerase
II and integrins; and iii) altered glutathione levels (40). P-glycoprotein, a product of the MDR1
gene, is an ATP-dependent drug efflux pump that extrudes drugs from
cells and eventually results in the resistance of cells to
chemotherapy (41). Numerous studies
have also shown that doxorubicin is involved in acquired multidrug
resistance mediated by the expression of P-glycoprotein in
drug-resistant tumor cells of osteosarcoma (42,43). The
results of the present study implied that the enhanced resistance
of osteosarcoma CSCs to doxorubicin could be associated with the
upregulation of P-glycoprotein.
MTX is another widely-used anticancer drug for
chemotherapy against osteosarcoma (14,44,45).
Several lines of evidence have demonstrated that DHFR is critical
in the regulation of chemoresistance to MTX in human osteosarcoma
cells (7,8). However, the causal association of DHFR
with chemoresistance to MTX has been extensively investigated;
therefore, the present study did not examine additional data
addressing the mechanisms by which DHFR may affect the drug
responsiveness in human osteosarcoma patients. In agreement with
the previous reports, the present study also validated the
increased DHFR gene and protein expression in the enriched OS-TICs
that were MTX-resistant. These results also indicate that
DHFR-mediated MTX resistance in osteosarcoma may be associated with
the phenotype of CSCs. Additional studies will be required to
clarify the molecular mechanisms responsible for the DHFR-mediated
MTX resistance in OS-TICs in more detail.
The findings of the present study demonstrate that
stemness/self-renewal genes, including Oct-4 and Nanog could be
important for osteosarcoma-derived stem-like cells. The present
study also found that CSCs/TICs were involved in multidrug
resistance of osteosarcoma, and that the upregulation of MDR1 and
DHFR were responsible for the resistance to anticancer drugs in
human OS-TICs. These findings may be beneficial to the development
of novel anticancer therapeutic strategies, since these identified
genes may represent attractive targets against human
osteosarcoma.
Acknowledgements
The present study was supported by research grants
from Chung Shan Medical University Hospital, Taichung, Taiwan,
R.O.C. (grant no. CSH-2010-C-025).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fan YL, Zheng M, Tang YL and Liang XH: A
new perspective of vasculogenic mimicry: EMT and cancer stem cells
(Review). Oncol Lett. 6:1174–1180. 2013.PubMed/NCBI
|
|
3
|
Gidekel S, Pizov G, Bergman Y and Pikarsky
E: Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer
Cell. 4:361–370. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nichols J, Zevnik B, Anastassiadis K, Niwa
H, Klewe-Nebenius D, Chambers I, Schöler H and Smith A: Formation
of pluripotent stem cells in the mammalian embryo depends on the
POU transcription factor Oct4. Cell. 95:379–391. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chambers I, Colby D, Robertson M, Nichols
J, Lee S, Tweedie S and Smith A: Functional expression cloning of
Nanog, a pluripotency sustaining factor in embryonic stem cells.
Cell. 113:643–655. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Costi MP and Ferrari S: Update on
antifolate drugs targets. Curr Drug Targets. 2:135–166. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hattinger CM, Reverter-Branchat G,
Remondini D, Castellani GC, Benini S, Pasello M, Manara MC,
Scotlandi K, Picci P and Serra M: Genomic imbalances associated
with methotrexate resistance in human osteosarcoma cell lines
detected by comparative genomic hybridization-based techniques. Eur
J Cell Biol. 82:483–493. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Serra M, Reverter-Branchat G, Maurici D,
Benini S, Shen JN, Chano T, Hattinger CM, Manara MC, Pasello M,
Scotlandi K and Picci P: Analysis of dihydrofolate reductase and
reduced folate carrier gene status in relation to methotrexate
resistance in osteosarcoma cells. Ann Oncol. 15:151–160. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jordan CT, Guzman ML and Noble M: Cancer
stem cells. N Engl J Med. 355:1253–1261. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chiou SH, Yu CC, Huang CY, Lin SC, Liu CJ,
Tsai TH, Chou SH, Chien CS, Ku HH and Lo JF: Positive correlations
of Oct-4 and Nanog in oral cancer stem-like cells and high-grade
oral squamous cell carcinoma. Clin Cancer Res. 14:4085–4095. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang JC: Evaluating therapeutic efficacy
against cancer stem cells: New challenges posed by a new paradigm.
Cell Stem Cell. 1:497–501. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dean M: ABC transporters, drug resistance,
and cancer stem cells. J Mammary Gland Biol Neoplasia. 14:3–9.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Marina N, Gebhardt M, Teot L and Gorlick
R: Biology and therapeutic advances for pediatric osteosarcoma.
Oncologist. 9:422–441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Basu-Roy U, Seo E, Ramanathapuram L, Rapp
TB, Perry JA, Orkin SH, Mansukhani A and Basilico C: Sox2 maintains
self renewal of tumor-initiating cells in osteosarcomas. Oncogene.
31:2270–2282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Di Fiore R, Santulli A, Ferrante RD,
Giuliano M, De Blasio A, Messina C, Pirozzi G, Tirino V, Tesoriere
G and Vento R: Identification and expansion of human
osteosarcoma-cancer-stem cells by long-term 3-aminobenzamide
treatment. J Cell Physiol. 219:301–313. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Adhikari AS, Agarwal N, Wood BM, Porretta
C, Ruiz B, Pochampally RR and Iwakuma T: CD117 and Stro-1 identify
osteosarcoma tumor-initiating cells associated with metastasis and
drug resistance. Cancer Res. 70:4602–4612. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tirino V, Desiderio V, d'Aquino R, De
Francesco F, Pirozzi G, Graziano A, Galderisi U, Cavaliere C, De
Rosa A, Papaccio G and Giordano A: Detection and characterization
of CD133+ cancer stem cells in human solid tumours. PLoS One.
3:e34692008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tirino V, Desiderio V, Paino F, De Rosa A,
Papaccio F, Fazioli F, Pirozzi G and Papaccio G: Human primary bone
sarcomas contain CD133+ cancer stem cells displaying high
tumorigenicity in vivo. FASEB J. 25:2022–2030. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Saini V, Hose CD, Monks A, Nagashima K,
Han B, Newton DL, Millione A, Shah J, Hollingshead MG, Hite KM, et
al: Identification of CBX3 and ABCA5 as putative biomarkers for
tumor stem cells in osteosarcoma. PLoS One. 7:e414012012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pascal LE, Oudes AJ, Petersen TW, Goo YA,
Walashek LS, True LD and Liu AY: Molecular and cellular
characterization of ABCG2 in the prostate. BMC Urol. 7:62007.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bunting KD: ABC transporters as phenotypic
markers and functional regulators of stem cells. Stem Cells.
20:11–20. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Park IH, Zhao R, West JA, Yabuuchi A, Huo
H, Ince TA, Lerou PH, Lensch MW and Daley GQ: Reprogramming of
human somatic cells to pluripotency with defined factors. Nature.
451:141–146. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Okita K, Ichisaka T and Yamanaka S:
Generation of germline-competent induced pluripotent stem cells.
Nature. 448:313–317. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Seigel GM, Hackam AS, Ganguly A, Mandell
LM and Gonzalez-Fernandez F: Human embryonic and neuronal stem cell
markers in retinoblastoma. Mol Vis. 13:823–832. 2007.PubMed/NCBI
|
|
26
|
Santagata S, Ligon KL and Hornick JL:
Embryonic stem cell transcription factor signatures in the
diagnosis of primary and metastatic germ cell tumors. Am J Surg
Pathol. 31:836–845. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ma S, Chan KW, Hu L, Lee TK, Wo JY, Ng IO,
Zheng BJ and Guan XY: Identification and characterization of
tumorigenic liver cancer stem/progenitor cells. Gastroenterology.
132:2542–2556. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Freberg CT, Dahl JA, Timoskainen S and
Collas P: Epigenetic reprogramming of OCT4 and NANOG regulatory
regions by embryonal carcinoma cell extract. Mol Biol Cell.
18:1543–1553. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hoei-Hansen CE, Kraggerud SM, Abeler VM,
Kaern J, Rajpert-De Meyts E and Lothe RA: Ovarian dysgerminomas are
characterised by frequent KIT mutations and abundant expression of
pluripotency markers. Mol Cancer. 6:122007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Trosko JE: From adult stem cells to cancer
stem cells: Oct-4 Gene, cell-cell communication, and hormones
during tumor promotion. Ann N Y Acad Sci. 1089:36–58. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pan G and Thomson JA: Nanog and
transcriptional networks in embryonic stem cell pluripotency. Cell
Res. 17:42–49. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chiou SH, Wang ML, Chou YT, Chen CJ, Hong
CF, Hsieh WJ, Chang HT, Chen YS, Lin TW, Hsu HS and Wu CW:
Coexpression of Oct4 and Nanog enhances malignancy in lung
adenocarcinoma by inducing cancer stem cell-like properties and
epithelial-mesenchymal transdifferentiation. Cancer Res.
70:10433–10444. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang J, Espinoza LA, Kinders RJ, Lawrence
SM, Pfister TD, Zhou M, Veenstra TD, Thorgeirsson SS and Jessup JM:
NANOG modulates stemness in human colorectal cancer. Oncogene.
32:4397–4405. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jeter CR, Liu B, Liu X, Chen X, Liu C,
Calhoun-Davis T, Repass J, Zaehres H, Shen JJ and Tang DG: NANOG
promotes cancer stem cell characteristics and prostate cancer
resistance to androgen deprivation. Oncogene. 30:3833–3845. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ibrahim EE, Babaei-Jadidi R, Saadeddin A,
Spencer-Dene B, Hossaini S, Abuzinadah M, Li N, Fadhil W, Ilyas M,
Bonnet D and Nateri AS: Embryonic NANOG activity defines colorectal
cancer stem cells and modulates through AP1- and TCF-dependent
mechanisms. Stem Cells. 30:2076–2087. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang L, Zhang X, Zhang M, Zhang J, Sheng
Y, Sun X, Chen Q and Wang LX: Increased Nanog expression promotes
tumor development and Cisplatin resistance in human esophageal
cancer cells. Cell Physiol Biochem. 30:943–952. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Serra M, Scotlandi K, Manara MC, Maurici
D, Lollini PL, De Giovanni C, Toffoli G and Baldini N:
Establishment and characterization of multidrug-resistant human
osteosarcoma cell lines. Anticancer Res. 13:323–329.
1993.PubMed/NCBI
|
|
38
|
Scotlandi K, Serra M, Manara MC, Lollini
PL, Maurici D, Del Bufalo D and Baldini N: Pre-treatment of human
osteosarcoma cells with N-methylformamide enhances P-glycoprotein
expression and resistance to doxorubicin. Int J Cancer. 58:95–101.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Taylor CW, Dalton WS, Parrish PR, Gleason
MC, Bellamy WT, Thompson FH, Roe DJ and Trent JM: Different
mechanisms of decreased drug accumulation in doxorubicin and
mitoxantrone resistant variants of the MCF7 human breast cancer
cell line. Br J Cancer. 63:923–929. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rajkumar T and Yamuna M: Multiple pathways
are involved in drug resistance to doxorubicin in an osteosarcoma
cell line. Anticancer Drugs. 19:257–265. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen CJ, Chin JE, Ueda K, Clark DP, Pastan
I, Gottesman MM and Roninson IB: Internal duplication and homology
with bacterial transport proteins in the mdr1 (P-glycoprotein) gene
from multidrug-resistant human cells. Cell. 47:381–389. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Baldini N, Scotlandi K, Barbanti-Bròdano
G, Manara MC, Maurici D, Bacci G, Bertoni F, Picci P, Sottili S,
Campanacci M, et al: Expression of P-glycoprotein in high-grade
osteosarcomas in relation to clinical outcome. N Engl J Med.
333:1380–1385. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Baldini N, Scotlandi K, Serra M, Picci P,
Bacci G, Sottili S and Campanacci M: P-glycoprotein expression in
osteosarcoma: A basis for risk-adapted adjuvant chemotherapy. J
Orthop Res. 17:629–632. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ferrari S and Palmerini E: Adjuvant and
neoadjuvant combination chemotherapy for osteogenic sarcoma. Curr
Opin Oncol. 19:341–346. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chou AJ and Gorlick R: Chemotherapy
resistance in osteosarcoma: Current challenges and future
directions. Expert Rev Anticancer Ther. 6:1075–1085. 2006.
View Article : Google Scholar : PubMed/NCBI
|