Introduction
Hepatocellular carcinoma (HCC) accounts for between
85–90% of primary liver cancers, which is the fifth most common
cancer and the third leading cause of cancer-associated mortality
worldwide (1). Hepatitis B virus
(HBV) infection is an important factor for HCC occurrence (2). The association between HBV and HCC has
been demonstrated by multiple epidemiological studies: High HBV
prevalence may result in a high frequency of occurrence of HCC, and
the risk of HCC in HBV carriers is significantly increased compared
with that of non-carriers (3,4). The main reason for poor prognosis of HCC
is invasion of the portal vein, leading to the formation of a tumor
thrombus and intrahepatic spread, which results in a high
intrahepatic metastatic rate and recurrence rate following surgery
(5).
Hepatitis B virus X protein (HBx), an important
regulatory protein, may enhance the transcription and replication
of HBV (6) and regulate biological
processes, including host gene transcription, cell cycle, apoptosis
and oxidative stress (7,8). Previous studies have demonstrated that
HBx is an important factor in the development of HBV-associated HCC
(9–11). Thus far, mechanisms of HCC metastasis
promotion by HBx include: Promotion of epithelial-mesenchymal
transition (EMT) (12,13); degradation of the extracellular matrix
(ECM) (14,15); reduction of cell-ECM interactions
(16); alteration of the cellular
morphology conductive to migration and motility (17,18);
repression of miRNA-148a (19) or
upregulation of miRNA-143 (20). HBx
may also upregulate intracellular ROS levels (21,22), and
ROS have been demonstrated to regulate the expression of EMT and
metastasis-associated genes, including E-cadherin, integrin and
matrix metalloproteinases in HCC cells, which contribute to HCC
metastasis (23,24).
Thioredoxin interacting protein (TXNIP), also known
as vitamin D3 upregulated protein 1, is involved in a wide range of
cellular processes, including proliferation, apoptosis, lipid and
glucose metabolism and redox regulation (25). TXNIP may also be involved in the
metastasis of a variety of tumors (26–29) and
may act as a key mediator of intracellular ROS levels through the
TXNIP/thioredoxin/ROS axis (25),
which contributes to the progression of tumors. However, the
function of TXNIP in the metastasis of HCC remains to be
investigated, and it is unclear whether TXNIP serves a function in
HBx-mediated metastasis of HCC.
In the present study, the expression levels of HBx
and TXNIP were investigated in HBV-associated HCC tissues, and the
association between their expression levels and the metastasis of
HBV-associated HCC was analyzed. The function of TXNIP in
HBx-induced migration and invasion of HepG2 cells was also
investigated. It was revealed that TXNIP expression in
HBV-associated HCC tissues may be an independent risk factor for
metastasis, and HBx may mediate metastasis of HBV-associated HCC
through upregulating TXNIP expression.
Materials and methods
Patients and tissue specimens
In the present study, 62 HBV-associated HCC tissue
samples were collected between October 2010 and August 2012 from
West China Hospital, Sichuan University (Chengdu, China). The age
of the patients ranged from 26 to 76 years (median, 46 years). Of
the 62 patients, 54 were male and 8 were female. All the patients
were confirmed by pathological diagnosis of HCC and had not
received irradiation or chemotherapy prior to surgical operation.
The patients were divided into two groups as follows: A metastatic
group with portal vein thrombus or peripheral metastasis, and a
non-metastatic group without portal vein thrombus and peripheral
metastasis. The present study was approved by the Ethics Committee
of West China Hospital, Sichuan University. Informed consent was
obtained from all the patients or their relatives prior to
analysis.
Tissue microarray construction and
immunohistochemical staining
All 62 samples were used to construct tissue
microarrays. Paraffin sections of the tissue microarrays were
deparaffinized in xylene and rehydrated in graded ethanol. Antigen
retrieval was performed in boiled 0.01 M citrate buffer (pH 6.0)
for 2 min. Endogenous peroxidase was blocked by 3%
H2O2 in PBS for 30 min at 37°C. Sections were
then incubated with rabbit anti-TXNIP antibody (dilution, 1:400;
cat. no. SAB2108250; Sigma-Aldrich, St. Louis, MO, USA) overnight
at 4°C. Subsequently, sections were visualized using the EnVision
G|2 System/AP (cat. no. K535521-2; Agilent Technologies, Inc.,
Santa Clara, CA, USA) and counterstained with Hematoxylin Staining
Solution (cat. no. C0107; Beyotime Institute of Biotechnology,
Haimen, China) for 5 min at room temperature, according to the
manufacturer's instructions. A total of five randomly selected
optical microscopic fields at ×400 magnification for each sample
were evaluated by two pathologists who were blind to the clinical
data according to the Axiotis score standard (30). The intensity and percentage of
positive cells were used to evaluate each section. Intensity was
scored as follows: 0) No detectable staining; 1) weak staining; 2)
moderate staining; and 3) strong staining. The scores for the
percentage of positive cells were as follows: 0) Rate of 0–25%
scored; 1) rate of 26–50%; 2) rate of 51–75%; and 3) rate of
>75%. The total score was calculated by multiplying the
intensity and positivity scores.
Cell culture and transfection or
infection
The human HCC HepG2 cell line was purchased from
American Type Culture Collection (ATCC; Manassas, VA, USA) and
cultured at 37°C in a 5% CO2 atmosphere, in Dulbecco's
modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). The HEK
293T cell line was purchased from ATCC and cultured at 37°C in a 5%
CO2 atmosphere, in RPMI-1640 medium (Invitrogen; Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS. Cells were
seeded onto 6-well plates 24 h before transfection or infection.
Cells were transfected with purified plasmids constructed in the
present study [pTRE-Tight-HBx, pEGFP-N1-TXNIP, pSicoR-TXNIP short
hairpin (sh) RNA] or preserved in our lab (pBabe-puro, pEGFP-N1,
pNKF, pNKF-HBx, pSicoR, psPAX2 and pMD2G) using X-tremeGENE HP DNA
transfection reagent (Roche Diagnostics GmbH, Mannheim, Germany),
according to the manufacturer's protocol, or infected with
lentivirus constructed in the present study as previously described
(31).
Construction of the HBx expression
stable cell line
pNKF-HBx and pTRE-Tight plasmids from the Tet-On
Advanced Inducible Gene Expression System (Clontech Laboratories,
Inc., Mountain View, CA, USA) were digested with Not 1 and Xba 1
(Takara Bio, Inc., Otsu, Japan). The digested HBx gene with FLAG
sequence was inserted into the digested pTRE-Tight plasmid.
pTRE-Tight-HBx and pBabe-puro plasmid with puromycin resistant was
preserved in our lab, they were then co-transfected into HepG2
Tet-On Advanced cells (Clontech Laboratories, Inc.) in the
proportion of 10:1. Puromycin-resistant clones were treated with
gradient concentrations (0.1–0.25 mg/l) of doxycycline (Dox;
Sigma-Aldrich) and tested for HBx expression, as well as inducing
efficiency, by western blot analysis as described below. Clones
with low constitutive expression and high inducing efficiency were
selected for subsequent investigation.
Overexpression of TXNIP in HepG2
cells
pEGFP-N1 was preserved in our lab. Total RNA was
extracted with TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) from HepG2 cells and reverse transcribed
into cDNA using PrimeScript RT Master mix (Takara Bio, Inc.),
according to the manufacturer's protocol, and used as a template
for TXNIP gene amplification by polymerase chain reaction (PCR)
using PrimeSTAR HS DNA Polymerase (Takara Bio, Inc.). The cycling
conditions for PCR were: 5 min incubation at 95°C, followed by 30
cycles of 98°C for 5 sec, 60°C for 5 sec and 72°C for 90 sec. The
primer sequences used were as follows: TXNIP forward (F),
5′-GACGCGCTCGAGATGGTGATGTTCAAGAAG-3′ and reverse (R),
5′-CACCTGGTCGACTGCTGCACATTGTTGTTG-3′. The PCR products were
digested with Xho 1 and Sal 1 (Takara Bio, Inc.) ligated into
pEGFP-N1 (also digested with Xho 1 and Sal 1). HepG2 cells were
transfected with pEGFP-N1-TXNIP or pEGFP-N1 as a negative control.
TXNIP expression was detected by western blot analysis as described
below.
Knockdown of TXNIP in HepG2 cells
Lentivirus system pSicoR, pxPAX2 and pMD2.G were
preserved in our lab. The oligonucleotides
(5′-TGCCACACTTACCTTGCCAATGTCAAGAGCATTGGCAAGGTAAGTGTGGCTTTTTTC-′3
and
5′-TCGAGAAAAAAGCCACACTTACCTTGCCAATGCTCTTGACATTGGCAAGGTAAGTGTGGCA-3′)
encoding shRNA targeting TXNIP were digested with Hpa 1 and Xho 1
(Takara Bio, Inc.) and ligated into pSicoR (also digested with Hpa
and Xho 1) using the DNA Ligation kit Ver.2.1 (Takara Bio, Inc.)
according to manufacturer's protocol. pSicoR-TXNIP shRNA or pSicoR,
psPAX2 and pMD2 G were co-transfected into HEK 293T cells in the
proportion of 4:3:1 as previously described (32), lentivirus-containing supernatant were
harvested 48 h post transfection. HepG2 cells were infected with
TXNIP shRNA lentivirus or pSicoR lentivirus. Knockdown efficiency
of TXNIP was evaluated by western blot analysis.
Reverse transcription-quantitative PCR
(RT-qPCR)
For RT-qPCR, total RNA was extracted with TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) from frozen
HCC tissues and reverse transcribed into cDNA using PrimeScript RT
Master Mix (Takara Bio, Inc.), according to the manufacturer's
protocol. RT-qPCR was then performed using a Light Cycler 96 with
Fast Start Universal SYBR-Green Master mix (Roche Diagnostics
GmbH). The cycling conditions for PCR were: initial denaturation
95°C for 120 sec, then 40 cycles of 95°C at 10 sec, 62°C at 15 sec
and 72°C at 30 sec. Glyceraldehyde 3-phosphate dehydrogenase
(GAPDH) was used as the internal reference. Relative gene
expression level was analyzed using the 2-ΔΔcq method (33). The primer sequences were as follows:
TXNIP, F, 5′-CTCTGCTCGAATTGACAGAAAAGGATT-3′ and R,
5′-CGCATGTCCCTGAGATAATATGATTGC-3′; HBx, F,
5′-GACTCCCCGTCTGTGCCTTCTCATC-3′ and R,
5′-AGACCAATTTATGCCTACAGCCTCC-3′; GAPDH, F,
5′-AGGAGCGAGATCCCTCCAAAATCAAGT-3′ and R,
5′-TGAGTCCTTCCACGATACCAAAGTTGT-3′.
Western blot analysis
For detection of Flag-HBx, HBx expression stable
cells were cultured at 37°C in a 5% CO2 atmosphere, in
DMEM supplemented with 10% FBS and gradient concentrations
(0.1–0.25 mg/l) of Dox for 48 h. Proteasome inhibitor MG132 (10 µM,
Sigma-Aldrich) was added to the medium 12 h prior to extracting
total protein with SDS-PAGE loading buffer (Beyotime Institute of
Biotechnology). For detection of TXNIP, cells were lysed in
M2 buffer (34). Equal
amounts of protein extracts (60 µg) were separated by 12% SDS-PAGE
and transferred to polyvinylidene fluoride membranes (Merck
Millipore, Darmstadt, Germany). Membranes were then blocked at room
temperature in 5% non-fat milk in Tris-buffered saline with 0.05%
Tween-20 (TBS-T) for 1 h, followed by incubation with primary
antibody against Flag-HBx (dilution, 1:1,000; cat. no. A2220;
Sigma-Aldrich) or TXNIP antibody (dilution, 1:100; cat. no.
sc-33099; Santa Cruz Biotechnology, Inc., Dallas, TX, USA)
overnight at 4°C. The membranes were then incubated with
horseradish peroxidase-conjugated secondary antibody (dilution,
1:5,000; cat. no. ZDR-5307; ZSGB-BIO, Beijing, China) at room
temperature for 1 h and visualized with enhanced chemiluminescence
western blotting substrate (Pierce; Thermo Fisher Scientific,
Inc.).
Scratch assay
HepG2 cells transfected and/or infected with
pEGFP-N1, pEGFP-N1-TXNIP, pSicoR lentivirus, TXNIP shRNA
lentivirus, pNKF, pNKF-HBx, pNKF-HBx in combination with TXNIP
shRNA lentivirus were collected and seeded onto 12-well plates at a
density of 3×106 cells per well. Monolayer cultures were
scratched with 200 µl tips. Serum-free DMEM medium was added and
optical microscopic images of the scratches at 0 and 24 h were
captured at ×100 magnification. Each experimental group took three
scratches, and each experiment repeated three times. Scratch space
was analyzed with Image J software (version 1.41; National
Institutes of Health, Bethesda, MD, USA). Relative cell migration
distance (%) = 100 A - B)/A, where A is the width of the scratch at
0 h and B is the width of the scratch at 24 h (35).
Matrigel invasion assay
Matrigel invasion assays were performed using
Transwell chambers (24-well insert; pore size, 8 µm; BD
Biosciences, Franklin Lakes, NJ, USA). HepG2 cells transfected
and/or infected with pEGFP-N1, pEGFP-N1-TXNIP, pSicoR lentivirus,
TXNIP shRNA lentivirus, pNKF, pNKF-HBx, pNKF-HBx in combination
with TXNIP shRNA lentivirus were collected in serum-free DMEM
medium and seeded into the top chamber coated with Matrigel (BD
Biosciences) at a density of 1×105 cells per well. The
lower chamber was filled with DMEM medium supplemented with 10%
FBS. Following incubation for 24 h, the upper layer of Matrigel was
removed. The cells on the underside of the filter were fixed by
anhydrous methanol and stained with hematoxylin (Beyotime Institute
of Biotechnology). Cell number was counted in five random optical
microscopic fields at ×200 magnification. Three chambers were used
per experimental condition.
Statistical analysis
Data are presented as the mean ± standard deviation.
Statistical analysis was performed using SPSS 20 software (IBM
SPSS, Armonk, NY, USA). Receiver operating characteristic (ROC)
curve analysis was applied to determine the cut-off score for TXNIP
expression in HBV-associated HCC tissues as previously described
(36). The independent sample
Student's t-test was performed to analyze the variables between two
groups. The χ2 test was used to analyze the associations
between TXNIP expression and clinicopathological features of
patients with HBV-associated HCC. Univariate and multivariate
logistics regression analysis was used to analyze the risk factors
associated with metastasis of HBV-associated HCC. Spearman's rank
correlation coefficient tests were used to assess the correlations
between TXNIP and HBx in HBV-associated HCC tissues.
Results
TXNIP expression in HBV-associated HCC
tissues and its association with clinicopathological features of
patients with HBV-associated HCC
TXNIP expression of the tissue microarray was
detected by immunohistochemical staining and evaluated according to
the Axiotis score standard. TXNIP expression in representative
samples of HBV-associated HCC tissues is depicted in Fig. 1A. The results of statistical analysis
revealed that TXNIP scores of the metastatic group (6.000±3.185)
increased compared with those of the non-metastatic group
(4.244±2.634; P=0.024; Fig. 1B).
According to the cut-off score determined by the ROC curve analysis
(Fig. 1C), low expression of TXNIP in
HCC tissues was defined when the TXNIP score was <4.9 and high
expression was defined when the TXNIP score was ≥4.9. The
χ2 test demonstrated that TXNIP expression levels were
positively associated with metastasis of HBV-associated HCC
(Table I). Additional multivariate
logistic regression analysis revealed that besides age, tumor size
and histological grade, high expression of TXNIP in HCC tissues was
also an independent risk factor for metastasis (hazard ratio,
1.533; confidence interval, 1.094–2.150; P=0.013; Table II) of HBV-associated HCC.
Histological grade of HCC tissues was based on the
Edmondson-Steiner criteria (37). The
ISHAK score of patients with HBV-associated HCC was based on the
ISHAK scoring system (38).
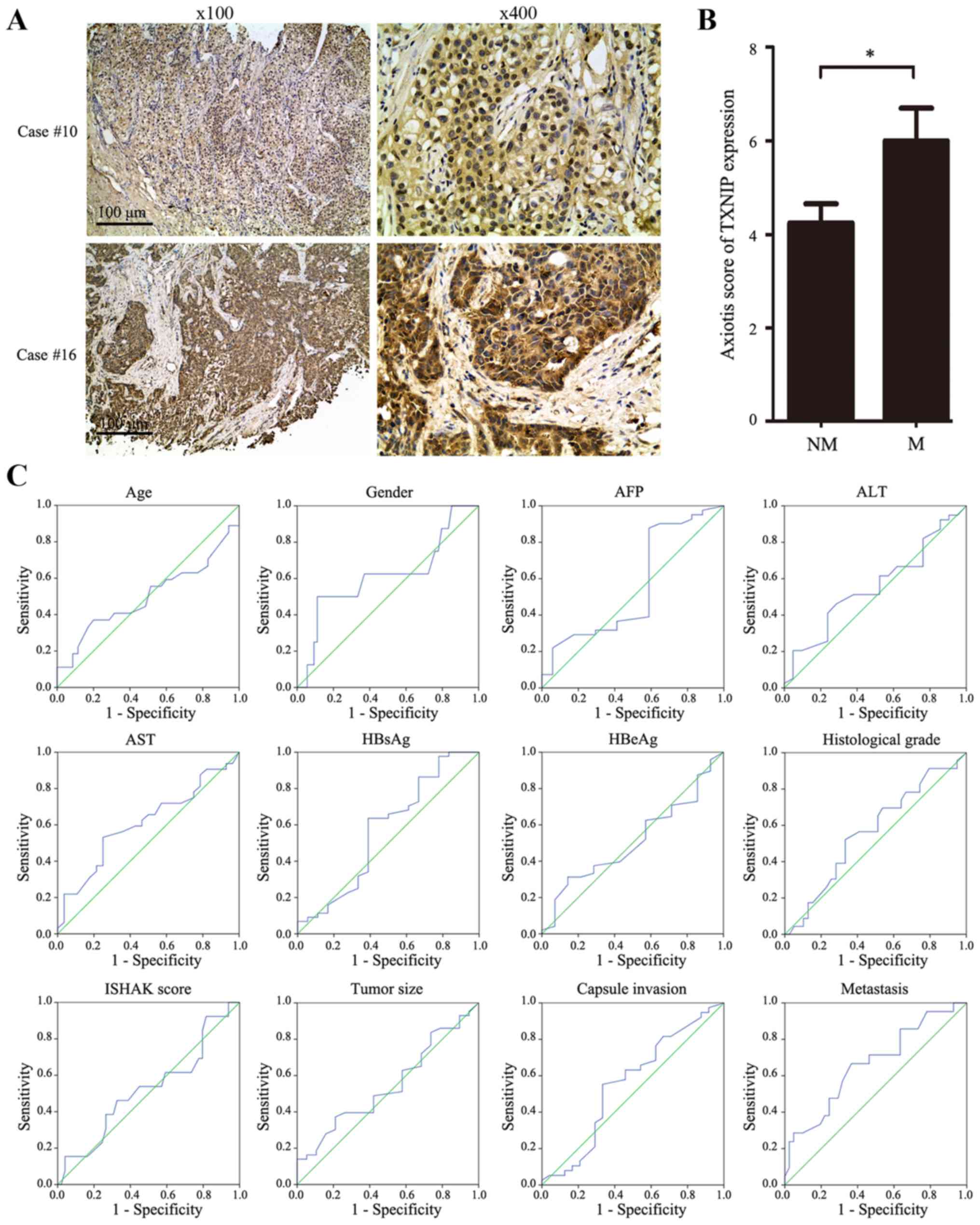 | Figure 1.Expression levels of TXNIP in
HBV-associated HCC tissues. (A) TXNIP expression in representative
tissues. Case 10, tissue without metastasis; case 16, tissue with
metastasis; scale bar, 100 µm. (B) Statistical analysis of TXNIP
expression levels in NM and M HBV-associated HCC tissues. (C) The
cut-off score for TXNIP expression was determined by receiver
operating characteristic curve analysis. The sensitivity and
specificity for each clinicopathological feature at different TXNIP
scores were plotted. *P<0.05 vs. NM. TXNIP, thioredoxin
interacting protein; HBV, hepatitis B virus; HCC, hepatocellular
carcinoma; NM, non-metastatic; M, metastatic; AFP, α-fetoprotein;
ALT, alanine transaminase; AST, aspartate transaminase; HBsAg,
surface antigen of hepatitis B virus; HBeAg, hepatitis B viral
protein. |
 | Table I.Results of χ2 tests to
investigate the association between thioredoxin interacting protein
expression level and clinicopathological features of patients with
hepatitis B virus-associated hepatocellular carcinoma. |
Table I.
Results of χ2 tests to
investigate the association between thioredoxin interacting protein
expression level and clinicopathological features of patients with
hepatitis B virus-associated hepatocellular carcinoma.
|
Characteristics | Low expression,
n | High expression,
n | χ2
value | P-value |
|---|
| Age, years |
|
|
|
|
|
<50/≥50 | 18/15 | 17/12 | 0.104 | 0.747 |
| Sex |
|
|
|
|
|
Female/male |
3/30 |
5/24 | 0.912 | 0.339 |
| AFP (ng/ml) |
|
|
|
|
|
<8/≥8 |
7/25 | 10/16 | 1.905 | 0.168 |
| ALT (IU/l) |
|
|
|
|
|
<55/≥55 | 19/13 | 20/8 | 0.954 | 0.329 |
| AST (IU/l) |
|
|
|
|
|
<46/≥46 | 14/18 | 18/10 | 2.53 | 0.112 |
| HBsAg (S/CO) |
|
|
|
|
|
<3000/>3000 | 11/22 |
7/22 | 0.633 | 0.426 |
| HBeAg (S/CO) |
|
|
|
|
|
<1/≥1 | 26/7 | 22/7 | 0.076 | 0.783 |
| ISHAK score |
|
|
|
|
|
<4/≥4 |
6/27 |
7/22 | 0.33 | 0.565 |
| Tumor size, cm |
|
|
|
|
|
<5/≥5 | 11/22 |
8/21 | 0.24 | 0.624 |
| Capsule
invasion |
|
|
|
|
|
No/yes | 16/17 |
8/21 | 2.841 | 0.092 |
| Histological
grade |
|
|
|
|
|
Moderate/high | 23/10 | 16/13 | 1.395 | 0.237 |
| Metastasis |
|
|
|
|
|
No/yes | 26/7 | 15/14 | 5.047 | 0.025 |
 | Table II.Univariate and multivariate logistics
regression analysis of risk factors associated with metastasis of
patients with hepatitis B virus-associated hepatocellular
carcinoma. |
Table II.
Univariate and multivariate logistics
regression analysis of risk factors associated with metastasis of
patients with hepatitis B virus-associated hepatocellular
carcinoma.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age, years |
|
|
|
|
| <50
vs. ≥50 | 0.184
(0.053–0.644) | 0.008 | 0.044
(0.004–0.443) | 0.008 |
| Gender |
|
|
|
|
| Female
vs. male | 0.972
(0.217–4.348) | 0.971 |
|
|
| AFP (ng/ml) |
|
|
|
|
| <8
vs. ≥8 | 2.692
(0.664–10.913) | 0.165 |
|
|
| ALT (IU/l) |
|
|
|
|
| <55
vs. ≥55 | 1.385
(0.455–4.213) | 0.566 |
|
|
| AST (IU/l) |
|
|
|
|
| <46
vs. ≥46 | 1.654
(0.561–4.875) | 0.362 |
|
|
| HBsAg (S/CO) |
|
|
|
|
|
<3000 vs. ≥3000 | 1.486
(0.447–4.935) | 0.518 |
|
|
|
|
| HBeAg (S/CO) |
|
|
|
|
| <1
vs. ≥1 | 0.729
(0.198–2.681) | 0.635 |
|
|
| ISHAK score |
|
|
|
|
| <4
vs. ≥4 | 0.515
(0.148–1.793) | 0.297 |
|
|
| Tumor size, cm |
|
|
|
|
| <5
vs. ≥5 | 6.729
(1.381–32.8) | 0.018 | 22.25
(2.002–247.259) | 0.012 |
| Capsule
invasion |
|
|
|
|
| No vs.
yes | 6.3
(1.606–24.72) | 0.008 | 6.561
(0.877–49.081) | 0.067 |
| Histological
grade |
|
|
|
|
|
Moderate vs. high | 5.037
(1.622–15.642) | 0.005 | 12.185
(1.877–79.09) | 0.009 |
| TXNIP
expression |
|
|
|
|
| Low vs.
high | 1.241
(1.021–1.508) | 0.03 | 1.533
(1.094–2.15) | 0.013 |
HBx upregulates TXNIP expression in
HepG2 cells
Previous studies have demonstrated that HBx may
upregulate intracellular ROS levels (21), and TXNIP expression may be induced by
ROS stimulation (39). However, to
the best of our knowledge, no previous studies have investigated
the association between HBx and TXNIP expression in HBV-associated
HCC tissues or HCC cells. To investigate their clinicopathological
implication and association, TXNIP and HBx mRNA levels were first
investigated in HBV-associated HCC tissues. The results revealed
that the mRNA levels of HBx and TXNIP were positively correlated
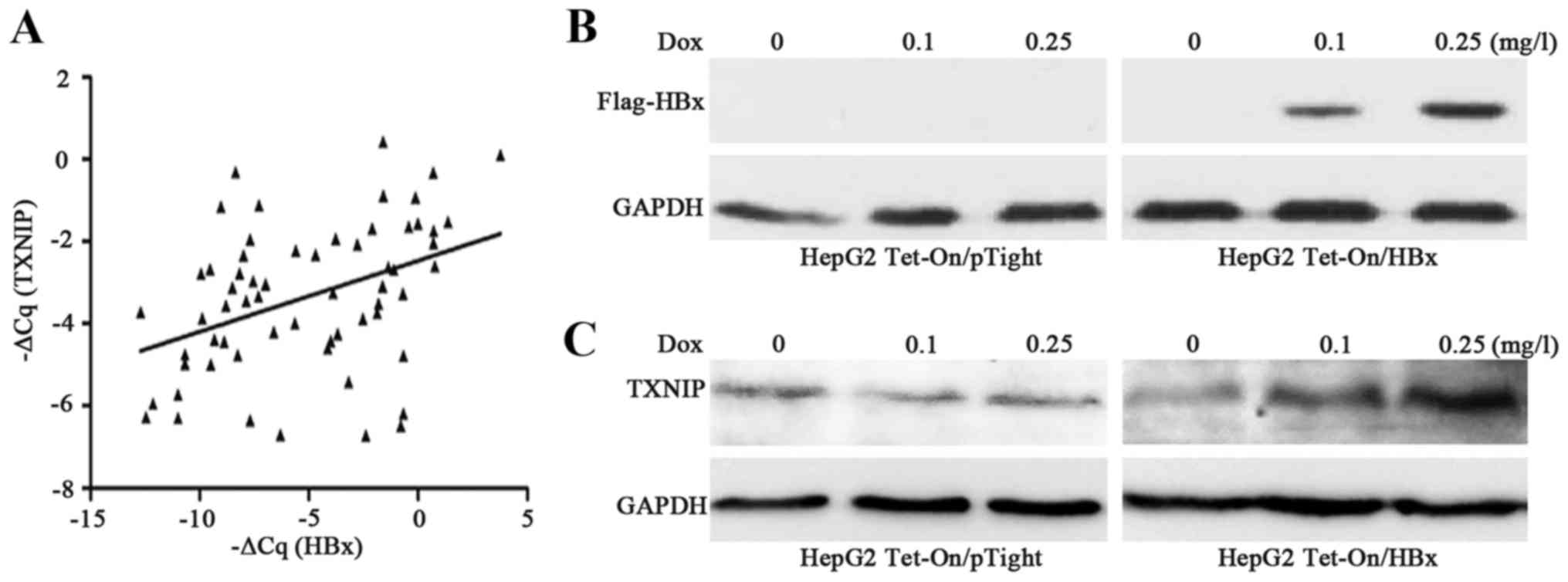
(r=0.42; P=0.0007; Fig. 2A). The
association between HBx and TXNIP was then investigated by
constructing a HBx expression stable cell line with HBx expression
induced and controlled by Dox. The results revealed that HBx
expression was increased as Dox concentration increased (Fig. 2B), and the expression of TXNIP was
upregulated as HBx expression increased (Fig. 2C). These results indicated that HBx
may upregulate TXNIP expression in HepG2 cells.
The function of TXNIP in HepG2 cell
migration and invasion
In the present study, high TXNIP expression was
demonstrated to be positively associated with metastasis of
HBV-associated HCC. The function of TXNIP in HCC cell migration and
invasion was investigated by scratch migration assays and Matrigel
invasion assays. The impact of TXNIP overexpression or knockdown on
HepG2 cell migration and invasion were studied. Firstly, the
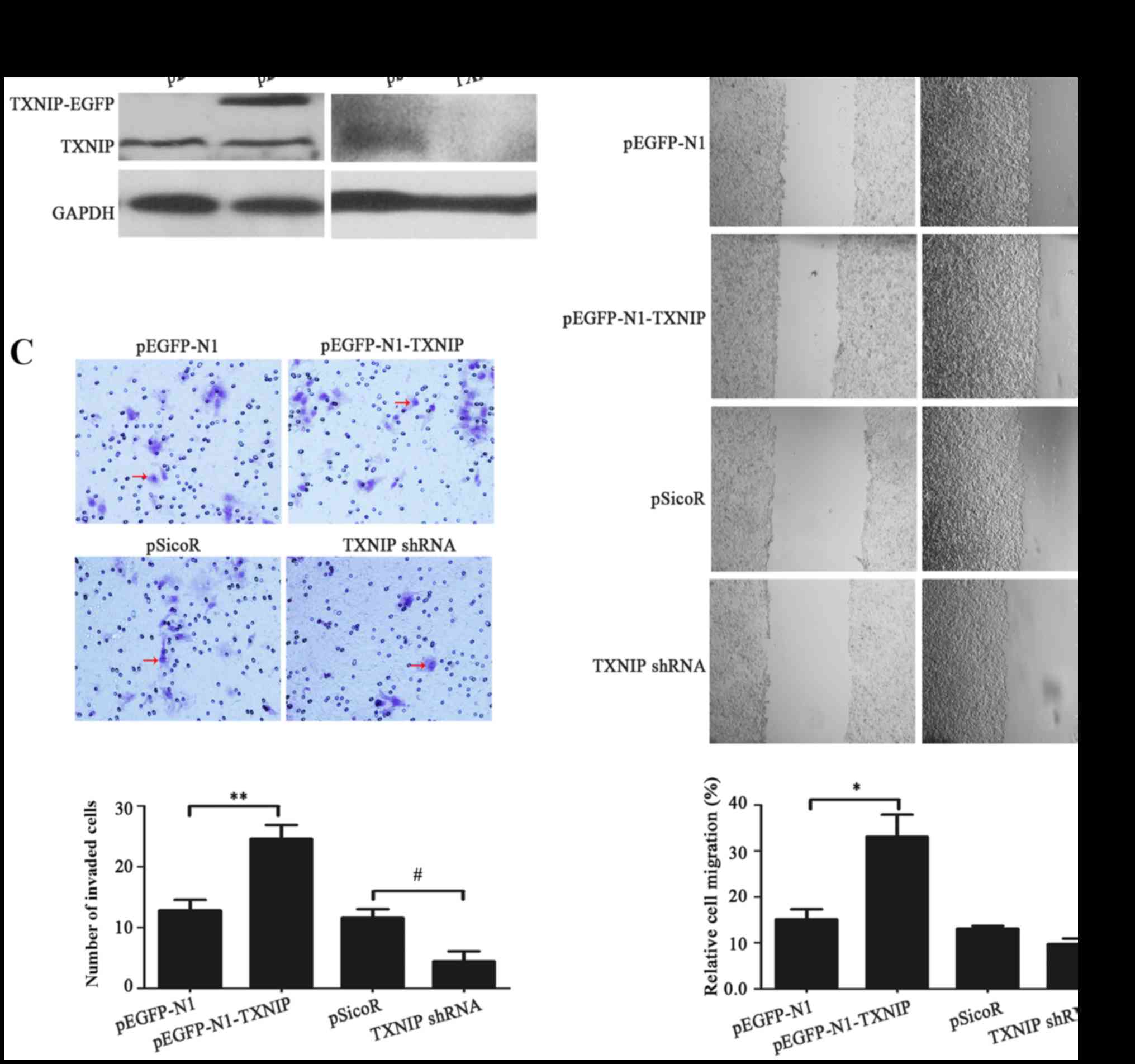
results of western blot analysis revealed that TXNIP-EGFP fusion
protein was efficiently expressed and that TXNIP expression was
effectively silenced by TXNIP shRNA in HepG2 cells (Fig. 3A). The results of scratch and Matrigel
assays demonstrated that TXNIP-EGFP expression may lead to
increased migration (P=0.02) and invasion (P=0.004) in HepG2 cells
compared with the control, while knockdown of TXNIP expression did
not significantly suppress migration (P<0.05) but did
significantly suppress invasion (P=0.012) of HepG2 cells (Fig. 3B and C, respectively). Therefore,
TXNIP overexpression may facilitate the promotion of migration and
invasion of HepG2 cells.
HBx promotes the migration and
invasion of HepG2 cells through upregulating TXNIP expression
The aforementioned results indicated that HBx may
upregulate TXNIP expression in HepG2 cells, and that TXNIP may have
a reinforcing effect on migration and invasion of HepG2 cells. To
investigate the function of TXNIP in HBx-induced migration and
invasion of HCC cells, HBx was ectopically expressed with or
without knockdown of TXNIP in HepG2 cells, and scratch and Matrigel
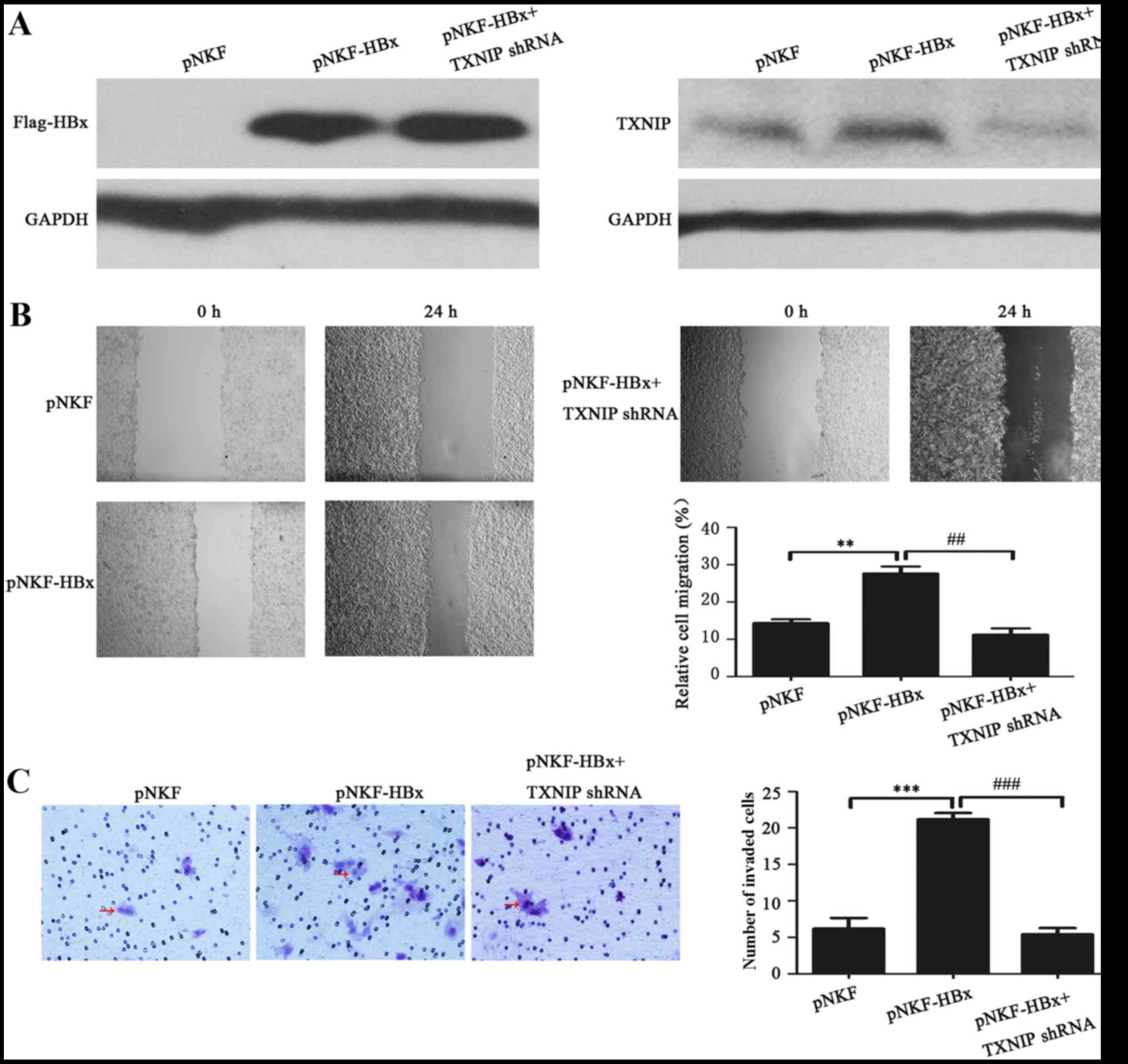
assays were performed. The results of western blot analysis
revealed that expression of HBx may upregulate TXNIP in HepG2
cells, while knockdown of TXNIP expression may attenuate this
effect (Fig. 4A). The results of
scratch and Matrigel assays revealed that HBx may promote migration
(P=0.003) and invasion (P<0.001) of HepG2, respectively, while
knockdown of TXNIP expression significantly attenuated HBx-mediated
enhancement of migratory (P=0.003) and invasive (P<0.001)
potential of HepG2 cells (Fig. 4B and
C, respectively). These data indicated that HBx, potentially
through upregulation of TXNIP expression, may promote the migration
and invasion of HepG2 cells.
Discussion
HBx is involved in metastasis of HBV-associated HCC
by various means, including upregulating intracellular ROS levels
(23,24). Acting as a key mediatory factor of
intracellular ROS level, TXNIP is involved in the progression of
numerous types of tumor (26).
However, the involvement of TXNIP in the development of
HBV-associated HCC remains unclear.
TXNIP was proposed as a tumor suppressor gene in
various tumors (40), including HCC
(41), but its function in metastasis
remains controversial. TXNIP expression levels in tumor cells of
hypoxic perinecrotic areas of glioblastoma and conventional RCC are
increased compared with non-hypoxic tumor cells or peritumoral
tissues (42), while hypoxia often
promotes cancer metastasis and leads to a poor prognosis (43,44). In an
in vitro intravasation model study, overexpression of TXNIP
in melanoma cells increased trans-endothelial intravasation
compared with the control (26),
while overexpression of TXNIP was reported to inhibit metastasis
through the gene KiSS-1 metastasis-suppressor in melanoma cells
(27). In the present study, TXNIP
was detected in HBV-associated HCC tissues. TXNIP expression levels
in HBV-associated HCC tissues were revealed to be positively
associated with tumor metastasis, and high expression levels of
TXNIP may be an independent risk factor of metastasis. The
involvement of TXNIP in regulating HCC cell migration and invasion
was then studied by scratch migration assays and Matrigel invasion
assays. The results demonstrated that TXNIP may enhance HepG2 cell
migration and invasion. Consistent with the present findings, a
previous study has demonstrated that TXNIP overexpression may
increase motility and invasion of HCC cells, and may give a
selective advantage to HCC cells (45). Similarly, a study on gene expression
profiles of three HCC lines with different organ-tropism (accession
no. GSE38945) indicated that TXNIP expression in HCC cell lines
with metastasis ability to the lung and celiac lymph node is
increased compared with HCC cell lines with only metastatic ability
to the lung or low metastatic ability (46). The aforementioned results indicated
that TXNIP may perform different functions in the occurrence and
development of HBV-associated HCC. However, additional studies and
more substantial investigations are required to study the
involvement of TXNIP in HBV-associated HCC.
HBx was reported to increase intracellular ROS level
by targeting the peroxisome or mitochondria (21,22). TXNIP
was reported to be induced by stress stimulation, including
infection, H2O2, ultraviolet and ROS
(39,47). It is therefore a reasonable hypothesis
that HBx may impact TXNIP expression in HBV-associated HCC tissues
and HCC cells. To confirm this hypothesis, HBx and TXNIP mRNA was
detected in HBV-associated HCC tissues. The results revealed that
mRNA levels of HBx and TXNIP in HBV-associated HCC tissues were
positively associated.
Previous studies have demonstrated that the function
of HBx is associated with its expression level and subcellular
distribution (48,49). Expression levels of HBx were difficult
to control by transient transfection due to different experimental
conditions, promoters and cell types. Therefore, a HBx expression
stable cell line was constructed based on the Tet-On Advanced
inducible system for additional investigation of the association
between TXNIP and HBx. The results revealed that HBx may increase
TXNIP expression levels in a gradient-dependent manner in HepG2
cells. These data supported the hypothesis that HBx may impact
TXNIP expression in HBV-associated HCC tissues and HCC cells.
Previous studies have indicated that ROS induced
TXNIP may impair the ability of cells to remove intracellular ROS
(23,50,51), while
increased intracellular ROS may contribute to the metastatic
process of HBV-associated HCC (23,24,52).
Whether TXNIP is involved in HBx-induced metastasis of
HBV-associated HCC is worthy of further study. Scratch and Matrigel
assays were employed to study the involvement of TXNIP in HBx
regulation of HCC cell migration and invasion. Results revealed
that HBx expression in HepG2 cells markedly enhanced cell migration
and invasion ability, which is consistent with the results of
previous studies (18,53). The increase of cell migration and
invasion was attenuated by knockdown of TXNIP expression. These
data indicated that HBx, is involved in promoting migration and
invasion of HCC cells, potentially through upregulating TXNIP
expression.
As a non-structural protein, HBx has a short
half-life of only ~30 min; and its concentration in cells or
tissues is too low to detect (54,55).
Previous studies have indicated that HBx is degraded in a
ubiquitin-proteasome pathway (56,57). In
addition, previous studies have demonstrated that HBx may be highly
expressed in HepG2 cells transiently transfected with HBx
expression plasmids, but its expression in the HBx expression
stable cell line is weak (54,58). In
the present study, HBx expression stable cell lines were treated
with proteasome inhibitors MG132 prior to western blot
analysis.
In summary, to the best of our knowledge, the
present study was the first to identify that high expression of
TXNIP in HBV-associated HCC tissues may be an independent risk
factor for metastasis. Utilizing scratch migration assays and
Matrigel invasion assays, it was confirmed that overexpression of
TXNIP in HepG2 cells enhanced cell migration and invasion, and
TXNIP may be involved in HBx-induced enhancement of cell migration
and invasion. Additional investigations designed to study the
mechanisms of HBx-induced upregulation of TXNIP and TXNIP-induced
migration and invasion of HepG2 cells are required.
Acknowledgements
The present study was supported by research grants
from the National Basic Research Program of China (grant no.
2013CB911300) and the National Natural Science Foundation of China
(grant no. 81371796).
References
|
1
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shlomai A, de Jong YP and Rice CM: Virus
associated malignancies: The role of viral hepatitis in
hepatocellular carcinoma. Semin Cancer Biol. 26:78–88. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Weinmann A, Koch S, Niederle IM,
Schulze-Bergkamen H, König J, Hoppe-Lotichius M, Hansen T, Pitton
MB, Düber C, Otto G, et al: Trends in epidemiology, treatment, and
survival of hepatocellular carcinoma patients between 1998 and
2009: An analysis of 1066 cases of a German HCC Registry. J Clin
Gastroenterol. 48:279–289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Paranagua-Vezozzo DC, Ono SK,
Alvarado-Mora MV, Farias AQ, Cunha-Silva M, França JI, Alves VA,
Sherman M and Carrilho FJ: Epidemiology of HCC in Brazil: Incidence
and risk factors in a ten-year cohort. Annals of hepatology.
13:386–393. 2014.PubMed/NCBI
|
|
5
|
Ye QH, Qin LX, Forgues M, He P, Kim JW,
Peng AC, Simon R, Li Y, Robles AI, Chen Y, et al: Predicting
hepatitis B virus-positive metastatic hepatocellular carcinomas
using gene expression profiling and supervised machine learning.
Nat Med. 9:416–423. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tang H, Delgermaa L, Huang F, Oishi N, Liu
L, He F, Zhao L and Murakami S: The transcriptional transactivation
function of HBx protein is important for its augmentation role in
hepatitis B virus replication. J Virol. 79:5548–5556. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tang H, Oishi N, Kaneko S and Murakami S:
Molecular functions and biological roles of hepatitis B virus ×
protein. Cancer Sci. 97:977–983. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tarocchi M, Polvani S, Marroncini G and
Galli A: Molecular mechanism of hepatitis B virus-induced
hepatocarcinogenesis. World J Gastroenterol. 20:11630–11640. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Buendia MA and Neuveut C: Hepatocellular
carcinoma. Cold Spring Harb Perspect Med. 5:a0214442015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bharadwaj M, Roy G, Dutta K, Misbah M,
Husain M and Hussain S: Tackling hepatitis B virus-associated
hepatocellular carcinoma-the future is now. Cancer Metastasis Rev.
32:229–268. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tian Y, Yang W, Song J, Wu Y and Ni B:
Hepatitis B virus X protein-induced aberrant epigenetic
modifications contributing to human hepatocellular carcinoma
pathogenesis. Mol Cell Biol. 33:2810–2816. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang SZ, Zhang AQ, Chen G, Zhang LD, Zhu
J, Li XW and Dong JH: Experimental study of epithelial-mesenchymal
transition induced by HBx protein in liver cancer cell. Zhonghua Yi
Xue Za Zhi. 90:818–821. 2010.PubMed/NCBI
|
|
13
|
Teng J, Wang X, Xu Z and Tang N:
HBx-dependent activation of Twist mediates STAT3 control of
epithelium-mesenchymal transition of liver cells. J Cell Biochem.
114:1097–1104. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lara-Pezzi E, Gómez-Gaviro MV, Gálvez BG,
Mira E, Iñiguez MA, Fresno M, Martínez-A C, Arroyo AG and
López-Cabrera M: The hepatitis B virus X protein promotes tumor
cell invasion by inducing membrane-type matrix metalloproteinase-1
and cyclooxygenase-2 expression. J Clin Invest. 110:1831–1838.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ou DP, Tao YM, Tang FQ and Yang LY: The
hepatitis B virus X protein promotes hepatocellular carcinoma
metastasis by upregulation of matrix metalloproteinases. Int J
Cancer. 120:1208–1214. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lara-Pezzi E, Majano PL, Yáñez-Mó M,
Carretero M, Moreno-Otero R, Sánchez-Madrid F and López-Cabrera M:
Effect of the hepatitis B virus HBx protein on integrin-mediated
adhesion to and migration on extracellular matrix. J Hepatol.
34:409–415. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lara-Pezzi E, Serrador JM, Montoya MC,
Zamora D, Yáñez-Mó M, Carretero M, Furthmayr H, Sánchez-Madrid F
and López-Cabrera M: The hepatitis B virus X protein (HBx) induces
a migratory phenotype in a CD44-dependent manner: Possible role of
HBx in invasion and metastasis. Hepatology. 33:1270–1281. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang
Y, Tang GN, Zhou WP and Sun SH: Hepatitis B virus X protein
(HBx)-related long noncoding RNA (lncRNA) down-regulated expression
by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by
targeting the intermediate filament protein vimentin. Hepatology.
57:1882–1892. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu X, Fan Z, Kang L, Han J, Jiang C, Zheng
X, Zhu Z, Jiao H, Lin J, Jiang K, et al: Hepatitis B virus X
protein represses miRNA-148a to enhance tumorigenesis. J Clin
Invest. 123:630–645. 2013.PubMed/NCBI
|
|
20
|
Zhang X, Liu S, Hu T, Liu S, He Y and Sun
S: Up-regulated microRNA-143 transcribed by nuclear factor kappa B
enhances hepatocarcinoma metastasis by repressing fibronectin
expression. Hepatology. 50:490–499. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zou LY, Zheng BY, Fang XF, Li D, Huang YH,
Chen ZX, Zhou LY and Wang XZ: HBx co-localizes with COXIII in
HL-7702 cells to upregulate mitochondrial function and ROS
generation. Oncol Rep. 33:2461–2467. 2015.PubMed/NCBI
|
|
22
|
Waris G, Huh KW and Siddiqui A:
Mitochondrially associated hepatitis B virus X protein
constitutively activates transcription factors STAT-3 and NF-kappa
B via oxidative stress. Mol Cell Biol. 21:7721–7730. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li W, Wu Z, Ma Q, Liu J, Xu Q, Han L, Duan
W, Lv Y, Wang F, Reindl KM and Wu E: Hyperglycemia regulates
TXNIP/TRX/ROS axis via p38 MAPK and ERK pathways in pancreatic
cancer. Curr Cancer Drug Targets. 14:348–356. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu WS: The signaling mechanism of ROS in
tumor progression. Cancer Metastasis Rev. 25:695–705. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yoshihara E, Masaki S, Matsuo Y, Chen Z,
Tian H and Yodoi J: Thioredoxin/Txnip: Redoxisome, as a redox
switch for the pathogenesis of diseases. Front Immunol. 4:5142014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cheng GC, Schulze PC, Lee RT, Sylvan J,
Zetter BR and Huang H: Oxidative stress and thioredoxin-interacting
protein promote intravasation of melanoma cells. Exp Cell Res.
300:297–307. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Goldberg SF, Miele ME, Hatta N, Takata M,
Paquette-Straub C, Freedman LP and Welch DR: Melanoma metastasis
suppression by chromosome 6: Evidence for a pathway regulated by
CRSP3 and TXNIP. Cancer Res. 63:432–440. 2003.PubMed/NCBI
|
|
28
|
Masaki S, Masutani H, Yoshihara E and
Yodoi J: Deficiency of thioredoxin binding protein-2 (TBP-2)
enhances TGF-b signaling and promotes epithelial to mesenchymal
transition. PloS One. 7:e399002012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wei J, Shi Y, Hou Y, Ren Y, Du C, Zhang L,
Li Y and Duan H: Knockdown of thioredoxin-interacting protein
ameliorates high glucose-induced epithelial to mesenchymal
transition in renal tubular epithelial cells. Cell Signal.
25:2788–2796. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu DB, Liu FW, Li J, Liu C, Liu L, Chen
EQ, Zhao LS, Tang H and Zhou TY: Intrahepatic IFN-alpha expression
in liver specimens from HBV-infected patients with different
outcomes. Eur Rev Med Pharmacol Sci. 17:2474–2480. 2013.PubMed/NCBI
|
|
31
|
Xia L, Huang W, Tian D, Zhang L, Qi X,
Chen Z, Shang X, Nie Y and Wu K: Forkhead box Q1 promotes
hepatocellular carcinoma metastasis by transactivating ZEB2 and
VersicanV1 expression. Hepatology. 59:958–973. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen W, Li M, Su GZ, Cao J, Sang W, Zhao
K, Wu QY, Zhu F and Xu KL: Establishment of mouse mesenchymal stem
cells overexpressing CXCR4 gene and evaluation of their functions.
Zhongguo Shi Yan Xue Ye Xue Za Zhi. 22:1391–1395. 2014.PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C (T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bai L, Xu X, Wang Q, Xu S, Ju W, Wang X,
Chen W, He W, Tang H and Lin Y: A superoxide-mediated
mitogen-activated protein kinase phosphatase-1 degradation and
c-Jun NH(2)-terminal kinase activation pathway for luteolin-induced
lung cancer cytotoxicity. Mol Pharmacol. 81:549–555. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dasari VR, Kaur K, Velpula KK, Gujrati M,
Fassett D, Klopfenstein JD, Dinh DH and Rao JS: Upregulation of
PTEN in glioma cells by cord blood mesenchymal stem cells inhibits
migration via downregulation of the PI3K/Akt pathway. PloS One.
5:e103502010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shi R, Zhao Z, Zhou H, Wei M, Ma WL, Zhou
JY and Tan WL: Reduced expression of PinX1 correlates to
progressive features in patients with prostate cancer. Cancer Cell
Int. 14:462014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Edmondson HA and Steiner PE: Primary
carcinoma of the liver: A study of 100 cases among 48,900
necropsies. Cancer. 7:462–503. 1954. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ishak K, Baptista A, Bianchi L, Callea F,
De Groote J, Gudat F, Denk H, Desmet V, Korb G, MacSween RN, et al:
Histological grading and staging of chronic hepatitis. J Hepatol.
22:696–699. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang X, Zhang JH, Chen XY, Hu QH, Wang
MX, Jin R, Zhang QY, Wang W, Wang R, Kang LL, et al: Reactive
oxygen species-induced TXNIP drives fructose-mediated hepatic
inflammation and lipid accumulation through NLRP3 inflammasome
activation. Antioxid Redox Signal. 22:848–870. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhou J, Yu Q and Chng WJ: TXNIP (VDUP-1,
TBP-2): A major redox regulator commonly suppressed in cancer by
epigenetic mechanisms. Int J Biochem Cell Biol. 43:1668–1673. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sheth SS, Bodnar JS, Ghazalpour A,
Thipphavong CK, Tsutsumi S, Tward AD, Demant P, Kodama T, Aburatani
H and Lusis AJ: Hepatocellular carcinoma in Txnip-deficient mice.
Oncogene. 25:3528–3536. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Le Jan S, Le Meur N, Cazes A, Philippe J,
Le Cunff M, Léger J, Corvol P and Germain S: Characterization of
the expression of the hypoxia-induced genes neuritin, TXNIP and
IGFBP3 in cancer. FEBS Lett. 580:3395–3400. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Semenza GL: The hypoxic tumor
microenvironment: A driving force for breast cancer progression.
Biochim Biophys Acta. 1863:382–391. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fraga A, Ribeiro R, Príncipe P, Lopes C
and Medeiros R: Hypoxia and prostate cancer aggressiveness: A tale
with many endings. Clin Genitourin Cancer. 13:295–301. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gunes A, Iscan E, Topel H, Avci ST,
Gumustekin M, Erdal E and Atabey N: Heparin treatment increases
thioredoxin interacting protein expression in hepatocellular
carcinoma cells. Int J Biochem Cell Biol. 65:169–181. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tao ZH, Wu WZ, Wang XL, Wan JL, Sun HC,
Wang L, Xia JL and Fan J: The establishment of a systematic
site-specific metastasis model of human hepatocellular carcinoma in
nude mouse. Zhonghua Gan Zang Bing Za Zhi. 19:110–113.
2011.PubMed/NCBI
|
|
47
|
Junn E, Han SH, Im JY, Yang Y, Cho EW, Um
HD, Kim DK, Lee KW, Han PL, Rhee SG and Choi I: Vitamin D3
up-regulated protein 1 mediates oxidative stress via suppressing
the thioredoxin function. J Immunol. 164:6287–6295. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ma J, Sun T, Park S, Shen G and Liu J: The
role of hepatitis B virus X protein is related to its differential
intracellular localization. Acta Biochim Biophys Sin (Shanghai).
43:583–588. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Henkler F, Hoare J, Waseem N, Goldin RD,
McGarvey MJ, Koshy R and King IA: Intracellular localization of the
hepatitis B virus HBx protein. J Gen Virol. 82:871–882. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Singh LP: Thioredoxin interacting protein
(TXNIP) and pathogenesis of diabetic retinopathy. J Clin Exp
Ophthalmol. 4:2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li X, Rong Y, Zhang M, Wang XL, LeMaire
SA, Coselli JS, Zhang Y and Shen YH: Up-regulation of thioredoxin
interacting protein (Txnip) by p38 MAPK and FOXO1 contributes to
the impaired thioredoxin activity and increased ROS in
glucose-treated endothelial cells. Biochem Biophys Res Commun.
381:660–665. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Han JM, Kang JA, Han MH, Chung KH, Lee CR,
Song WK, Jun Y and Park SG: Peroxisome-localized hepatitis Bx
protein increases the invasion property of hepatocellular carcinoma
cells. Arch Virol. 159:2549–2557. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ryu SH, Jang MK, Kim WJ, Lee D and Chung
YH: Metastatic tumor antigen in hepatocellular carcinoma: Golden
roads toward personalized medicine. Cancer Metastasis Rev.
33:965–980. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Schek N, Bartenschlager R, Kuhn C and
Schaller H: Phosphorylation and rapid turnover of hepatitis B virus
X-protein expressed in HepG2 cells from a recombinant vaccinia
virus. Oncogene. 6:1735–1744. 1991.PubMed/NCBI
|
|
55
|
Kim JH, Kang S, Kim J and Ahn BY:
Hepatitis B virus core protein stimulates the proteasome-mediated
degradation of viral X protein. J Virol. 77:7166–7173. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang Z, Sun E, Ou JH and Liang TJ:
Inhibition of cellular proteasome activities mediates
HBX-independent hepatitis B virus replication in vivo. J Virol.
84:9326–9331. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hu Z, Zhang Z, Doo E, Coux O, Goldberg AL
and Liang TJ: Hepatitis B virus X protein is both a substrate and a
potential inhibitor of the proteasome complex. J Virol.
73:7231–7240. 1999.PubMed/NCBI
|
|
58
|
McClain SL, Clippinger AJ, Lizzano R and
Bouchard MJ: Hepatitis B virus replication is associated with an
HBx-dependent mitochondrion-regulated increase in cytosolic calcium
levels. J Virol. 81:12061–12065. 2007. View Article : Google Scholar : PubMed/NCBI
|