Introduction
Cholangiocarcinoma (CCA), anatomically classified
into intrahepatic CCA (IHCC) and extrahepatic CCA (EHCC), is a
highly aggressive malignancy due to early invasion, widespread
metastasis and a lack of effective therapeutic approaches (1–3). Complete
resection is the only way to cure the disease at present, but the
recurrence rates of the patients that receive this treatment are
high (4). Clinically, the prognostic
factors of CCA have not been well established to date. Certain
molecular biomarkers have been reported to be associated with poor
survival and tumor progression, mucin (MUC)1, MUC4, fascin and
epidermal growth factor receptor (EGFR), but the majority of
biomarkers are not used routinely in clinical practice (5). Therefore, novel biomarkers for
prognostic stratification and individualized therapy are
required.
Cullin (Cul)-really interesting new gene ubiquitin
ligase (CRL) complexes represent the largest known class of
ubiquitin ligases, and are involved in a wide variety of
physiological and developmental process, such as cell cycle
progression, DNA damage response and gene expression regulation
(6–8).
In mammals, there are two Cul4 proteins, Cul4A and Cul4B, which
share 82% identity in protein sequences. Notably, Cul4B, as opposed
to Cul4A, exhibits a nuclear localization signal, in its N
terminus, and is localized in the nucleus, suggesting that Cul4B
may be involved in nucleus-based functions (7). Previously, the present and other authors
have reported that Cul4B is overexpressed in numerous solid tumor,
types, such as lung, colon and liver cancer (9–11). Hu
et al (11) suggested that
Cul4B exerts an oncogenic effect by contributing to the epigenetic
silencing of tumor suppressors. However, the expression and role of
Cul4Bin the progression of CCA remain unknown.
Epithelial-mesenchymal transition (EMT), which
involves the downregulation of epithelial markers and the
upregulation of mesenchymal markers, has been shown to participate
in the progression and metastasis of multiple types of cancer
(12–14). However, the link between Cul4B and the
EMT process is unclear in CCA.
The present study demonstrated that Cul4B is
overexpressed in CCA, which may serve as an unfavorable prognostic
factor in patients with EHCC, but not in patients with IHCC. The
present study demonstrated that Cul4B expression is oncogenic and
may promote the EMT process in CCA cells. Cul4B+/EGFR+ defines a
subset of EHCC patients with poor prognosis.
Patients and methods
Patients and tissue microarray (TMA)
construction
The present study consisted of 219 CCA patients, 79
with IHCC and 140 with EHCC, who were treated between January 2007
and December 2013 at the Qilu Hospital of Shandong University
(Jinan, China), Shandong Provincial Hospital (Jinan, China) and
Affiliated Hospital of Qingdao University (Qingdao, China).
Informed written consent was obtained from the patients. The
present study was approved by the Institutional Review Board at the
School of Medicine of Shandong University (Jinan, China). Patients
that exhibited other types of malignancy or had succumbed to
illness within 1 month subsequent to surgery were excluded from the
study. Follow-up data were available for 194 patients, ranging
between 4 and 92 months subsequent to the surgery (mean, 27
months). A total of three TMAs were constructed. Two cores of 1.0
mm in diameter were obtained from each representative tumor focus,
and their morphology was confirmed by two pathologists. Detailed
clinical and pathological profiles were obtained from the medical
records of the patients and maintained in a secure relational
database with TMA data. Patient demographics are shown in Table I.
 | Table I.Summary of demographics of patients
with cholangiocarcinoma. |
Table I.
Summary of demographics of patients
with cholangiocarcinoma.
| Parameters | IHCC, n (%) | EHCC, n (%) |
|---|
| Age, years |
|
|
|
<60 | 49 (62.0) | 76 (54.3) |
| ≥60 | 30 (38.0) | 64 (45.7) |
| Gender |
|
|
| Male | 40 (50.6) | 93 (66.4) |
|
Female | 39 (49.4) | 47 (33.6) |
| Tumor size, cm |
|
|
|
<5 | 27 (34.2) | 79 (58.5) |
| ≥5 | 52 (65.8) | 56 (41.5) |
| Histological
differentiation |
|
|
| Well and
moderate | 60 (75.9) | 122 (87.1) |
| Poorly
erately | 19 (24.1) | 18 (12.9) |
| T stage |
|
|
| I+II | 58 (73.4) | 63 (45.0) |
|
III+IV | 21 (26.6) | 77 (55.0) |
| N stage |
|
|
|
Negative | 56 (70.9) | 104 (74.3) |
|
Positive | 23 (29.1) | 36 (25.7) |
| UICC stage |
|
|
|
I+II | 43 (54.4) | 76 (54.3) |
|
III+IV | 36 (45.6) | 64 (45.7) |
Immunohistochemistry (IHC)
IHC was performed as previously described (15). IHC staining was performed on TMA
slides using the standardized labeled streptavidin biotin kit (Dako
North America, Inc., Carpinteria, CA, USA) according to the
protocol of the manufacturer. The 4-µm slides were then
deparaffinized in xylene and rehydrated through graded ethanol and
in deionized water in the final wash. The sections were submerged
in an antigenic retrieval buffer (citric acid, pH 6.0) for
heat-mediated retrieval by high pressure for 3 min. The slides were
then incubated overnight at 4°C with anti-Cul4B primary antibody
(cat. no. c9995; 1:500 dilution; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany). The slides were blindly evaluated by two
independent pathologists. A previously described scoring system was
utilized to validate Cul4B expression (15). The scores of two parameters were
multiplied by the staining intensity (0–3) and the percentage of
positive cells, which ranged between 0 and 4; (0, 0–10; 1, 11–25;
2, 26–50; 3, 51–75; and 4, 76–100%). Final scores of ≥8 were
classified as overexpressed, while slides with scores <8
classified as non-overexpressed. For EGFR, the membrane
immunostaining was scored following a 4-step scale (scores 0, 1+,
2+ and 3+). Slides with a score of 2+ and 3+ were classified as
positive or overexpressed, in contrast to slides with a score of 0
or 1+, which were defined as negative or non-overexpressed.
Cell culture
The CCA rat brain endothelial (RBE) and QBC939 cell
lines were purchased from the Cell Bank of the Chinese Academy of
Sciences (Shanghai, China). The CCA HUCCT1 cell line was obtained
from RIKEN BioResource Center (Tsukuba, Japan). All lines were
cultured in RPMI-1640 medium (Hyclone; GE Healthcare Life Sciences,
Logan, UT, USA) supplemented with 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA). The cells were
maintained at 37°C in a humidified atmosphere with 5%
CO2.
In vitro overexpression of Cul4B
Human plasmids expressing Flag-tagged Cul4B
(Flag-Cul4B) and negative control vector have been described
previously (11). Cul4B and empty
control plasmids were independently transfected into RBE cells
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the protocol of the manufacturer.
Small interfering RNA (siRNA)-mediated
Cul4B knockdown
Three Cul4B-specific siRNAs were designed and
synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China), and
the most effective type of single siRNA (forward,
5′-GGCAGCACUAUUGUAAUUATT-3′ and reverse,
5′-UAAUUACAAUAGUGCUGCCT-3′) was used in additional experiments. A
non-specific negative control siRNA was also designed and
synthesized (forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′).
Cell proliferation, migration and
invasion assays
Cell proliferation was measured using a
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
(MTS) assay with CellTiter 96® AQueous One
Solution Reagent (Promega GmbH, Mannheim, Germany) following the
protocol of the manufacturer. Migration and invasion assays were
performed as previously described (15). To quantify the number of invading
cells, the cells were counted in five randomly selected microscopic
fields (magnification, ×200). A total of three independent
experiments were performed.
Soft agar colony assay
Soft agar colony assay was performed as previously
described (16). HUCCT1 cells
transfected with the indicated siRNAs or Flag-Cul4B were
trypsinized and suspended in RPMI-1640 medium containing 0.3%
lukewarm agar at a cell concentration of 5×103 cells/ml.
The suspension was spread on top of 0.5% solidified agar plates.
Colony formation was observed subsequent to a 2-week culture. The
colonies were counted and photographed using a CKX41 microscope
(Olympus Corporation, Tokyo, Japan).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the CCA cell lines
using TRIzol (cat no. 15596026; Thermo Fisher Scientific, Inc.) and
complementary DNA was synthesized by RT. A SYBR Green®
Real-Time PCR Master Mix (Toyobo Co., Ltd., Osaka, Japan) and an
ABI PRISM® 7700 Sequence Detection System (Applied
Biosystems; Thermo Fisher Scientific, Inc.) were used in the
present study. The following thermocycling conditions were
maintained: 50°C for 2 min; 95°C for 10 min; and 40 cycles of 95°C
for 15 sec and 60°C for 1 min. The primers for Cul4B were as
follows: Forward, 5′-TGGAAGTTCATTTACCACCAGAGATG-3′, and reverse,
5′-TTCTGCTTTTAACACACAGTGTCCTA-3′. The relative Cul4B expression was
normalized to the messenger RNA (mRNA) expression of GAPDH, which
was amplified with the following primers: Forward,
5′-GAGTCAACGGATTTGGTCGT-3′, and reverse, 5′-TTGATTTTGGAGGGATCTC-3′.
PCR assays were performed in triplicate, and fold induction was
calculated using the 2−ΔΔCq method (17).
Western blot analysis
Western blot analysis was performed as previously
described (15). Briefly, total
protein was extracted using RIPA buffer (cat. no. 20–188; EMD
Millipore, Billerica, MA, USA) and measured using a bicinchoninic
acid protein assay kit (Beyotime Institute of Biotechnology,
Haimen, China). Protein (20 µg/lane) was directly electrophoresed
using 10% SDS-PAGE and then transferred to an Immobilon-P
polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA,
USA). The membranes were blocked at room temperature for 1 h in
blocking buffer containing 3% bovine serum albumin (Sigma-Aldrich;
Merck KGaA), then incubated overnight at 4°C with the primary
antibody against Cul4B (1:1,000; Sigma-Aldrich; Merck KGaA).
Membranes were subsequently incubated with a horseradish
peroxidase-conjugated anti-rabbit IgG secondary antibody (cat. no.
7074, 1:1500, Cell Signaling Technology, Inc., Danvers, MA, USA) at
37°C for 30 min. Primary antibody directed against β-actin was used
as a loading control (cat. no. sc-47778;, 1:5,000; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA). The signals were detected
with an enhanced chemiluminescence reagent (Merck KGaA). Three
independent experiments were performed.
Statistical analysis
The software used for statistical analyses was SPSS
version 19.0 (IBM SPSS, Armonk, NY, USA). Two-sided Student's t
test and Mann-Whitney U test were used for statistical comparisons,
while Spearman's rank correlation test was used to evaluate the
correlations between Cul4B overexpression and a number of
clinicopathological parameters. The Kaplan-Meier method and Cox
regression hazard tests were applied for the analysis of follow-up
data, and hazard ratio (HR) with 95% confidence intervals (CI) were
calculated. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression of Cul4B and associations
between Cul4B expression and clinicopathological factors in
CCA
The present study performed IHC staining of Cul4B in
a cohort of 219 patients with CCA to define the role of Cul4B in
the progression of CCA. Predominantly nuclear staining of Cul4B was
observed. Cul4B protein levels were greater in the cancer cells
than in the adjacent benign ductal epithelia (data not shown).
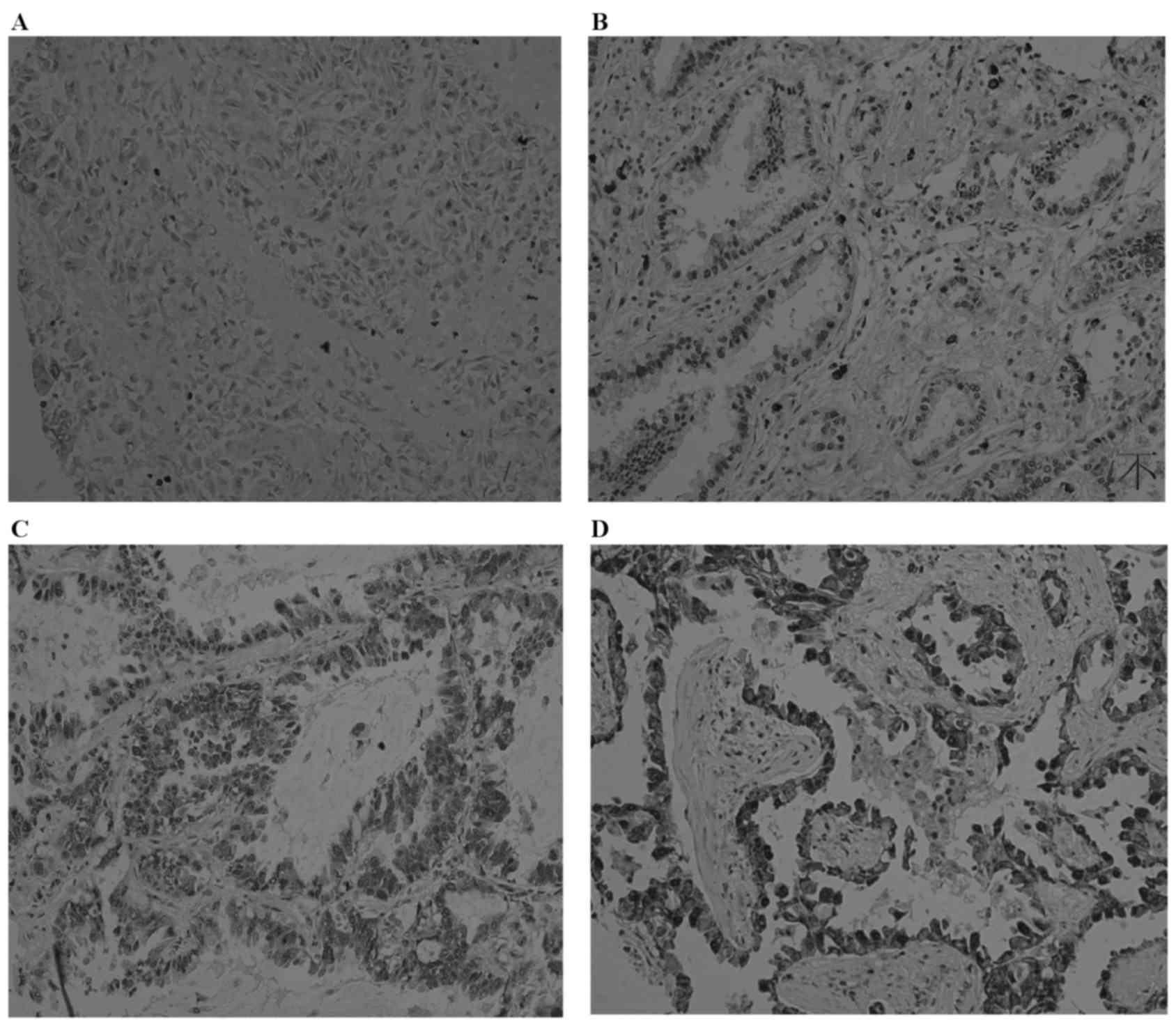
Representative images of Cul4B expression are shown in Fig. 1. Overall, Cul4B was overexpressed in
28.6% (40/140) of the patients with EHCC and in 26.6% (21/79) of
the patients with IHCC. The associations between Cul4B
overexpression and various clinicopathological factors are
summarized in Table II. Among the
patients with EHCC, Cul4B overexpression was significantly
associated with higher pathological tumor stage (P=0.024) and lymph
node metastasis (P=0.013). No associations were identified between
Cul4B overexpression and gender (P=0.821), age (P=0.520), tumor
size (P=0.650) or histological differentiation (P=0.299). By
contrast, as shown in Table II,
Cul4B expression was not associated with any clinicopathological
variables in patients with IHCC. The overexpression of EGFR was
present in 19 (25.0%) of the 76 patients with IHCC and in 23
(16.7%) of the 138 patients with EHCC. Notably, there was a
marginal association between Cul4B overexpression and EGFR
expression (P=0.093) in patients with EHCC, but not in patients
with IHCC.
 | Table II.Association of Cul4B expression level
with clinicopathological parameters in cholangiocarcinoma. |
Table II.
Association of Cul4B expression level
with clinicopathological parameters in cholangiocarcinoma.
|
| Cul4B in IHCC |
| Cul4B in EHCC |
|
|---|
|
|
|
|
|
|
|---|
| Parameters | Not overexpressed,
n (%) | Over expressed, n
(%) | P-value | Not overexpressed,
n (%) | Over expressed, n
(%) | P-value |
|---|
| Age, years |
|
| 0.989 |
|
| 0.520 |
|
<60 | 36 (73.5) | 13 (26.5) |
| 56 (71.8) | 20 (28.2) |
|
|
≥60 | 22 (73.3) | 8 (26.7) |
| 44 (68.8) | 20 (31.2) |
|
| Gender |
|
| 0.486 |
|
| 0.821 |
|
Male | 28 (70.0) | 12 (30.0) |
| 67 (72.0) | 26 (28.0) |
|
|
Female | 30 (76.9) | 9 (23.1) |
| 33 (70.3) | 14 (29.7) |
|
| Tumor size, cm |
|
| 0.328 |
|
| 0.650 |
|
<5 | 18 (66.7) | 9 (33.3) |
| 55 (69.6) | 24 (30.4) |
|
| ≥5 | 40 (76.9) | 12 (23.1) |
| 41 (73.2) | 15 (26.8) |
|
| Histological
differentiation |
|
| 0.531 |
|
| 0.299 |
| Well
and moderate | 43 (71.7) | 17 (28.3) |
|
|
|
|
|
Poor | 15 (78.9) | 4 (21.1) |
| 11 (61.1) | 7 (38.9) |
|
| T stage |
|
| 0.630 |
|
| 0.024 |
|
I+II | 41 (71.9) | 16 (28.1) |
| 39 (61.9) | 24 (38.1) |
|
|
III+IV | 17 (77.3) | 5 (22.7) |
| 61 (79.2) | 16 (20.8) |
|
| Lymph node
metastasis |
|
| 0.422 |
|
| 0.013 |
|
Negative | 42 (75.0) | 14 (25.0) |
| 68 (65.4) | 36 (34.6) |
|
|
Positive | 16 (69.6) | 7 (30.4) |
| 32 (88.8) | 4 (11.2) |
|
| UICC stage |
|
| 0.619 |
|
| 0.217 |
|
I+II | 30 (69.8) | 13 (30.2) |
| 51 (67.1) | 25 (32.9) |
|
|
III+IV | 28 (77.8) | 8 (22.2) |
| 49 (76.6) | 15 (23.4) |
|
| EGFR
expression |
|
| 0.139 |
|
| 0.093 |
| Not
overexpressed | 44 (77.2) | 13 (22.8) |
| 85 (73.9) | 30 (26.1) |
|
|
Overexpressed | 11 (57.9) | 8 (42.1) |
| 13 (56.5) | 10 (43.5) |
|
Prognostic role of CUl4B expression in
CCA
To assess the possible association between Cul4B
expression and patient survival, Kaplan-Meier curves with a
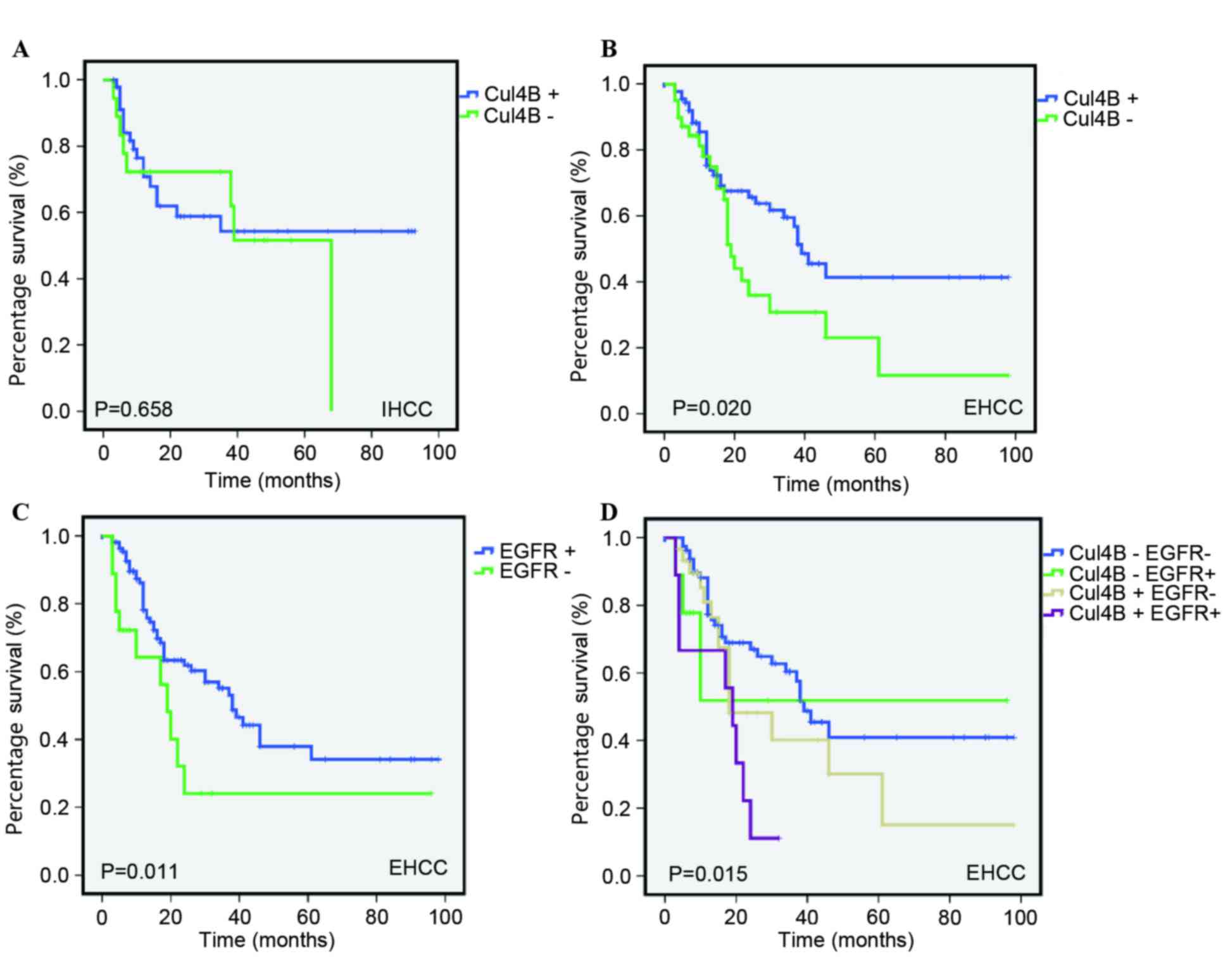
log-rank test for overall survival (OS) were performed. In EHCC, as
shown in Fig. 2, patients with Cul4B
overexpression had a lower OS rate than patients who did not
exhibit overexpression. The estimated mean OS time was
significantly different between patients with Cul4B-overexpressed
and patients with Cul4B-non-overexpressed tumors (55.029±2.595 and
86.974±0.882 months, respectively; P<0.001). By contrast, no
statistical significance (P=0.658) was identified between Cul4B
overexpression and OS in IHCC.
In univariate Cox regression analysis, Cul4B
overexpression was a prognostic factor for cancer mortality
(HR=1.779, 95% CI=1.102–2.690, P=0.028; Table III) in EHCC. Additionally,
histological differentiation (P=0.037), tumor stage (P=0.023),
Union for International Cancer Control (UICC) stage (P<0.010)
and EGFR expression were also significantly associated with OS. The
classification of tumor stage and UICC stage is based on the 7th
edition of the UICC-American Joint Committee on Cancer (UICC/AJCC)
(18). In a multivariate analysis,
UICC stage and EGFR expression exhibited predictive value, whereas
Cul4B expression did not (Table
III). In IHCC, Cul4B expression was not observed to be
associated with the OS (P=0.768) of patients with CCA. Four
factors, including tumor size, lymph node metastasis, UICC stage
and EGFR expression, were identified as prognostic factors by
univariate analysis. In the multivariate analysis, as shown in
Table IV, only UICC stage and lymph
node metastasis were independent prognostic factors. These results
suggested that Cul4B was an unfavorable prognostic indicator in
Chinese patients with EHCC.
 | Table III.Univariate and multivariate analysis
for overall survival in extrahepatic cholangiocarcinoma. |
Table III.
Univariate and multivariate analysis
for overall survival in extrahepatic cholangiocarcinoma.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Parameters | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age, years |
|
|
|
|
|
|
|
<60 | 1 | (Reference) |
| 1 | (Reference) |
|
|
≥60 | 1.533 | 0.947–2.481 | 0.082 | – | – | – |
| Gender |
|
|
|
|
|
|
|
Male | 1 | (Reference) |
| 1 | (Reference) |
|
|
Female | 0.831 | 0.496–1.394 | 0.484 | – | – | – |
| Tumor size, cm |
|
|
|
|
|
|
|
<3 | 1 | (Reference) |
| 1 | (Reference) |
|
| ≥3 | 1.06 | 0.642–1.749 | 0.820 | – | – | – |
| Histological
differentiation |
|
|
|
|
|
|
| Well
and moderate | 1 | (Reference) |
| 1 | (Reference) |
|
|
Poor | 0.54 | 0.303–0.962 | 0.037 | 0.920 | 0.478–1.796 | 0.821 |
| T stage |
|
|
|
|
|
|
|
I+II | 1 | (Reference) |
| 1 | (Reference) |
|
|
III+IV | 1.782 | 1.084–2.929 | 0.023 | 1.199 | 0.703–2.045 | 0.506 |
| Lymph node
metastasis |
|
|
|
|
|
|
|
Negative | 1 | (Reference) |
| 1 | (Reference) |
|
|
Positive | 1.288 | 0.734–2.263 | 0.378 |
|
|
|
| UICC stage |
|
|
|
|
|
|
|
I+II | 1 | (Reference) |
| 1 | (Reference) |
|
|
III+IV | 3.141 | 1.931–5.110 | <0.010 | 3.083 | 1.894–5.018 | <0.010 |
| Cul4B
expression |
|
|
|
|
|
|
| Not
overexpressed | 1 | (Reference) |
| 1 | (Reference) |
|
|
Overexpressed | 1.779 | 1.102–2.690 | 0.028 | 1.358 | 0.809–2.423 | 0.162 |
| EGFR
expression |
|
|
|
|
|
|
| Not
overexpressed | 1 | (Reference) |
| 1 | (Reference) |
|
|
Overexpressed | 1.989 | 1.100–3.600 | 0.023 | 1.876 | 1.038–3.390 | 0.037 |
 | Table IV.Univariate and multivariate analysis
for overall survival in intrahepatic cholangiocarcinoma. |
Table IV.
Univariate and multivariate analysis
for overall survival in intrahepatic cholangiocarcinoma.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Parameters | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age, years |
|
|
|
|
|
|
|
<60 | 1 | (Reference) |
| 1 | (Reference) |
|
|
≥60 | 1.003 | 0.518–1.941 | 0.993 | – | – | – |
| Gender |
|
|
|
|
|
|
|
Male | 1 | (Reference) |
| 1 | (Reference) |
|
|
Female | 0.555 | 0.289–1.067 | 0.177 | – | – | – |
| Tumor size, cm |
|
|
|
|
|
|
|
<5 | 1 | (Reference) |
| 1 | (Reference) |
|
| ≥5 | 2.136 | 1.006–4.536 | 0.048 | 1.711 | 0.778–3.764 | 0.181 |
| Histological
differentiation |
|
|
|
|
|
|
| Well
and moderate | 1 | (Reference) |
| 1 | (Reference) |
|
|
Poor | 0.558 | 0.285–1.094 | 0.089 | – | – | – |
| T stage |
|
|
|
|
|
|
|
I+II | 1 | (Reference) |
| 1 | (Reference) |
|
|
III+IV | 1.532 | 0.791–2.968 | 0.206 | – | – | – |
| Lymph node
metastasis |
|
|
|
|
|
|
|
Negative | 1 | (Reference) |
| 1 | (Reference) |
|
|
Positive | 4.362 | 2.210–8.610 | <0.010 | 2.402 | 1.065–5.418 | 0.035 |
| UICC stage |
|
|
|
|
|
|
|
I+II | 1 | (Reference) |
| 1 | (Reference) |
|
|
III+IV | 3.935 | 1.969–7.867 | <0.010 | 2.566 | 1.110–5.935 | 0.028 |
| Cul4B
expression |
|
|
|
|
|
|
| Not
overexpressed | 1 | (Reference) |
| 1 | (Reference) |
|
|
Overexpressed | 1.112 | 0.550–2.246 | 0.768 | – | – | – |
| EGFR
expression |
|
|
|
|
|
|
| Not
overexpressed | 1 | (Reference) |
| 1 | (Reference) |
|
|
Overexpressed | 1.866 | 1.215–3.505 | 0.030 | 1.453 | 1.206–2.815 | 0.285 |
Cul4B+/EGFR+ defines a subset of EHCC
patients with poor prognosis
The present study then determined whether combining
Cul4B and EGFR further improved their prognostic value in patients
with EHCC, by grouping all patients with EHCC according to Cul4B
and EGFR overexpression status. Kaplan-Meier analyses were then
conducted using the group exhibiting neither Cul4B overexpression
nor EGFR overexpression as the reference. As shown in Fig. 2D, the group possessing EGFR and Cul4B
overexpression exhibited the worst cancer-associated survival
compared with the three other groups.
Cul4B promotes proliferation,
migration and invasion in CCA cells
To explore the biological role of Cul4B in CCA in
vitro, the present study first evaluated the endogenous
expression of Cul4B in different CCA cell lines. As shown in
Fig. 3A, QBC939 cells exhibited the
highest level of Cul4B, whereas HUCCT1 and RBE cells exhibited
relatively low levels of Cul4B (QBC939>HUCCT1>RBE). Using an
MTS assay, the present study revealed that the proliferation of
QBC939 cells was significantly reduced subsequent to the silencing
of Cul4B compared with that of the negative control (P=0.033;
Fig. 3B). Subsequent to 48 and 72 h
of Cul4B siRNA treatment, the number of QBC939 cells was reduced by
20.7±3.5 and 27.9±5.6%, respectively (Fig. 3B). In addition, the present study also
performed overexpression of Cul4B in RBE cells. The growth of RBE
cells increased significantly subsequent to transfection with a
Cul4B expression plasmid (48 h, P=0.034; 96 h, P=0.024; Fig. 3B). The overexpression of Cul4B
significantly increased the level of colony formation of HUCCT1
cells compared with that of the negative control (P=0.006; Fig. 3C). Transwell experiments were then
performed to determine the migration and invasive abilities of CCA
cells either transfected with siRNA or with the Cul4B expression
plasmid. The data of the present study revealed that the migration
and invasive capacities of QBC939 and HUCCT1 cells decreased
subsequent to the knocking down of Cul4B. By contrast,
overexpression of Cul4B significantly increased the migration and
invasive capacities of HUCCT1and RBE cells (HUCCT1, P=0.014; RBE,
P=0.017; Fig. 3D).
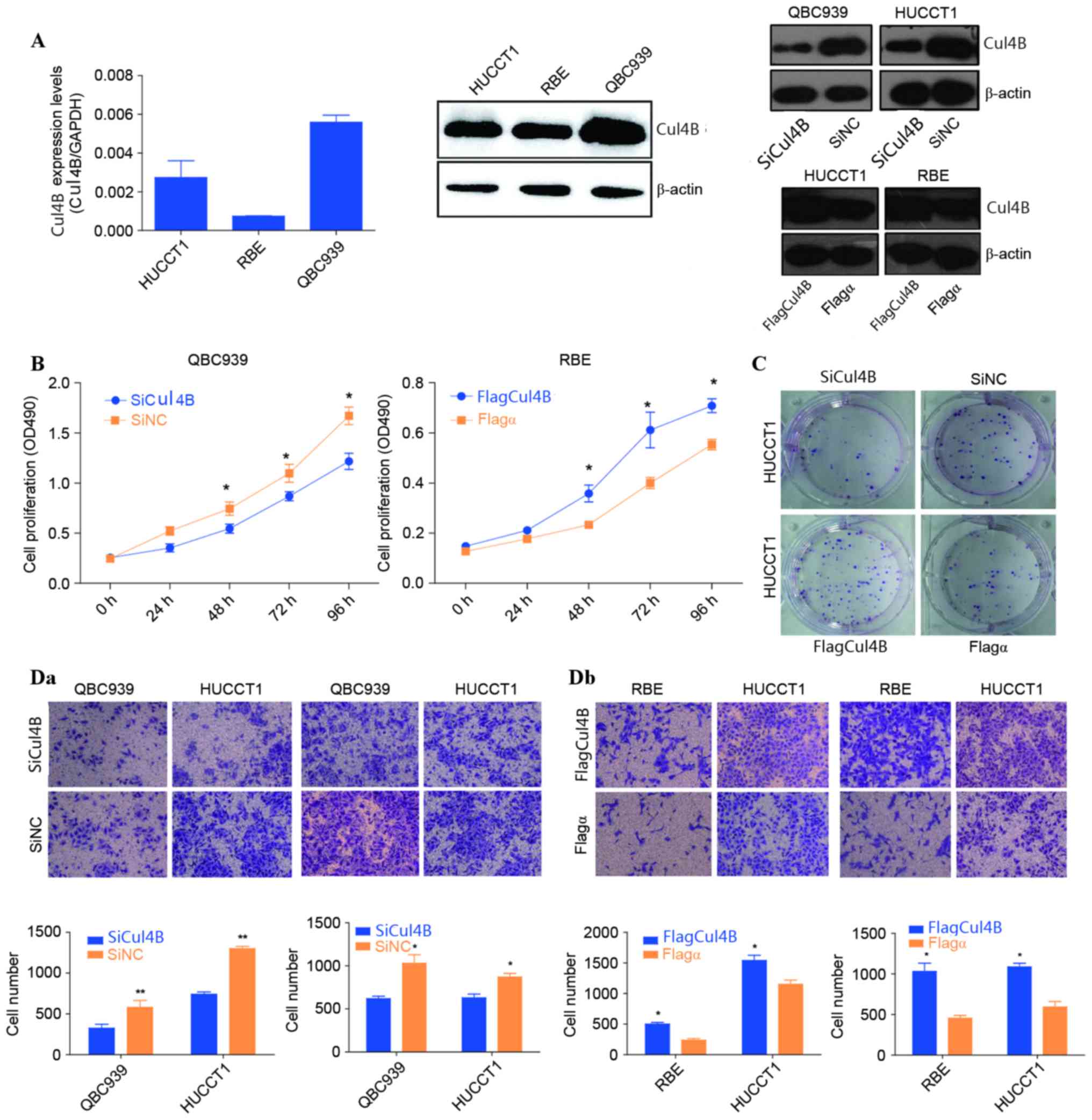 | Figure 3.Biological roles of Cul4B on CCA in
vitro. (A) The messenger RNA and protein levels of Cul4B in
HUCCT1, RBE and QBC939 cell lines were determined by reverse
transcription-quantitative polymerase chain reaction and western
blot analysis, respectively. Protein expression levels of Cul4B
following the overexpression and siRNA knockdown of the gene are
also shown. β-Actin and GAPDH were used as internal references. (B)
3-(4,5-Dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
assays were performed to examine growth rates at different time
points ranging between 0 and 96 h in QBC939 and RBE cells. (C) A
colony formation assay was performed to examine the colony
formation ability of HUCCT1 cells following siRNA knockdown and
overexpression of Cul4B. (Da) Cellular migration and (Db) invasion
capacities of CCA cells were evaluated by Transwell assays.
Representative images of migratory and invasive cells were shown.
Statistical results of cell numbers at the bottom of the membrane
were visualized and are presented as the mean ± standard deviation.
*P<0.05, **P<0.01 vs. SiCul4B or Flagα. SiCul4B, siRNA
knockdown of Cul4B; SiNC, negative control siRNA; CCA,
cholangiocarcinoma; RBE, rat brain endothelial; siRNA, small
interfering RNA; Cul, cullin; OD, optical density. |
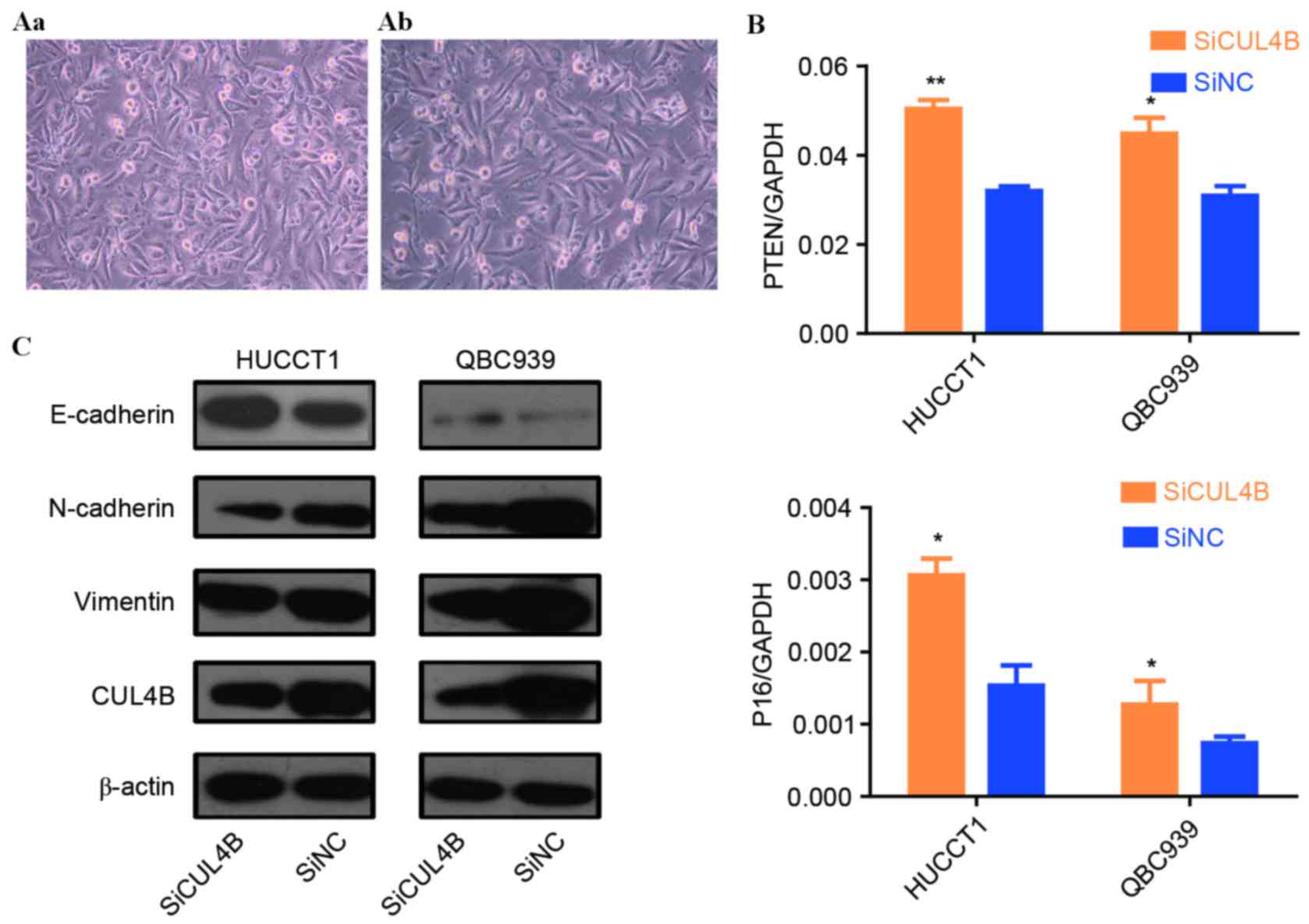
In vitro effect of Cul4B on EMT
The present study subsequently investigated whether
Cul4B is a regulator of EMT in CCA in vitro. Of note, Cul4B
overexpression induced an elongated fibroblast-like morphology with
scattered distribution in cultured RBE cells (Fig. 4A). Cul4B was then transiently knocked
down, resulting in the suppression of the EMT of CCA cells, as
shown by the inhibition of E-cadherin expression (an epithelial
marker) and an increase in vimentin and N-cadherin (mesenchymal
markers) at the protein level (Fig.
4B).
Modulation of p16 and phosphatase and
tensin homolog (PTEN) by Cul4B in CCA cell lines
Hu et al (11)
reported that Cul4B may contribute to the transcriptional
regulation of tumor-suppressor genes in human breast adenocarcinoma
cell lines. The present study therefore investigated the modulation
of two known tumor suppressor genes, P16 and PTEN, by Cul4B. As
shown in Fig. 4C, siRNA knockdown of
Cul4B led to a significant upregulation of p16 and PTEN expression
at the mRNA level in QBC939 cells (P16, P=0.049; PTEN, P=0.041),
suggesting that Cul4B promotes tumor progression partially through
the repression of the aforementioned tumor-suppressor genes.
Discussion
Cul4B is a scaffold protein of the CRL complex, and
is involved in the regulation of a broad spectrum of biological
processes, including cell cycle progression, DNA replication and
DNA damage response (6,7,19). Cul4B
has previously been shown to be overexpressed in various types of
solid malignancy (9–11,16,19,20).
However, the precise role of Cul4B in CCA is unknown. The present
study provides support for the hypothesis that Cul4B serves an
oncogenic role in CCA progression. Firstly, the data of the present
study clearly demonstrated that siRNA knockdown of Cul4B
significantly inhibits the proliferation and soft agar growth of
CCA cells in vitro. Secondly, siRNA knockdown of Cul4B
significantly decreased the migration and invasive capacities of
CCA cells, which are two critical events in the progression of
cancer towards metastasis. Therefore, Cul4B overexpression is
significantly associated with the presence of lymph node metastasis
and higher clinical tumor stages in clinical CCA cases. Thirdly
Cul4B is an unfavorable prognostic factor in a subset of patients
with EHCC. In concordance with these findings, Jiang et al
(9) reported that high Cul4B
expression was associated with the depth of tumor invasion, lymph
node metastasis, distant metastasis, histological differentiation,
vascular invasion and advanced tumor stage of numerous types of
colon cancer.
The overexpression of Cul4B in a subset of patients
with CCA may result from the activation of Cul4B by increased
transcription, possibly through amplification or promoter
mutations. Alternatively, Cul4B may be activated by an unknown
upstream genetic event. It has been reported that Cul4A, another
member of CRL4, was amplified in primary breast cancer, squamous
cell carcinoma, adrenocortical carcinoma and hepatocellular
carcinoma (19). Previously,
genome-wide high-density single nucleotide polymorphism arrays
further revealed a high Cul4A gene copy number in a subset of lung
and ovarian carcinoma patients (21).
Although mutations of the Cul4B gene are causally associated with
human X-linked mental retardation (19), the data are limited regarding
aberrations of Cul4B at the DNA level in cancer. Therefore,
additional investigation into the genetic characterization of Cul4B
in CCA using clinical samples is required.
The mechanism by which Cul4B contributes to cancer
invasion and progression remains undefined. Cul4B may exert
oncogenicity in several ways. Previously, Hu et al (11) demonstrated that, by catalyzing histone
(H) 2AK119 monoubiquitination and coordinating with
polycomb-repressive complex 2, Cul4B can promote the
transcriptional silencing of an array of tumor-suppressor genes.
Additionally, Yang et al (16)
reported that Cul4B promotes tumorigenesis by coordinating with
suppressor of variegation 3–9 homolog 1/heterochromatin
protein1/DNA (cytosine-5)-methyltransferase 3A in DNA
methylation-based epigenetic silencing. The depletion of Cul4B
resulted in H3K9 trimethylation and DNA methylation, leading to the
repression of a collection of genes, including the tumor suppressor
insulin-like growth factor-binding protein 3. Consistently with
these findings, the present study noticed that siRNA knockdown of
Cul4B significantly downregulated the expression of P16 and PTEN in
CCA cells. It was recently reported that Cul4B upregulates
Wnt/β-catenin signaling in hepatocellular carcinoma (HCC) through
transcriptionally repressing Wnt antagonists, thus contributing to
the malignancy of HCC (10).
The present study demonstrated that Cul4B promoted
EMT of CCA cells. EMT is a developmental program, and is essential
for tumor cells to disseminate to adjacent tissues and establish
new tumors in distant sites (15).
The data of the present study suggest that siRNA knockdown of Cul4B
results in the upregulation of E-cadherin and the downregulation of
vimentin in QBC939 and HUCCT1 cells, respectively. As EMT is an
important process implicated in invasion and metastasis, these
results may partially explain the mechanism by which Cul4B promotes
CCA progression.
Clinically, the precise stratification of CCA
patients according to their clinical prognosis in alignment with
therapeutic options remains a challenge (3). A number of studies have identified
multiple biomarkers that appear to possess prognostic significance.
Of these, p53 mutation, cyclins, cancer antigen19-9 and connective
tissue growth factor appeared to serve as predictors of outcome
(5). The present study demonstrated
that Cul4B expression is an unfavorable prognostic factor in
Chinese patients with CCA. A negative correlation between Cul4B
expression and OS in patients with EHCC, but not in patients with
IHCC, was also revealed. The lack of association of Cul4B
expression with poor survival in patients with IHCC may be
attributed to the small number of patients, included in the study,
or the fact that patients with CCA at different sites may present
different pathogenic features. It has previously been reported that
CCA differentially expresses cell cycle regulatory proteins based
on tumor location and morphology (22).
The present study demonstrated that the
co-overexpression of Cul4B and EGFR defines a subset of EHCC
patients with poor prognosis. A marginally positive correlation
between Cul4B and EGFR overexpression in EHCC patients was
revealed. EGFR has been suggested to be an important prognostic
factor and a potential therapeutic target in CCA (23). Wang et al (24) reported that Cul4A, another member of
Cul4, may be a promising therapy target and a potential biomarker
for prognosis and EGFR target therapy in patients with lung cancer.
Therefore, the ability of Cul4B to regulate EGFR expression in CCA
requires additional investigation.
In conclusion, the present study is the first to
demonstrate that the overexpression of Cul4B is an unfavorable
prognostic factor in Chinese patients with EHCC. Determining Cul4B
overexpression in surgically excised CCA tissues may aid to predict
the OS of patients. Additionally, these findings suggest that Cul4B
and EGFR expression may define a subset of patients with CCA with
poor prognosis.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (Beijing, China; grant nos.,
81072110, 81171951, 81330050 and 81672554).
References
|
1
|
Blechacz B, Komuta M, Roskams T and Gores
GJ: Clinical diagnosis and staging of cholangiocarcinoma. Nat Rev
Gastroenterol Hepatol. 8:512–522. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Patel T: Cholangiocarcinoma-controversies
and challenges. Nat Rev Gastroenterol Hepatol. 8:189–200. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Malhi H and Gores GJ: Cholangiocarcinoma:
Modern advances in understanding a deadly old disease. J Hepatol.
45:856–867. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hezel AF and Zhu AX: Systemic therapy for
biliary tract cancers. Oncologist. 13:415–423. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Briggs CD, Neal CP, Mann CD, Steward WP,
Manson MM and Berry DP: Prognostic molecular markers in
cholangiocarcinoma: A systematic review. Eur J Cancer. 45:33–47.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Petroski MD and Deshaies RJ: Function and
regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol.
6:9–20. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sarikas A, Hartmann T and Pan ZQ: The
cullin protein family. Genome Biol. 12:2202011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zou Y, Mi J, Cui J, Lu D, Zhang X, Guo C,
Gao G, Liu Q, Chen B, Shao C and Gong Y: Characterization of
nuclear localization signal in the N terminus of CUL4B and its
essential role in cyclin E degradation and cell cycle progression.
J Biol Chem. 284:33320–33332. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang T, Tang HM, Wu ZH, Chen J, Lu S,
Zhou CZ, Yan DW and Peng ZH: Cullin 4B is a novel prognostic marker
that correlates with colon cancer progression and pathogenesis. Med
Oncol. 30:5342013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yuan J, Han B, Hu H, Qian Y, Liu Z, Wei Z,
Liang X, Jiang B, Shao C and Gong Y: CUL4B activates Wnt/β-catenin
signalling in hepatocellular carcinoma by repressing Wnt
antagonists. J Pathol. 235:784–795. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hu H, Yang Y, Ji Q, Zhao W, Jiang B, Liu
R, Yuan J, Liu Q, Li X, Zou Y, et al: CRL4B catalyzes H2AK119
monoubiquitination and coordinates with PRC2 to promote
tumorigenesis. Cancer Cell. 22:781–795. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nauseef JT and Henry MD:
Epithelial-to-mesenchymal transition in prostate cancer: Paradigm
or puzzle? Nat Rev Urol. 8:428–439. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Thompson EW, Newgreen DF and Tarin D:
Carcinoma invasion and metastasis: A role for
epithelial-mesenchymal transition? Cancer Res. 65:5991–5995. 2015.
View Article : Google Scholar
|
|
14
|
Kong D, Banerjee S, Ahmad A, Li Y, Wang Z,
Sethi S and Sarkar FH: Epithelial to mesenchymal transition is
mechanistically linked with stem cell signatures in prostate cancer
cells. PLoS One. 5:e124452010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang L, Zhang J, Yang X, Chang YW, Qi M,
Zhou Z, Zhang J and Han B: SOX4 is associated with poor prognosis
in prostate cancer and promotes epithelial-mesenchymal transition
in vitro. Prostate Cancer Prostatic Dis. 16:301–307. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang Y, Liu R, Qiu R, Zheng Y, Huang W, Hu
H, Ji Q, He H, Shang Y, Gong Y and Wang Y: CRL4B promotes
tumorigenesis by coordinating with SUV39H1/HP1/DNMT3A in DNA
methylation-based epigenetic silencing. Oncogene. 34:104–118. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Method. 25:402–408. 2011.
View Article : Google Scholar
|
|
18
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee J and Zhou P: Pathogenic role of the
CRL4 ubiquitin ligase in human disease. Front Oncol. 2:212012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mok MT and Cheng AS: CUL4B: A novel
epigenetic driver in Wnt/β-catenin-dependent hepatocarcinogenesis.
J Pathol. 236:1–4. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Beroukhim R, Mermel CH, Porter D, Wei G,
Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J,
Urashima M, et al: The landscape of somatic copy-number alteration
across human cancers. Nature. 463:899–905. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jarnagin WR, Klimstra DS, Hezel M, Gonen
M, Fong Y, Roggin K, Cymes K, DeMatteo RP, D'Angelica M, Blumgart
LH and Singh B: Differential cell cycle-regulatory protein
expression in biliary tract adenocarcinoma: Correlation with
anatomic site, pathologic variables, and clinical outcome. J Clin
Oncol. 24:1152–1160. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang H, Berezov A, Wang Q, Zhang G,
Drebin J, Murali R and Greene MI: ErbB receptors: From oncogenes to
targeted cancer therapies. J Clin Invest. 117:2051–2058. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Y, Zhang P, Liu Z, Wang Q, Wen M,
Wang Y, Yuan H, Mao JH and Wei G: CUL4A overexpression enhances
lung tumor growth and sensitizes lung cancer cells to erlotinib via
transcriptional regulation of EGFR. Mol Cancer. 13:2522014.
View Article : Google Scholar : PubMed/NCBI
|