Introduction
Laryngeal carcinoma, the second most common type of
head and neck malignancy, accounts for ~2.4% of all newly diagnosed
malignant tumors (1). In the United
States, it is estimated that there will be 13,360 new cases of
laryngeal carcinoma and 3,660 associated mortalities in 2017. Among
these estimated new cases and mortalities, males account for 79.12%
of the new cases and 80.33% of the mortalities (2). This is primarily due to tobacco and
alcohol abuse, and results in substantial annual morbidity and
mortality (3). Laryngeal squamous
cell carcinoma (LSCC), the most common subtype of laryngeal
carcinoma, accounts for >95% of laryngeal carcinoma cases
(4). Currently, the primary
therapeutic strategy for LSCC is surgical intervention or total
laryngectomy, followed by radiation therapy and chemotherapy
(5). These treatments have a
favorable curative effect on early-stage patients; however, are
associated with a poor prognosis in more advanced-stage cases
(6). Despite improvements in the
diagnosis and treatment of LSCC, the 5-year survival rate has not
significantly increased in the past 20 years (7). Therefore, an improved understanding of
the molecular mechanisms of LSCC carcinogenesis and progression is
essential in the development of novel diagnostic and therapeutic
targets for patients with LSCC.
MicroRNAs (miRNAs) have been demonstrated to be a
novel method of gene regulation mechanism (8). miRNAs are a large family of small (22–25
nucleotides in length) non-protein-coding, endogenous and
single-stranded RNAs, which regulate post-transcriptional gene
expression through association with the 3′-untranslated region
(UTR) of target mRNAs, resulting in mRNA degradation or
translational inhibition (9,10). Currently, >30% of human
protein-coding genes are considered to be accommodated by miRNAs
(11). Increasing evidence suggested
that miRNAs serve a critical role in a number of physiological and
pathological processes, including cell growth, development,
differentiation, apoptosis, survival, migration and invasion
(12,13). The abnormal expression of miRNAs is
hypothesized to be involved in various types of human cancer,
including human LSCC (14–16). Dysregulated miRNAs may function as
tumor suppressors or oncogenes, in the carcinogenesis and
development of various types of human cancer (17). These results suggested that miRNAs may
be a novel target for LSCC therapy.
A majority of studies have indicated that miRNA-125b
(miR-125b) is aberrantly expressed in various types of cancer
(18–20). However, to the best of our knowledge,
there are currently no studies on miR-125b in LSCC. In the present
study, the expression and function of miR-125b in LSCC was
investigated. The results of the present study demonstrated that
miR-125b was markedly downregulated in LSCC tissues and cell lines.
The statistical analysis also indicated that the expression level
of miR-125b was significantly associated with clinical stage and
alcohol history in patients with LSCC. Upregulation of miR-125b
decreased cell growth, migration and invasion by directly targeting
signal transducer and activator of transcription 3 (STAT3). These
results suggested that miR-125b may be investigated for further
therapy in patients with LSCC.
Materials and methods
Clinical specimens
Tissue samples, LSCC tissues and their corresponding
adjacent non-neoplastic tissues, were obtained from 52 patients
with LSCC undergoing surgery resection at The First Affiliated
Hospital of Xinjiang Medical University (Urumqi, China). Patient
clinical features are presented in Table
I. All patients had not received other therapies, including
radiotherapy and chemotherapy, prior to surgery. Tissue samples
were immediately preserved in liquid nitrogen following excision
from patients and subsequently transferred to a −80°C refrigerator
until use. The present study was approved by the Human Research
Ethics Committee of The First Affiliated Hospital of Xinjiang
Medical University. Written informed consent was obtained from all
patients used in the present study, as well as their
clinicopathological features.
 | Table I.Association between the expression of
miRNA-125b and clinicopathological factors in patients with
laryngeal squamous cell carcinoma. |
Table I.
Association between the expression of
miRNA-125b and clinicopathological factors in patients with
laryngeal squamous cell carcinoma.
|
|
| Relative miR-125b
expression |
|
|---|
|
|
|
|
|
|---|
| Clinical
features | Case number | High | Low | P-value |
|---|
| Sex |
|
|
| 0.697 |
|
Male | 45 | 16 | 29 |
|
|
Female | 7 | 3 | 4 |
|
| Age, years |
|
|
| 0.559 |
|
<60 | 21 | 9 | 12 |
|
|
≥65 | 31 | 10 | 21 |
|
| Smoking
history |
|
|
| 0.510 |
|
Negative | 13 | 6 | 7 |
|
|
Positive | 39 | 13 | 26 |
|
| Alcohol
history |
|
|
| 0.010a |
|
Negative | 23 | 13 | 10 |
|
|
Positive | 29 | 6 | 23 |
|
| T stage |
|
|
| 0.389 |
|
T0/1/2 | 25 | 11 | 14 |
|
|
T3/4 | 27 | 8 | 19 |
|
| N stage |
|
|
| 1.000 |
|
Negative | 34 | 12 | 22 |
|
|
Positive | 18 | 7 | 11 |
|
| M stage |
|
|
| 0.247 |
|
Negative | 29 | 13 | 16 |
|
|
Positive | 23 | 6 | 17 |
|
| Clinical stage |
|
|
| 0.043b |
| Low
(I–II) | 25 | 13 | 12 |
|
| High
(III–IV) | 27 | 6 | 21 |
|
Cell culture
The LSCC cell line (AMC-HN-8) and a normal human
keratinocyte cell line (HaCaT) were obtained from Shanghai
Institute of Biochemistry and Cell Biology (Shanghai, China). Cells
were maintained in Dulbecco's modified Eagle's medium (DMEM)
supplemented with 10% (v/v) fetal bovine serum (FBS), and 1% (v/v)
penicillin/streptomycin (all Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), at 37°C in a humidified atmosphere containing 5%
CO2.
Transfection
AMC-HN-8 cells were transfected with miR-125b mimics
and negative control (NC) miRNA, or cotransfected with luciferase
reporter plasmid [pGL3-STAT3-3′UTR-wild-type (Wt) or
pGL3-STAT3-3′UTR-mutated (Mut)] using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The sequence of the miR-125b mimic was
5′-UCCCUGAGACCCUAACUUGUGA-3′. The sequence of the NC mimic was
5′-UUCUCCGAACGUGUCACGUTT-3′. MicroRNA mimics and luciferase report
plasmid were synthesized and obtained from Shanghai GenePharma
Company (Shanghai, China).
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
Total RNA was isolated from LSCC tissues, adjacent
non-neoplastic tissues and cells using Trizol reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. Reverse transcription was performed using the Moloney
Murine Leukemia Virus Reverse Transcription system (Promega
Corporation, Madison, WI, USA). qPCR was subsequently performed
using SYBR® Premix Ex Taq™ II (Takara, China)
and the ABI 7300 Real-Time PCR detection system, according to the
manufacturer's protocol. The thermocycling conditions for qPCR was
as follows: 5 min at 95°C; followed by 40 cycles of 95°C for 30 sec
and 65°C for 45 sec. The primer sequences used for qPCR were:
miR-125b forward, 5′-GCUCCCUGAGACCCUAAC-3′ and reverse,
5′-CAGTGCAGGGTCCGAGGT-3′; U6 forward, 5′-CGCTTCGGCAGCACATATACTA-3′
and reverse, 5′-GCGAGCACAGAATTAATACGAC-3′; STAT3 forward,
5′-CCCATACCTGAAGACCAAGTTTATC-3′ and reverse,
5′-TGGAAATAATGGTGAAGGTGCTG-3′; GAPDH forward,
5′-TGCACCACCAACTGCTTA-3′ and reverse, 5′-GGATGCAGGGATGATGTTC-3′.
Results were quantified using the 2−ΔΔCq method
(21).
Cell Counting Kit-8 (CCK8) assay
The role of miR-125b on LSCC cell proliferation was
evaluated using the CCK8 assay (Dojindo Molecular Technologies,
Inc., Kumamoto, Japan). A total of 24 h following transfection,
3000 AMC-HN-8 cells were added into each well of 96-well plates in
triplicate. At various times (24, 48, 72 and 96 h) following
treatment, the CCK8 assay was performed. A total of 10 µl CCK8
solution was added into each well and incubated for 2 h at 37°C.
The optical density at 450 nm for each well was measured using an
ELISA reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The
suppression rate was calculated using the following formula:
Suppression rate=(1-ODmiR-125b/ODNC) ×100.
Each experiment was performed at least three times.
Migration and invasion assay
The function of miR-125b in cell motility was
measured using Transwell chambers (Costar; Corning Incorporated,
Corning, NY, USA) with 8-µm pore polycarbonate membranes. A total
of 48 h following transfection, 3×104 AMC-HN-8 cells in
200 µl DMEM without FBS were added into the upper chamber of the
Transwell chamber in a 24-well plate. In the lower chamber, 500 µl
DMEM containing 20% FBS was used as a chemoattractant. The
Transwell chamber was precoated with Matrigel (BD Biosciences, San
Jose, CA) for the invasion assay and without Matrigel for the
migration assay. Cells were incubated at 37°C for 24 h. The
chambers were subsequently fixed with 100% methanol for 10 min and
stained with 0.1% crystal violet for 5 min at room temperature.
Cells that did not migrate or invade through the membrane were
removed using a cotton swab. The membranes were subsequently viewed
under an inverted microscope and images were captured
(magnification, ×200). All the experiments were performed in
triplicate.
Bioinformatic analysis
The potential targets of miR-125b were analyzed
using miRanda (http://www.microrna.org) and TargetScan (http://www.targetscan.org/).
Western blot analysis
A total of 72 h following transfection, cells were
lysed in cold radioimmunoprecipitation assay buffer (Beyotime
Institute of Biotechnology, Haimen, China) containing protease and
phosphatase inhibitors. Bicinchoninic Acid Assay kit (Beyotime
Institute of Biotechnology) was used to determine the protein
concentrations. Equal amounts of protein were subjected to SDS-PAGE
and subsequently transferred to polyvinylidene difluoride membranes
(EMD Millipore, Billerica, MA, USA). The membranes were blocked
with 5% non-fat milk at room temperature for 2 h, and subsequently
incubated with primary antibodies against STAT3 (1:1,000 dilution;
catalog: #9139; Cell Signaling Technology, Inc., Danvers, MA, USA)
and β-actin (1:1,000 dilution; sc-130065; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) at 4°C overnight. Membranes were washed with
TBS containing 0.1% Tween-20 and incubated with the corresponding
horseradish peroxidase-conjugated secondary antibody (1:5,000
dilution; sc-2005; Santa Cruz Biotechnology, Inc.) for 2 h at room
temperature. The bound antibodies were visualized using an enhanced
chemiluminescence kit (GE Healthcare Life Sciences, Chalfont, UK)
and analyzed using Quantity One software (Bio-Rad Laboratories,
Inc.).
Dual-luciferase reporter assay
The human LSCC cells, plated in a 24-well plate at
~70% confluence, were transfected with miR-125b mimics, NC and
cotransfected with luciferase reporter plasmids
(pGL3-STAT3-3′UTR-Wt or pGL3-STAT3-3′UTR-Mut) using Lipofectamine
2000. A total of 48 h following transfection, Renilla and firefly
luciferase activities were measured using the Dual-Luciferase
Reporter assay system (Promega Corporation), according to the
manufacturer's protocol. Renilla luciferase activity was used as a
control. Each sample was assayed in triplicate.
Statistical analysis
Data were presented as the mean ± standard deviation
and compared using two-tailed Student's t-test or one-way analysis
of variance using SPSS software (version 17.0; SPSS, Inc., Chicago,
IL, USA). The SNK method was applied to compare between two groups
in multiple groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-125b expression is downregulated
in LSCC
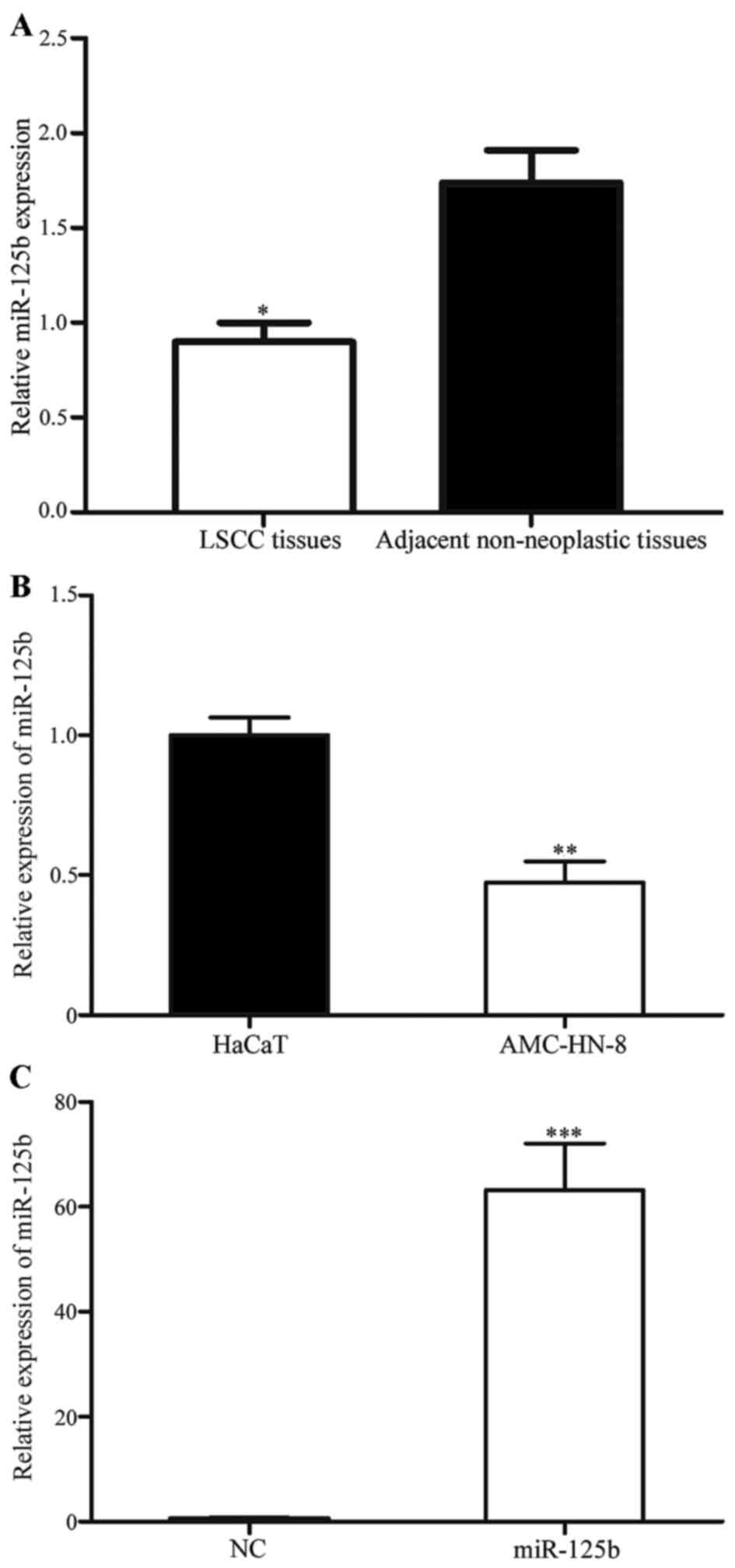
The expression of miR-125b in 52 paired LSCC tissues
and corresponding adjacent non-neoplastic tissues was studied using
quantitative RT-PCR. As presented in Fig.
1A, miR-125b was significantly downregulated in LSCC tissues
compared with the corresponding adjacent non-neoplastic tissues
(P<0.05). In addition, the expression level of miR-125b in an
LSCC and normal human keratinocyte cell line was detected. As
presented in Fig. 1B, miR-125b was
also significantly downregulated in AMC-HN-8 cells compared with
HaCaT cells (P<0.05). These results indicated that miR-125b may
serve important roles in LSCC.
To investigate the roles of miR-125b in LSCC cells,
miR-125b mimics were transfected into AMC-HN-8 cells using
Lipofectamine 2000. To assess the transfection efficiency, RT-qPCR
was performed following transfection. As presented in Fig. 1C, miR-125b was significantly
upregulated in cells transfected with miR-125b mimics compared with
cells transfected with the NC (P<0.05).
Association between miR-125b
expression and clinicopathological features of patients with
LSCC
To investigate whether miR-125b expression was
associated with the clinicopathological features of patients with
LSCC, statistical analysis was used. As presented in Table I, the statistical analysis
demonstrated that miR-125b expression was significantly associated
with clinical stage (P=0.043) and alcohol history (P=0.010).
However, there were no statistically significant associations
between miR-125b expression and other clinicopathological features
(P>0.05).
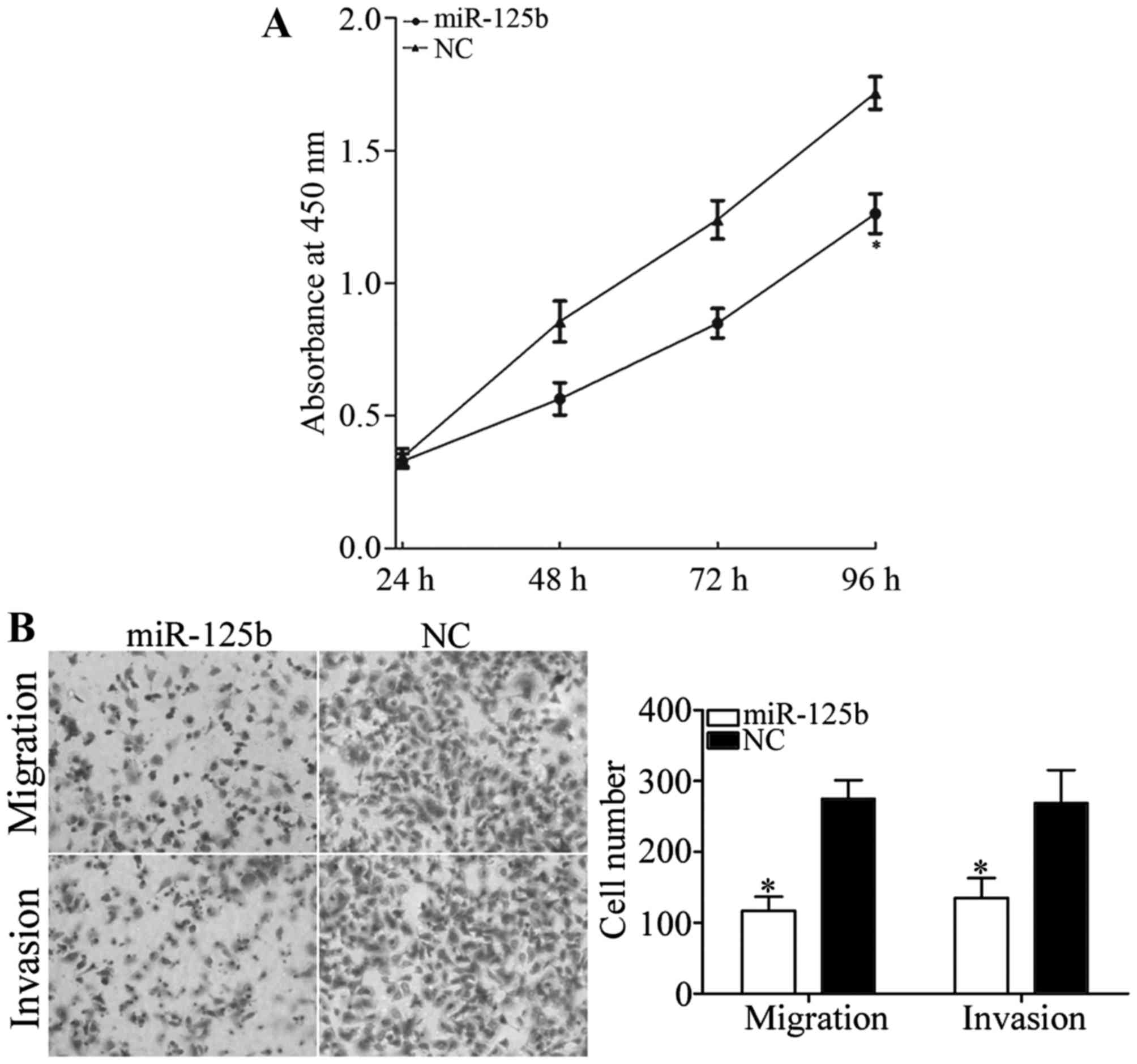
miR-125b decreases cell growth,
migration and invasion in AMC-HN-8 cells
The CCK8 assay was performed to investigate the
roles of miR-125b on cell growth. As presented in Fig. 2, miR-125b significantly decreased cell
growth in AMC-HN-8 cells compared with the NC (P<0.05). The
inhibition rate of miR-125b in AMC-HN-8 cells was 26.51±3.5%.
Transwell chambers were used to investigate the
roles of miR-125b in cellular migration and invasion. As presented
in Fig. 2B, miR-125b significantly
inhibited AMC-HN-8 cell migration and invasion compared with the NC
(P<0.05). The results revealed that miR-125b serves a
suppressive role in LSCC cell motility.
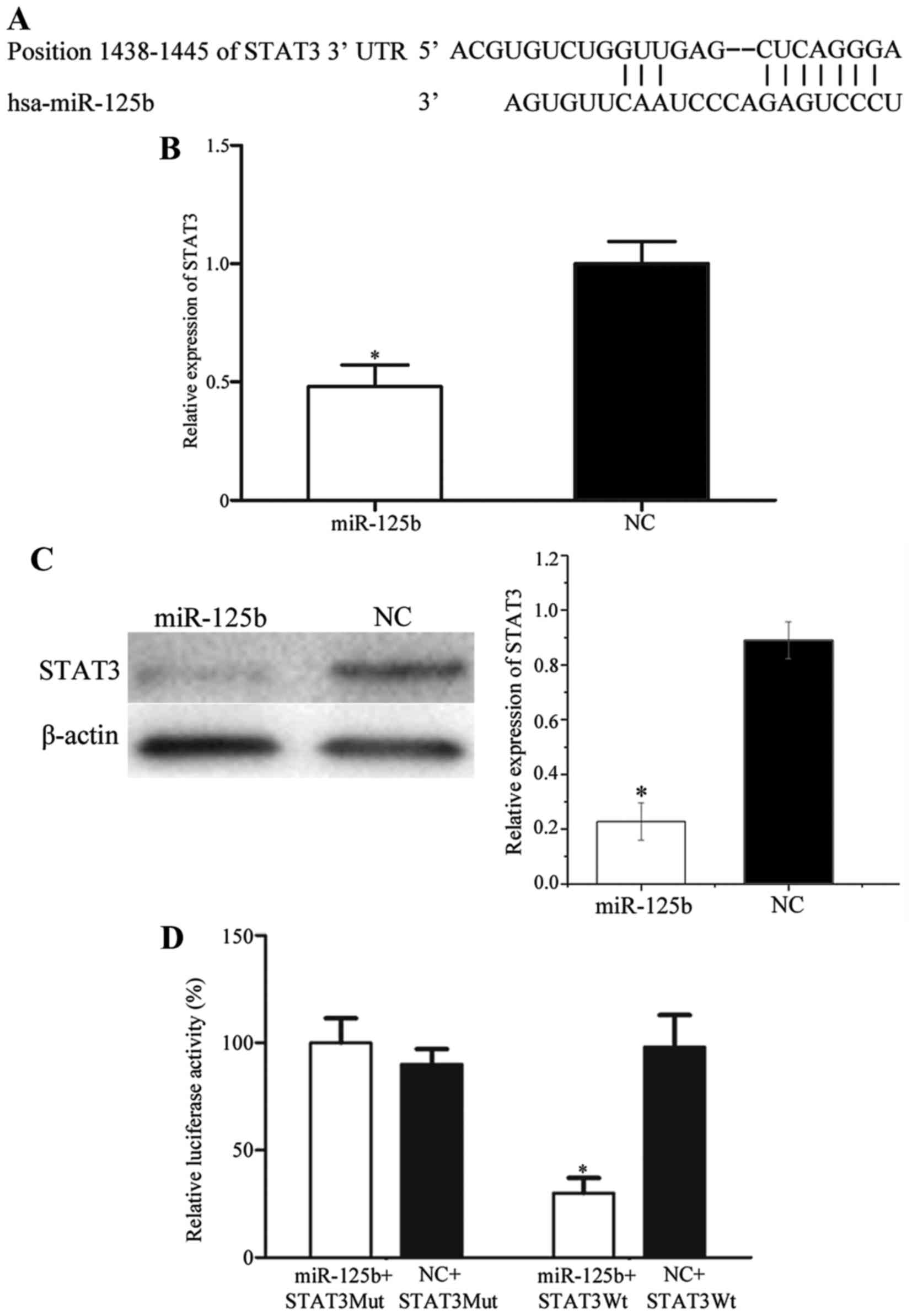
STAT3 is a direct target gene of
miR-125b in vitro
To investigate the target genes of miR-125b, public
databases (miRanda and TargetScan) were used. STAT3 was predicted
to be a potential target of miR-125b (Fig. 3A). Subsequently, RT-qPCR and western
blot analysis were performed to investigate the regulation of
miR-125b on STAT3 expression at the mRNA and protein level in LSCC
cells. As presented in Fig. 3B and C,
STAT3 was significantly downregulated in AMC-HN-8 cells transfected
with miR-125b compared with cells transfected with the NC
(P<0.05). Dual-luciferase reporter assays were also performed to
determine whether STAT3 was a direct target gene of miR-125b. As
presented in Fig. 3D, luciferase
activity was significantly downregulated in the miR-125b mimics and
pGL3-STAT3-3′UTR-Wt group, whereas there was no statistical
difference in the miR-125b mimics and pGL3-STAT3-3′UTR-Mut group,
compared with the respective NC groups. These results indicated
that STAT3 was a direct gene of miR-125b in vitro.
Discussion
miR-125 has two subtypes, miR-125a and miR-125b.
miR-125b is encoded by the miR-125b-1 and miR-125b-2 genes, which
are mapped to 11q24.1 and 21q21.1, respectively. However,
miR-125b-1 and miR-125b-2 precursors are processed to form the same
mature miRNA, miR-125b (22).
miR-125b has been observed to be downregulated in various types of
cancer, including oral cancer (23),
bladder cancer (18), liver cancer
(19), osteosarcoma (20) and endometrial cancer (24). Previous studies have also demonstrated
that miR-125b was upregulated in a number of types of cancer,
including prostate cancer (25),
glioblastoma (26) and pediatric
acute promyelocytic leukemia (27).
However, there are no studies investigating the expression of
miR-125b in LSCC. In the present study, it was observed that
miR-125b was downregulated in LSCC tissue samples and cell lines.
The statistical analysis also demonstrated that the expression
level of miR-125b was associated with the clinical stage and
alcohol history of patients with LSCC. The results suggested that
miR-125b may serve important roles in LSCC.
Currently, the majority of studies have demonstrated
that miR-125b functions as a tumor suppressor. For example, in
bladder cancer, miR-125b inhibited cell proliferation and motility,
and enhanced cell apoptosis by regulating the expression of
nicotinamide-adenine dinucleotide-dependent protein deacetylase
sirtuin-7 and matrix metalloproteinase-13 (22,28).
Furthermore, miR-125b decreased bladder cancer cell colony
formation efficiency in vitro and the development of tumors
in nude mice by targeting transcription factor E2F3 (18). In osteosarcoma, Liu et al
(20) reported that miR-125b
suppressed cell proliferation and migration via inhibition of
STAT3. In endometrial cancer, miR-125b targeted receptor
tyrosine-protein kinase erbB-2 to inhibit cell invasion (24). In human liver cancer, miR-125b was
demonstrated to inhibit cell proliferation, migration and invasion
by targeting LIN28B2 (19).
Upregulation of miR-125b increased human hepatocellular carcinoma
(HCC) cell sensitivity to 5-FU by regulating the expression of
hexokinase II (29). However,
miR-125b has also been verified to be an oncogene. In prostate
cancer, miR-125b enhanced prostatic xenograft tumors proliferation
through downregulation of p53 and Bcl-2-binding component 3
(30). In glioblastoma, Wu et
al (26) verified that miR-125b
was upregulated and significantly associated with poor prognosis.
miR-125b functioned as an oncogene by suppressing cellular
apoptosis and enhancing cellular proliferation. These opposing
studies indicated that the functions of miR-125b in cancers are
tissue-type dependent. In the present study, miR-125b was
demonstrated to inhibit LSCC cell proliferation, migration and
invasion. The present study expanded the expression and functions
of miR-125b in cancer.
Identification of miR-125b target genes is essential
for understanding its functions in LSCC carcinogenesis and cancer
development. It is also important for investigating novel targeted
therapies of LSCC. In the present study, an important molecular
association between miR-125b and STAT3 was verified. The STAT
family consists of seven members (STAT1, STAT2, STAT3, STAT4,
STAT5a, STAT5b and STAT6) and STAT3 is a central transcription
factor in the STAT family (31). In
1994, STAT3 was first verified to be an interleukin-6-activated
acute-phase response factor (32).
Multiple studies demonstrated that STAT3 serves essential roles in
a variety of human cancer types, including LSCC (33–35). STAT3
serves important roles in mediating a number of oncogenic signaling
pathways, including epidermal growth factor receptor signaling and
Janus kinase (JAK)/STAT signaling pathways, and contributes to
tumor physiological and pathological processes, including growth,
survival, resistance to apoptosis, cell-cycle progression,
angiogenesis and metastasis (36–38).
Inhibition of STAT3 by pharmacological agents and genetic
interference, suppressed cell growth, metastasis, enhanced
apoptosis and decreased carcinogenesis (39,40). These
studies suggested that STAT3 may be a therapeutic target in the
treatment of LSCC.
STAT3 has also been observed to be regulated by
multiple miRNAs in a number of kinds of cancer. In HCC, miR-637
suppressed cell proliferation and enhanced cellular apoptosis
through downregulation of STAT3 expression (41). miR-124 also inhibited HCC cell growth
by directly targeting STAT3 (42). In
pancreatic cancer, miR-130b decreased cell growth and invasion via
inhibition of STAT3 (43). In gastric
cancer, miR-874 targeted the STAT3/vascular endothelial growth
factor A signaling pathway and functioned as a tumor suppressor
(44). miR-375 was involved in
helicobacter pylori-induced gastric tumorigenesis by
inhibiting the activity of JAK2/STAT3 signaling (45). However, there are no studies about the
regulation of STAT3 by miRNA in LSCC. In the present study,
upregulation of miR-125b in LSCC revealed that miR-125b decreases
cell growth and motility through regulation of STAT3. These studies
suggested that miR-125b/STAT3-based targeted therapy may be a novel
treatment for LSCC.
In conclusion, to the best of our knowledge, this
was the first study to demonstrate that miR-125b was downregulated
in LSCC, and associated with clinical stage and alcohol history. In
addition, it was also demonstrated that miR-125b decreases cell
growth, migration and invasion by directly targeting STAT3 in LSCC.
The results revealed that the expression and function of miR-125b
and its target gene STAT3 in LSCC, may aid in understanding the
molecular mechanism of tumorigenesis and development, as well as
providing a theoretical basis to investigate miR-125b as a further
treatment target in LSCC.
References
|
1
|
Sun X, Song Y, Tai X, Liu B and Ji W:
MicroRNA expression and its detection in human supraglottic
laryngeal squamous cell carcinoma. Biomed Rep. 1:743–746.
2013.PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Luo J, Wu J, Li Z, Qin H, Wang B, Wong TS,
Yang W, Fu QL and Lei W: miR-375 suppresses IGF1R expression and
contributes to inhibition of cell progression in laryngeal squamous
cell carcinoma. Biomed Res Int. 2014:3745982014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guo Y, Fu W, Chen H, Shang C and Zhong M:
miR-24 functions as a tumor suppressor in Hep2 laryngeal carcinoma
cells partly through down-regulation of the S100A8 protein. Oncol
Rep. 27:1097–1103. 2012.PubMed/NCBI
|
|
6
|
Cosetti M, Yu GP and Schantz SP: Five-year
survival rates and time trends of laryngeal cancer in the US
population. Arch Otolaryngol Head Neck Surg. 134:370–379. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang W, Lin P, Han C, Cai W, Zhao X and
Sun B: Vasculogenic mimicry contributes to lymph node metastasis of
laryngeal squamous cell carcinoma. J Exp Clin Cancer Res.
29:602010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ying SY, Chang DC, Miller JD and Lin SL:
The microRNA: Overview of the RNA gene that modulates gene
functions. Methods Mol Biol. 342:1–18. 2006.PubMed/NCBI
|
|
10
|
Maroney PA, Yu Y and Nilsen TW: MicroRNAs,
mRNAs, and translation. Cold Spring Harb Symp Quant Biol.
71:531–535. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao Y and Srivastava D: A developmental
view of microRNA function. Trends Biochem Sci. 32:189–197. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yates LA, Norbury CJ and Gilbert RJ: The
long and short of microRNA. Cell. 153:516–519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tahara H, Kay MA, Yasui W and Tahara E:
MicroRNAs in cancer: The 22nd Hiroshima cancer Seminar/the 4th
Japanese Association for RNA Interference Joint International
Symposium, 30 August 2012, Grand Prince Hotel Hiroshima. Jpn J Clin
Oncol. 43:579–582. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Karatas OF, Yuceturk B, Suer I, Yilmaz M,
Cansiz H, Solak M, Ittmann M and Ozen M: Role of miR-145 in human
laryngeal squamous cell carcinoma. Head Neck. 38:260–266. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tian L, Zhang J, Ge J, Xiao H, Lu J, Fu S,
Liu M and Sun Y: MicroRNA-205 suppresses proliferation and promotes
apoptosis in laryngeal squamous cell carcinoma. Med Oncol.
31:7852014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tian Y, Fu S, Qiu GB, Xu ZM, Liu N, Zhang
XW, Chen S, Wang Y, Sun KL and Fu WN: MicroRNA-27a promotes
proliferation and suppresses apoptosis by targeting PLK2 in
laryngeal carcinoma. BMC Cancer. 14:6782014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu D, Zhou Y, Pan H, Zhou J, Fan Y and Qu
P: microRNA-99a inhibiting cell proliferation, migration and
invasion by targeting fibroblast growth factor receptor 3 in
bladder cancer. Oncol Lett. 7:1219–1224. 2014.PubMed/NCBI
|
|
18
|
Huang L, Luo J, Cai Q, Pan Q, Zeng H, Guo
Z, Dong W, Huang J and Lin T: MicroRNA-125b suppresses the
development of bladder cancer by targeting E2F3. Int J Cancer.
128:1758–1769. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liang L, Wong CM, Ying Q, Fan DN, Huang S,
Ding J, Yao J, Yan M, Li J, Yao M, et al: MicroRNA-125b
suppressesed human liver cancer cell proliferation and metastasis
by directly targeting oncogene LIN28B2. Hepatology. 52:1731–1740.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu LH, Li H, Li JP, Zhong H, Zhang HC,
Chen J and Xiao T: miR-125b suppresses the proliferation and
migration of osteosarcoma cells through down-regulation of STAT3.
Biochem Biophys Res Commun. 416:31–38. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu D, Ding J, Wang L, Pan H, Zhou Z, Zhou
J and Qu P: microRNA-125b inhibits cell migration and invasion by
targeting matrix metallopeptidase 13 in bladder cancer. Oncol Lett.
5:829–834. 2013.PubMed/NCBI
|
|
23
|
Henson BJ, Bhattacharjee S, O'Dee DM,
Feingold E and Gollin SM: Decreased expression of miR-125b and
miR-100 in oral cancer cells contributes to malignancy. Genes
Chromosomes Cancer. 48:569–582. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shang C, Lu YM and Meng LR: MicroRNA-125b
down-regulation mediates endometrial cancer invasion by targeting
ERBB2. Med Sci Monit. 18:BR149–BR155. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Amir S, Ma AH, Shi XB, Xue L, Kung HJ and
White RW Devere: Oncomir miR-125b suppresses p14(ARF) to modulate
p53-dependent and p53-independent apoptosis in prostate cancer.
PLoS One. 8:e610642013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu N, Lin X, Zhao X, Zheng L, Xiao L, Liu
J, Ge L and Cao S: MiR-125b acts as an oncogene in glioblastoma
cells and inhibits cell apoptosis through p53 and
p38MAPK-independent pathways. Br J Cancer. 109:2853–2863. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang H, Luo XQ, Feng DD, Zhang XJ, Wu J,
Zheng YS, Chen X, Xu L and Chen YQ: Upregulation of microRNA-125b
contributes to leukemogenesis and increases drug resistance in
pediatric acute promyelocytic leukemia. Mol Cancer. 10:1082011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Han Y, Liu Y, Zhang H, Wang T, Diao R,
Jiang Z, Gui Y and Cai Z: Hsa-miR-125b suppresses bladder cancer
development by down-regulating oncogene SIRT7 and oncogenic long
non-coding RNA MALAT1. FEBS Lett. 587:3875–3882. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jiang JX, Gao S, Pan YZ, Yu C and Sun CY:
Overexpression of microRNA-125b sensitizes human hepatocellular
carcinoma cells to 5-fluorouracil through inhibition of glycolysis
by targeting hexokinase II. Mol Med Rep. 10:995–1002.
2014.PubMed/NCBI
|
|
30
|
Shi XB, Xue L, Ma AH, Tepper CG, Kung HJ
and White RW: miR-125b promotes growth of prostate cancer xenograft
tumor through targeting pro-apoptotic genes. Prostate. 71:538–549.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hennighausen L and Robinson GW:
Interpretation of cytokine signaling through the transcription
factors STAT5A and STAT5B. Genes Dev. 22:711–721. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lütticken C, Wegenka UM, Yuan J, Buschmann
J, Schindler C, Ziemiecki A, Harpur AG, Wilks AF, Yasukawa K, Taga
T, et al: Association of transcription factor APRF and protein
kinase Jak1 with the interleukin-6 signal transducer gp130.
Science. 263:89–92. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dai L, Liu T, Zhou X, Wei Q, Li Y, Cui C
and Li M: Expression and significance of phosphated signal
transducer and activator of transcription 3 and p53 protein in
laryngeal squamous cell carcinoma. Lin Chung Er Bi Yan Hou Tou Jing
Wai Ke Za Zhi. 27:991–994. 2013.(In Chinese). PubMed/NCBI
|
|
34
|
Zhao H, Wamg Y and He X: Expression of
p-STAT3 in laryngeal squamous carcinoma and its correlation with
PTEN. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 27:981–985.
2013.(In Chinese). PubMed/NCBI
|
|
35
|
Wang L, Duan X, Tang X, Shao Y, Zhao R,
Qian X and Gao X: Clinical significance of Stat3 and Cyclin D1
expression in laryngeal squamous cell carcinoma. Lin Chung Er Bi
Yan Hou Tou Jing Wai Ke Za Zhi. 25:966–969. 2011.(In Chinese).
PubMed/NCBI
|
|
36
|
Zhong Z, Wen Z and Darnell JE Jr: Stat3: A
STAT family member activated by tyrosine phosphorylation in
response to epidermal growth factor and interleukin-6. Science.
264:95–98. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bromberg J and Darnell JE Jr: The role of
STATs in transcriptional control and their impact on cellular
function. Oncogene. 19:2468–2473. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kamran MZ, Patil P and Gude RP: Role of
STAT3 in cancer metastasis and translational advances. Biomed Res
Int. 2013:4218212013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xiong A, Yang Z, Shen Y, Zhou J and Shen
Q: Transcription factor STAT3 as a novel molecular target for
cancer prevention. Cancers (Basel). 6:926–957. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Siveen KS, Sikka S, Surana R, Dai X, Zhang
J, Kumar AP, Tan BK, Sethi G and Bishayee A: Targeting the STAT3
signaling pathway in cancer: Role of synthetic and natural
inhibitors. Biochim Biophys Acta. 1845:136–154. 2014.PubMed/NCBI
|
|
41
|
Zhang JF, He ML, Fu WM, Wang H, Chen LZ,
Zhu X, Chen Y, Xie D, Lai P, Chen G, et al: Primate-specific
microRNA-637 inhibits tumorigenesis in hepatocellular carcinoma by
disrupting signal transducer and activator of transcription 3
signaling. Hepatology. 54:2137–2148. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lu Y, Yue X, Cui Y, Zhang J and Wang K:
MicroRNA-124 suppresses growth of human hepatocellular carcinoma by
targeting STAT3. Biochem Biophys Res Commun. 441:873–879. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhao G, Zhang JG, Shi Y, Qin Q, Liu Y,
Wang B, Tian K, Deng SC, Li X, Zhu S, et al: MiR-130b is a
prognostic marker and inhibits cell proliferation and invasion in
pancreatic cancer through targeting STAT3. PLoS One. 8:e738032013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang X, Tang J, Zhi X, Xie K, Wang W, Li
Z, Zhu Y, Yang L, Xu H and Xu Z: miR-874 functions as a tumor
suppressor by inhibiting angiogenesis through STAT3/VEGF-A pathway
in gastric cancer. Oncotarget. 6:1605–1617. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Miao L, Liu K, Xie M, Xing Y and Xi T:
miR-375 inhibits Helicobacter pylori-induced gastric carcinogenesis
by blocking JAK2-STAT3 signaling. Cancer Immunol Immunother.
63:699–711. 2014. View Article : Google Scholar : PubMed/NCBI
|