Introduction
Estrogen regulates numerous physiological processes
including normal cell growth, the central nervous and skeletal
systems and the development and regulation of tissue-specific genes
in the genital tract (1–3). Estrogen also affects the pathological
process of numerous hormone-dependent diseases including breast,
endometrial and ovarian cancer (1).
The biological actions of estrogen are mediated by the binding to
one of two specific estrogen receptors (ERs), ERα or ERβ, which
belong to the nuclear receptors superfamily (3). The binding of estrogen to its receptor
leads to a conformational change in the structure of the ER and to
the formation of estrogen receptor dimers that bind estrogen
response elements as homo- or hetero-dimers within the regulatory
sequences of estrogen-dependent genes (4). This conformational change, occurring as
a result of ligand binding, facilitates the association of
coactivator receptors and stabilizes the estrogen receptor complex
with estrogen response elements (4).
This promotes gene transcription and supports the stimulation of
cell growth in various tissues (5–7). ERα was
the first estrogen receptor to be isolated and cloned in 1980 from
a cell line of human breast cancer (MCF7) (8,9). In the
presence of estradiol (E2), this receptor may induce cell
proliferation via the regulation of certain genes including Myc,
cyclinD and Wnt11 (10). These genes
have several effects, including interfering with cadherins,
stimulating the cell cycle, promoting the transition from G1 to S
phase and altering apoptosis (10,11). This
leads to an increase in cell division, which may cause errors in
replication and promote cancer development (11).
In the 1990s, ERβ was the second estrogen receptor
discovered and identified in the rat prostate and ovaries, encoded
by 485 amino acids (1996) (12). In
the same year, ERβ was also isolated from human tissues, in this
case encoded by 477 amino acids (13). The role of ERβ in breast cancer
remains to be established (14,15). The
majority of previous studies have shown that ERβ functions as a
negative modulator of ERα and is associated with a good prognosis
and prolonged disease-free survival (16,17).
Previous studies have revealed that the expression
of ERβ is correlated with a poor prognosis, including accelerated
cell proliferation and distant metastasis (18,19).
However, Speirs et al (20)
have identified that co-expression of ERα and ERβ is associated
with high-grade tumors and metastases. In addition, Grober et
al (21) demonstrated that ERβ is
able to interfere with ERα in the regulation of target genes. It is
therefore necessary to understand the role of ERβ in breast cancer
and to elucidate the nature of its association with ERα. In this
context, the present study was conducted to investigate the
expression the of estrogen receptors ERα and ERβ in a series of
breast cancer tumors. This was achieved by comparing the results
with clinical and pathological parameters, as well as the
expression of the oncoprotein human epidermal growth factor
receptor 2 (HER2) and the proliferation index Ki-67. The objective
of this study was also to analyze the ERα and ERβ subgroups
according to the aforementioned parameters.
Materials and methods
Patients and samples
The tissue of malignant mammary tumors was excised
during tumorectomy from 32 females (mean age, 58.5 years; range,
32–85 years) was analyzed. Healthy tissues collected from patients
during the tumorectomy were used as controls. The patients all had
invasive ductal carcinoma and did not receive radiotherapy or
chemotherapy prior to surgery. The samples were subject to a
histological examination by a pathology specialist to determine the
presence of malignant cells. Each diagnosed sample was divided into
two portions: One portion was immediately processed for
immunohistochemistry and the other portion was frozen and
maintained at −80°C until RNA extraction. All pathological,
clinical and personal data were anonymized and separated from any
personal identifiers. All the procedures followed were examined and
approved by the Saleh Azaiez Oncology Institute (Tunis,
Tunisia).
Total RNA isolation and reverse
transcription
Total RNA was extracted from breast specimens using
a mechanical stirrer in the presence of a lysis buffer [4.5 M
guanidine-HCl, 50 ml Tris-Hcl, 30% Triton X-100 (w/v) pH 6.6
(25°C)], prior to the use of total RNA isolation and high pure RNA
isolation kits (Roche Diagnostics, Basel, Switzerland). Equal
amounts of total RNA (1 µg) were reverse transcribed. cDNA
synthesis was carried out using the PrimeScript™ 1st strand cDNA
Synthesis kit (Takara Biotechnology Co., Ltd., Dalian, China).
Primers and quantitative polymerase
chain reaction (qPCR)
All PCR reactions were performed using an ABI Prism
7700 Sequence Detection system (Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). cDNA was amplified using the
SYBR1-Green PCR Core Reagents kit (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The reaction mixes used for qPCR were as
follows: 10 µl SYBR-Green (Applied Biosystems; Thermo Fisher
Scientific, Inc.), 6 µl water; 1 µl forward primer; 1 µl reverse
primer; and 2 µl cDNA. The primers used to amplify ERα were as
follows: forward, 5′-TGCCAAGGAGACTCGCTA-3′; reverse,
5′-TCAACATTCTCCCTCCTC-3′. For ERβ, the primer sequences were
forward, 5′-TGTTACGAAGTGGGAATGTGA-3′ and reverse,
5′-TCTTGTTCTGGACAGGGATG-3 (40 cycles of: 94°C for 30 sec, 55°C for
30 sec, 72°C for 30 sec for both ERα and ERβ). 18S was used as an
endogenous control. The primer sequences used to amplify 18S were
forward, 5′GTAACCCGTTGAACCCCATT-3′ and reverse,
5′-CCATCCAATCGGTAGTAGCG-3′ (40 cycles of: 94°C for 30 sec, 56°C for
30 sec and 72°C for 30 sec). Relative mRNA levels were calculated
based on Cq values and corrected for the 18S expression according
to the equation 2−ΔΔCq (22). Relative mRNA levels in the control
tissue were equated to 1 and other values were expressed relative
to this.
All primer pairs were initially validated by testing
them for equal amplification efficiencies. The amplification
efficiency was close to 2 under these conditions. Experiments were
performed in triplicate for each data point.
Immunohistochemical staining
The immunohistochemistry expression of the
oncoprotein Her2/neu and the proliferation index Ki-67 were tested
on the same set of tumors. The primary antibodies used were as
follows: Mouse anti-human Her2 (#CB11) and mouse anti-human Ki-67
(#MM1) (Novocastra; Leica Biosystems GmbH, Wetzlar, Germany).
A total of 32 tissue samples were fixed for 24 h at
room temperature in 0.1 M phosphate buffered 10% formaldehyde,
dehydrated in a graded ethanol series of increasing concentrations
(70, 85, 90 and 100% for 10 min each), impregnated with xylene and
embedded in paraffin. Sections (4 µm thick) were processed
following the NovoLink™ Polymer Detection systems (Novocastra;
Leica Biosystems GmbH) method. Sections were deparaffinized by an
overnight incubation at 59°C, and subsequently placed in a xylene
bath for 15 min at room temperature. Sections were subsequently
hydrated, incubated for 30 min in 1% hydrogen peroxide to block
endogenous activity, and then antigen retrieval was performed by
incubating the sections in a 0.01 M citrate buffer (Epitope
Retrieval Solution pH 6.0; Leica Microsystems GmbH, Wetzlar,
Germany) for 30 min at 98°C. Subsequently, the primary antibodies
were applied for 1 h at 4°C, with a dilution of 1:40 for Her2 and
1:200 for Ki-67. The sections were then incubated at room
temperature with Post Primary Block for 30 min to block
non-specific polymer binding. The sections were incubated with a
NovoLink™ Polymer for 30 min at room temperature, followed by
incubations with 3,3′-diaminobenzidine (DAB) working solution for 5
min at room temperature to develop peroxidase activity. The slides
were counterstained with hematoxylin and mounted. Staining
specificity was checked using negative controls. Negative controls
were obtained by replacing the primary antibody with an antibody of
the same unrelated isotope during the immunohistochemistry
technique or by omission of the primary antibody. Primary breast
tissues were incubated in blocking peptides (Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) instead of primary
antibodies.
The Ki-67 assessment and the Her2 status tests were
performed by two experienced breast pathologists. The percentage of
positively stained cells obtained is an average following counting
of the stained cells and the total number of cells were counted in
four high-magnification fields using a light microscope
(magnification, ×400; Media Cybernetics, Inc., Rockville, MD, USA).
The staining of Ki-67 was scored for the percentage of positive
cells (0, 0–5%; 1, 6–25%; 2, 26–50%; 3, >50%). The optimal
cutoff value was identified as 1 for the low Ki-67 expression level
and >1 for high expression (23).
The scoring of Her2 was performed on a 0–3 scale (24). Positive (3+) was defined as intense
complete membranous staining in >30% of the tumor cell
population; borderline (2+) was defined as moderate membranous
staining in >10% of tumor cells; 1+ was defined as either weak
or barely perceptible membranous staining in >10% of the tumor
cells. Furthermore, a chromogenic in situ hybridization
(CISH) analysis was performed, as described previously (25) for Her2/neu gene amplification in all
2+ cases, as defined by IHC. Scores of 0, 1+ and 2+ according to
IHC but negative following CISH were considered as negative for the
Her2/neu expression, whereas 3+ scores and 2+ cases defined as
positive by CISH were considered as positive for Her2/neu
expression.
Statistical analysis
The analysis of the results involving the expression
of the estrogen receptors ERα and ERβ together with the various
histological and clinical parameters was performed using the
χ2 test with R (i386 3.2.1) software. Data are presented
as the mean ± standard deviation. P<0.05 was considered to
indicate a statistically significant difference.
Results
Expression of mRNA ERα and ERβ in
malignant human breast tissues and their association with clinical
parameters
In total, 32 breast tumor samples were analyzed and
compared for the expression of the two ER isoforms by reverse
transcription-qPCR. The expression of estrogen receptors in breast
cancer tumors had a positivity of ~68% for ERα and 65% for ERβ. As
presented in Table I, no significant
difference was identified between the levels of expression of the
two ER isoforms (P>0.05).
 | Table I.Association between mRNA ERα and ERβ
levels and standard clinicopathological factors and molecular
settings. |
Table I.
Association between mRNA ERα and ERβ
levels and standard clinicopathological factors and molecular
settings.
|
|
| ERα |
| ERβ |
|
|---|
|
|
|
|
|
|
|
|---|
|
Characteristics | Total
population | Negative (%) | Positive (%) | P-value | Negative (%) | Positive (%) | P-value |
|---|
| Total | 32 (100%) | 10 (31.28) | 22 (68.75) |
| 11 (34.37) | 21 (65.62) | >0.05 |
| Age |
|
|
|
|
|
|
|
| ≤50
years | 12 (37.5) | 4 (40) | 8 (36.36) |
| 5 (45.45) | 7 (33.33) |
|
| >50
years | 20 (62.5) | 6 (60) | 14 (63.63) | 0.13 | 6 (54.54) | 14 (66.66) | 0.06 |
| Grade |
|
|
|
|
|
|
|
|
SBRI | 7 (21.87) | 4 (40) | 3 (13.63) |
| 4 (36.36) | 3 (14.28) |
|
|
SBRII | 17 (53.12) | 3 (30) | 14 (63.63) |
| 5 (45.45) | 12 (57.1) |
|
|
SBRIII | 8 (25) | 3 (30) | 5 (22.72) | 0.01 | 2 (18.18) | 6 (28.57) | 0.01 |
| Lymph node
status |
|
|
|
|
|
|
|
|
Positive | 17 (53.12) | 6 (60) | 11 (50) |
| 4 (36.36) | 13 (61.90) |
|
|
Negative | 15 (46.87) | 4 (40) | 11 (50) | >0.05 | 7 (63.63) | 8 (38.09) | 0.20 |
| Tumor size |
|
|
|
|
|
|
|
| ≤30
mm | 22 (68.75) | 6 (60) | 16 (72.72) |
| 8 (72.72) | 14 (66.66) |
|
| >30
mm | 10 (31.25) | 4 (40) | 6 (27.27) | 0.006 | 3 (27.27) | 7 (33.33) | 0.06 |
| HER2 status |
|
|
|
|
|
|
|
|
Positive | 8 (25) | 3 (30) | 5 (22.72) |
| 6 (72.72) | 2 (57.14) |
|
|
Negative | 24 (75) | 7 (70) | 17 (77.27) | 0.0009 | 5 (27.27) | 19 (42.85) |
7.905−7 |
| Ki-67 |
|
|
|
|
|
|
|
|
≤5% | 9 (28.12) | 3 (44) | 6 (27.27) |
| 3 (27.27) | 6 (28.57) |
|
|
>5% | 23 (71.87) | 7 (56) | 16 (72.72) | 0.0066 | 8 (72.72) | 15 (71.42) | 0.01 |
In order to determine whether the expression of each
ER isoform in the breast tumors was associated with clinical
parameters, associations between tumor grade and size, lymph node
metastasis and menopausal status were examined (Table I). When compared with tumor grade,
there was a significant association between ERα and ERβ expression
and tumor grade (P=0.01; Table I).
Analysis of the mRNA expression of the receptors ERα and ERβ
revealed a significant increase of ERα in tumors that were grade 2
compared with grades 1 and 3. However, the highest ERβ expression
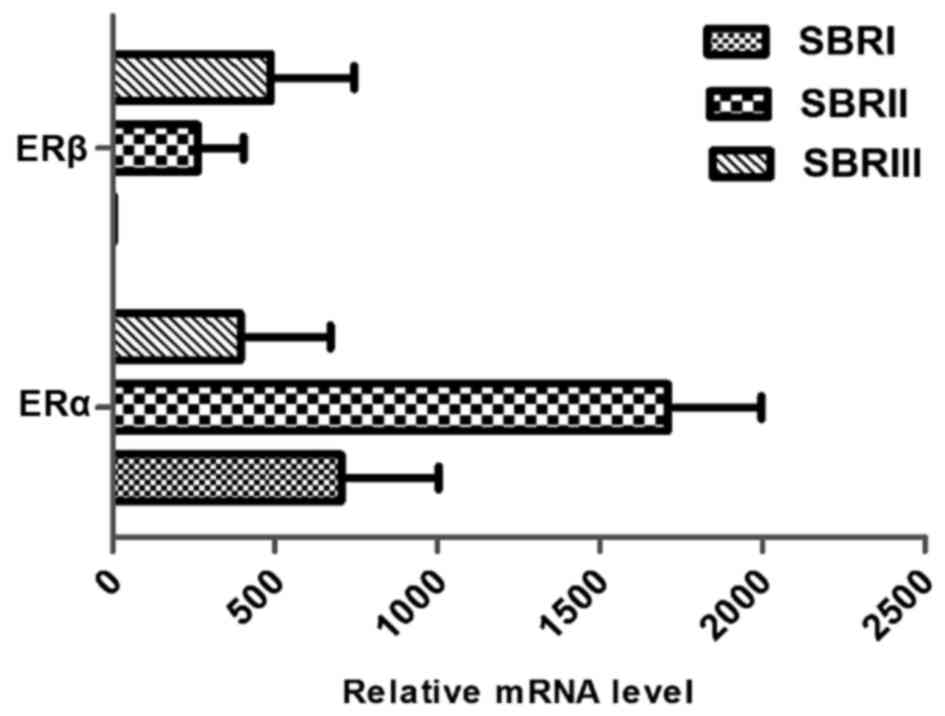
was found in the Scarff-Bloom-Richardson (SBR)3 grade (Fig. 1). According to the histopathological
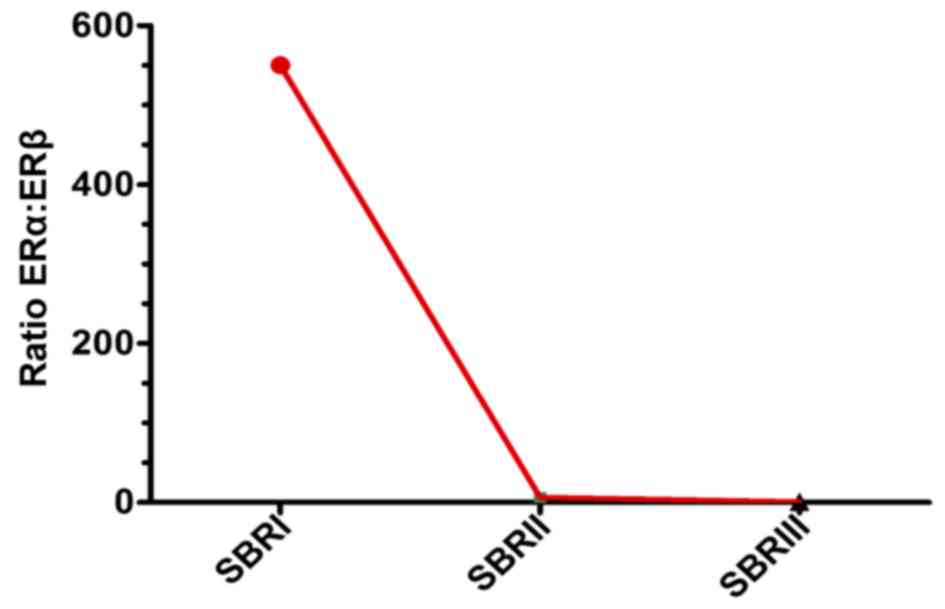
grade progress, the ERα: ERβ ratio declined from SBR1 to SBR2 and
SBR3 (Fig. 2).
Tumor size was correlated with ERα (P=0.006)
expression and did not correlate with ERβ expression (P=0.06;
Table I). In addition, there was no
significant difference in ERα and ERβ expression according to lymph
node metastasis status (P>0.05; Table
I). As presented in Fig. 3, the
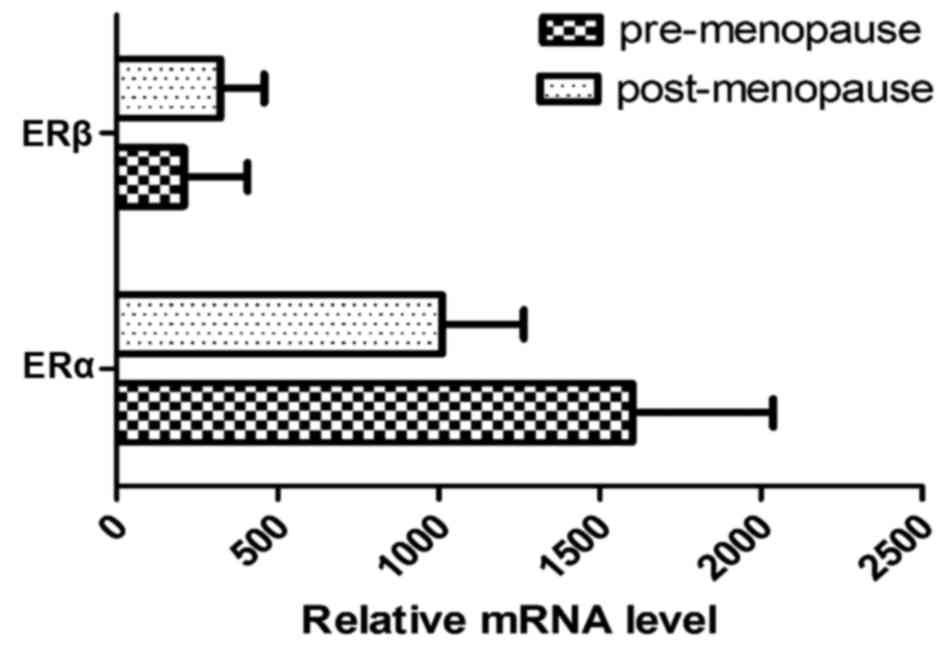
expression of the two ER isoforms differed with menopausal status.
The amount of mRNA ERα expression was 1.5X higher in premenopausal
breast tumors compared with postmenopausal breast tumors.
Inversely, mRNA expression of ERβ increased between the
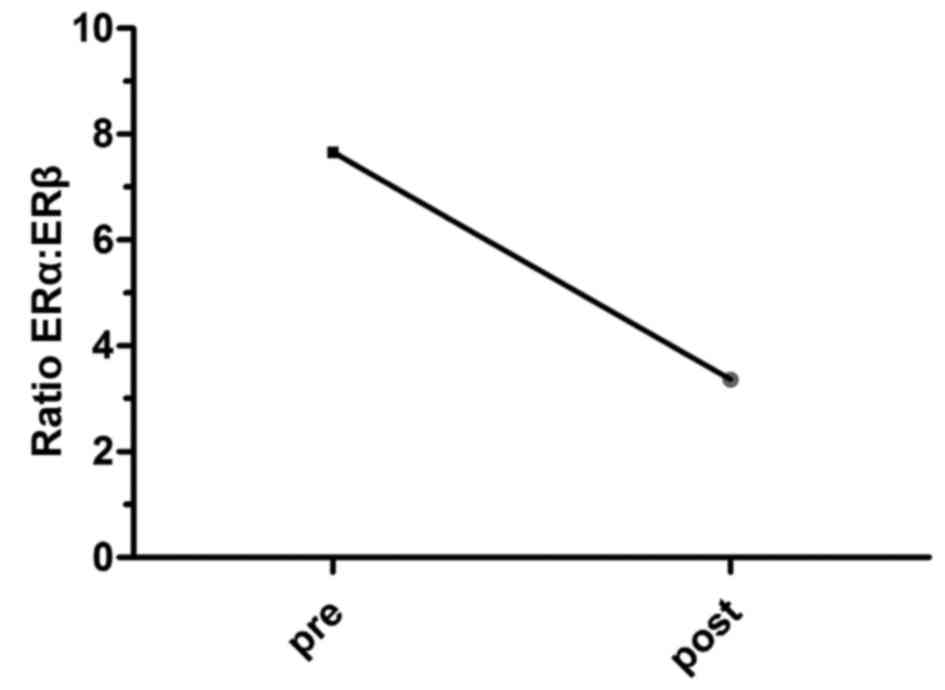
premenopausal and postmenopausal status (Fig. 3). Furthermore, a shift in the ERa:ERβ
ratio was noted and this was revealed to decline in the
postmenopausal vs. premenopausal status group (7.66 to 3.36)
(Fig. 4).
HER2/neu and Ki-67 in ER-positive vs.
ER-negative cases
Differences in the HER2 and Ki-67 status between
ER-positive and ER-negative cases were further analyzed.
Immunohistochemical analysis was performed to assess the expression
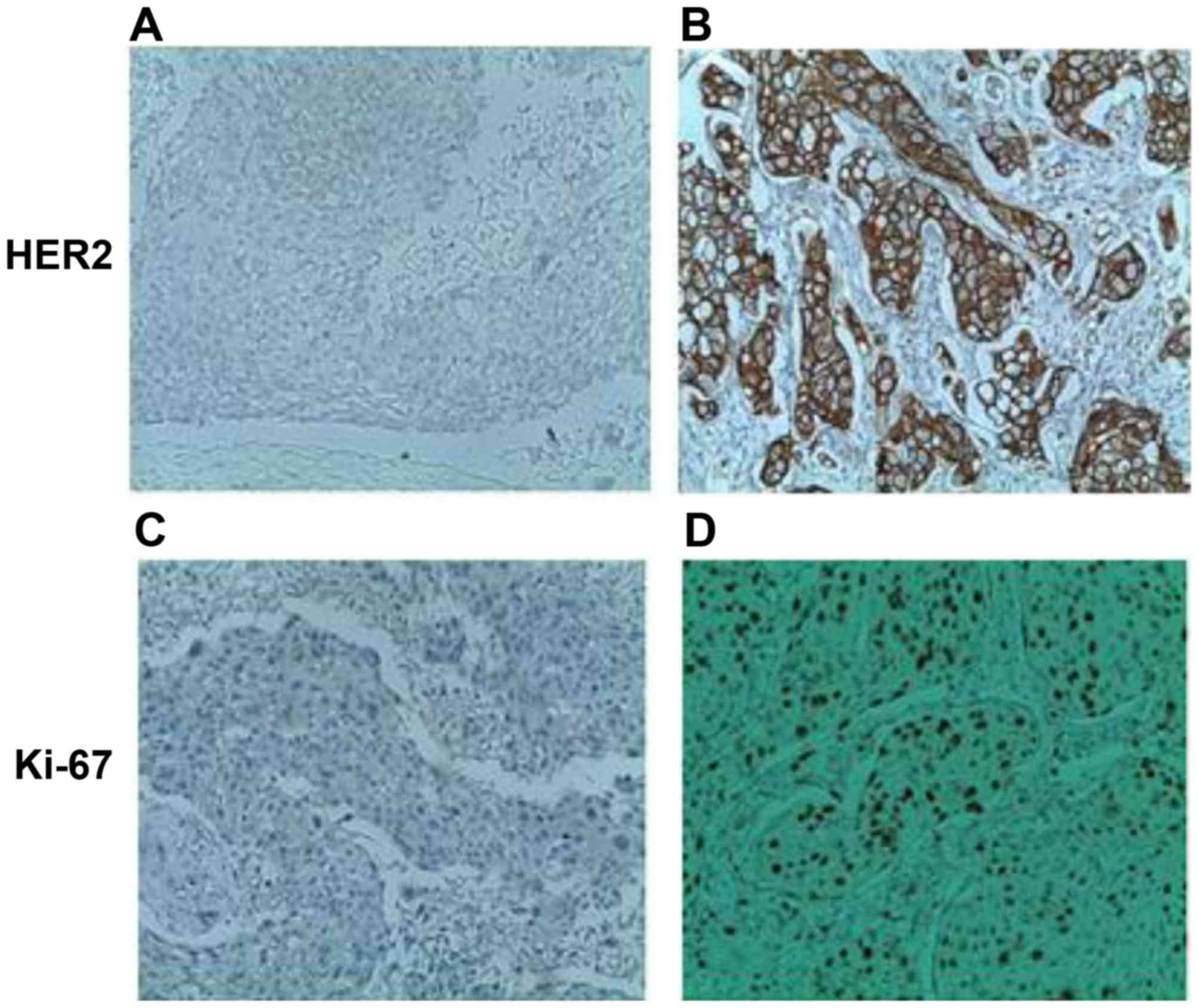
of HER2 and Ki-67 in breast cancer samples. Fig. 5 reveals membrane localization of the
HER2 protein in malignant breast cells. Ki-67 was primarily
localized to the nucleus of breast neoplastic cells (Fig. 5). As summarized in Table I, a marked negative association
between the two estrogen receptors and the oncoprotein HER2/neu was
observed. In total, 17/22 ERα positive cases were negative for
HER2. Similarly, 19/21 ERβ positive cases were negative for HER2.
This negative association was more significant for ERβ
(P=7.905×10−7) than ERα (P=0.0009; Table I).
For Ki-67, there was a discrepancy in the prognostic
importance of this factor between the ER-positive and ER-negative
cases. A high Ki-67 index of ≥5% was associated with the
ER-positive subgroup (P=0.006 for ERα and P=0.01 for ERβ; Table I).
Association between ERα and ERβ breast
cancer subgroups and clinical information
The expression of ER subtypes within the tumor group
was further analyzed. According to positive or negative expression
of the hormone receptors ERα and ERβ, ERα+ and ERβ+ was
significantly the most expressed ER subgroup in patients with
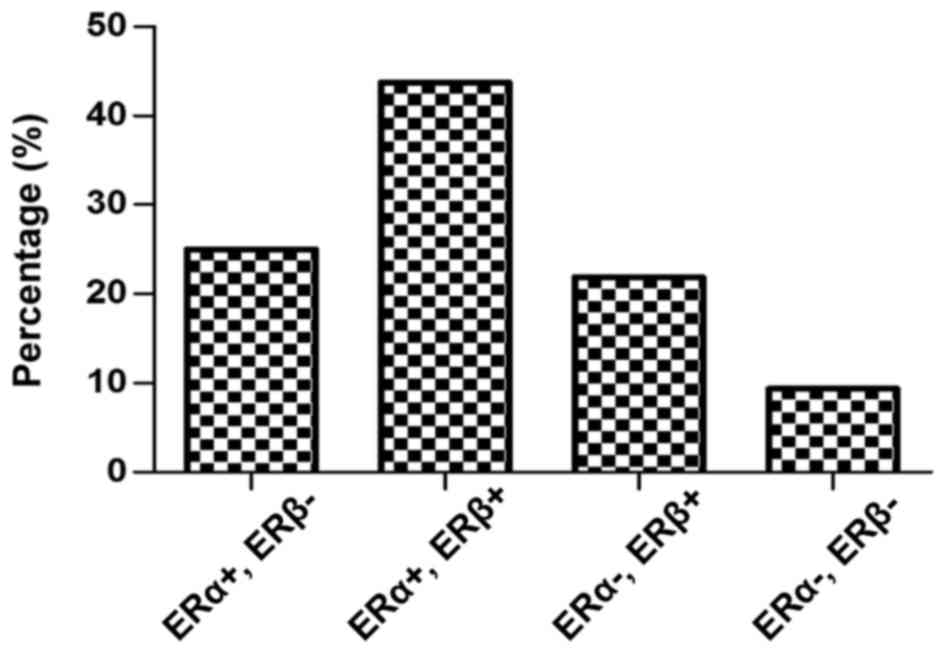
breast cancer (P=0.01; Fig. 6). A
total of 43.75% of the malignant breast samples co-expressed the
two ER subtypes, compared with only 25 and 21.87% of the breast
tumors which expressed either (ERα+, ERβ- or ERα-, ERβ+ subgroups,
respectively). A small number of breast cancer samples were in the
ERα-, ERβ- subgroup (9.37%; Fig.
6).
The distribution of ERα and ERβ expression groups
according to the menopausal status exhibited a significant
difference for the ERα+, ERβ+ subgroup (P=0.05). As presented in
Table II, 71.42% of the ERα and ERβ
co-expression cases occurred following the menopause stage, whereas
only 28.57% occur prior to menopause. The other ER subgroups
studied were distributed approximately homogeneously prior to and
following the menopause phase. When compared with tumor grade,
there was a significant association between the ERα+, ERβ+ subgroup
and SBR grade (P=0.005). This was not observed for the other ER
subgroups, but there was a non-significant association between the
ERα+, ERβ-subgroups and the primary stages of cancer (P=0.08).
Similarly, depending on the nodal status, there was an association
between the ERα+, ERβ+ subgroup with the infiltration of the
lymphatic ganglion (P=0.05; Table
II).
 | Table II.Association between ERα and ERβ
breast cancer subgroups with clinicopathological and molecular
parameters. |
Table II.
Association between ERα and ERβ
breast cancer subgroups with clinicopathological and molecular
parameters.
| Clinicopathological
and molecular parameters | ERα+, ERβ-
(n=8) | ERα+, ERβ+
(n=14) | ERα-, ERβ+
(n=7) | ERα-, ERβ-
(n=3) |
|---|
|
| n (%) | n (%) | n (%) | n (%) |
| Menopausal
status |
|
|
|
|
|
Premenopausal | 4 (50) | 4
(28.57) | 3 (42.85) | 1 (33.33) |
|
Postmenopausal | 4 (50) | 10 (71.42) | 4 (57.14) | 2 (66.66) |
|
P-value | >0.05 | 0.05 | >0.05 | >0.05 |
| Histological
grade |
|
|
|
|
|
SBRI | 2 (25) | 1(7.14) | 2 (28.57) | 2 (66.66) |
|
SBRII | 5 (62.5) | 9 (64.28) | 3 (42.85) | 0 |
|
SBRIII | 1 (12.5) | 4 (28.57) | 2 (28.57) | 1 (33.33) |
|
P-value | 0.08 | 0.01 | 0.80 | 0.22 |
| Lymph node
status |
|
|
|
|
|
Positive | 2 (25) | 10 (71.42) | 4 (57.14) | 2 (66.66) |
|
Negative | 6 (75) | 4
(28.57) | 3 (42.85) | 1 (33.33) |
|
P-value | 0.13 | 0.05 | >0.05 | >0.05 |
| HER2/neu |
|
|
|
|
|
Positive | 4 (50) | 1 (7.14) | 1 (14.28) | 2 (66.66) |
|
Negative | 4 (50) | 13 (92.85) | 6 (85.71) | 1 (33.33) |
|
P-value | >0.05 | 3.22×10–5 | 0.03 | >0.05 |
| Ki-67 |
|
|
|
|
|
≤5% | 6 (75) | 3 (21.42) | 3 (42.85) | 1 (33.33) |
|
>5% | 2 (25) | 11 (78.57) | 4 (57.14) | 2 (66.66) |
|
P-value | 0.10 | 0.01 | >0.05 | >0.05 |
Subset analyses of HER2/neu and Ki-67
in ERa, ERβ breast cancer subgroups
As presented in Table
II, the complex associations between various pathological
variables were considered in the current study. Using this
analysis, it is possible to visualize the association of biological
factors (ERα, ERβ, HER2 and Ki-67) with ER subgroups and study
their associations with conventional pathological factors. Breast
tumors with an ERα+, ERβ+ profile were characterized by negative
HER2 status. A statistically significant relationship between the
co-expression of estrogen receptors and HER2 negative was
identified in the tumor samples (P=3.21×10−5). A
statistically significant negative association was also observed
between the ERα-, ERβ+ profile and HER2 expression (P=0.03;
Table II).
Using a Ki-67 cut-off value of 5% (Table II), only the ERα+, ERβ+ breast cancer
subgroup was significantly associated with a Ki-67 index >5%
(P=0.008).
Discussion
Previous studies have been conducted to decipher the
role of ERs in breast carcinogenesis (16,18,19).
Nonetheless, the role of the ERβ isoform in this malignancy and its
association with ERα remains to be elucidated and the results of
earlier studies are somewhat contradictory (26,27). The
results of the current study indicate that ERα and ERβ expression
was retained in a majority of breast cancer cases (68 and 65%,
respectively). The associations between ERα and ERβ expression and
menopausal status revealed that each isoform was more significantly
expressed in postmenopausal patients than in premenopausal ones
(Table I), primarily for ERβ which
was ~2 times higher in post-menopausal cases (P=0.06). These data
are concordant with previous studies demonstrating that the two ER
isoforms are frequently positive in postmenopausal patients
(28–30). However, considering the relative
amount of mRNA, the ERα expression was found to be higher in
premenopausal phases (1.5-fold) compared with postmenopausal breast
tumors in the present study. Inversely, the mRNA expression level
of ERβ was often higher in postmenopausal patients. Consequently,
the ERα:ERβ ratio decreases from 7.66 to 3.36 (Fig. 4), which translates into the ERβ
activity increasing in malignant breast tumors of post-menopausal
patients. Furthermore, the expression levels of ERα and ERβ were
revealed to be associated with smaller tumors and significantly
associated with the SBR histopathological grade (P=0.01; Table I). In fact, ERβ was more highly
expressed than ERα in the SBRIII grade. In addition, the ERα mRNA
level reached a maximum in the SBRII grade and decreased in SBR
grade III. However, the ERβ mRNA level was associated with the
advancement of the SBR grade. ERβ expression reached its maximum in
the SBR III grade and was higher than that of ERα. Consequently,
the ERα:ERβ ratio was inversely associated with SBR grade
advancement, and the ratio tended to 0 at the SBR III grade
(Fig. 2).
These results demonstrate a slight association
between ERβ expression (but not ERα) and node-positive breast
cancer (P=0.06), which is corroborated by the findings of Hou et
al (31), which demonstrated that
ERβ exerts stimulatory effects on breast cancer development and
metastasis. Furthermore, Yan et al (23) revealed that the expression of ERβ2 is
also correlated with high-grade tumors, distant metastasis and
breast cancer mortality (23).
Estrogen-ER signaling is important in normal mammary
gland development and breast carcinogenesis (32). Due to the crosstalk between the ER and
epidermal growth factor receptor (EGFR)/HER2 signaling pathways
and/or their downstream effectors, the majority of patients develop
resistance to endocrine therapy (predominantly tamoxifen) (33). In fact, the relation of ERα and ERβ
with HER2 is of particular interest. In the current study, the
HER2-negative status was associated with positive ERα and ERβ
expression. This result is concordant with previous data that
demonstrated a significant negative correlation between the
expression of the hormone receptor and HER2/neu amplification
(24,34,35). The
combination of HER2 overexpression and a high Ki-67 index has been
suggested to be a prognostic molecular marker for breast cancer
(36). The present study revealed
that positive ER receptors (α and β) were associated with a high
Ki-67 index of ≥5% (P=0.006 and P=0.01 respectively) for breast
cancer (Table I). These results are
concordant with previous reports (18,37), and
imply that ERα and ERβ may have a role in breast cancer
development, metastasis and proliferation.
ERβ may be expressed alone (ERα-, ERβ+) or
co-expressed with ERα (ERα+, ERβ+); therefore, a comparison of the
previously cited clinical and molecular parameters referring to the
(ERα, ERβ) breast cancer subgroups is required. The present study
identified that ERα and ERβ co-expression was retained in the
majority of breast cancer cases (P=0.01; Fig. 6), hypothesizing an interaction between
the two nuclear receptors. In this context, Järvinen et al
(38) and Leung et al
(39) reported that positive ERβ
expression is associated with positive ERα in breast cancer tumors.
Given that ERα and ERβ were coexpressed in the majority of breast
tumors, this expression may occur in the form of their
heterodimers. ERα and ERβ heterodimers may also serve a significant
role in breast cancer, but this remains to be established (40).
The present study demonstrated that the
co-expression of ERα and ERβ occurs frequently following menopause,
which may be associated with the in situ synthesis of
steroid hormones (the epithelium of the mammary gland) from the
steroid precursors dehydroepiandrosterone (DHEA) and DHEA-sulfate.
Through an intracrinology mechanism, E2 may be synthesized locally
by the aromatization of the androgen by aromatase (41). Depending on the presence of ERα and
ERβ in certain cells, the receptors form functional homo- or
heterodimers on promoter elements (40). The results of the current study reveal
that the progression of breast cancer may be dependent on the
expression levels of estrogenic receptors, particularly the
co-expression of the ERα and ERβ isoforms (ERα+, ERβ+ subgroup).
Indeed, ERα and ERβ co-expression is associated with high-grade
tumors (grade II and III; P=0.005) and potentially lymph node
infiltration (P=0.05). This finding is concordant with the findings
of Speirs et al (20), which
demonstrated that tumors that expressed ERα and ERβ were
node-positive and tended to be of a higher grade. This implies that
ERα and ERβ may cooperate to generate a tumor phenotype with a
higher metastatic potential. The present study hypothesizes that
there is a synergic effect between ERα and ERβ, which exerts
stimulative effects on breast cancer development and
metastasis.
The potential stimulative effect exerted by the
co-expression of the ERα and ERβ receptors is supported by the
significantly high proliferation rate estimated by the Ki-67 index
in the (ERα+, ERβ+) breast cancer subgroup. Notably, it has been
reported that an increase in the proliferation rate occurs during
the progression towards invasive ductal carcinoma, implying a
significant role for Ki-67 in breast tumorigenesis (42). However, the (ERα+, ERβ-) subgroup is
associated with primary cancer grade (SBRI and II), non-infiltrated
lymph nodes and a lower proliferation index (<5%). This reveals
that the expression of ERα alone may be a marker of non-aggressive
tumors, as has been suggested by a previous study (28). Our results demonstrate a negative
association between the (ERα-, ERβ +) subgroup and HER2. Such
results are corroborated by a study by Marotti et al
(34), which demonstrated that the
co-expression of ERa and ERb was associated with a negative status
of HER2. In addition, Lindberg et al (35) revealed that ERβ is able to increase
phosphatase and tensin homolog levels and decrease HER2/HER3
signaling, thereby reducing protein kinase B signaling. The
co-expression of ERβ and ERα (ERα+, ERβ+ subgroup) was negatively
associated with HER2 expression in the present study. These
findings are consistent with a previous report, which concluded
that ERβ is significantly associated with ERα expression and
inversely associated with HER2 over-expression (34). As these two receptor systems (ER
isoforms and HER2) have the capacity to activate one another
(30), the present study hypothesizes
a synergistic effect between ERα and ERβ to abrogate HER2
activation as a part of the crosstalk between ER and growth factor
receptors. In this context, ER-induced signaling pathways,
demonstrated through in vitro cellular models, were found to
induce EGFR ligands, such as transforming growth factor α (43) and lead to downregulation of EGFR
(44) and HER2 (45). This growth factor receptor, HER2/neu,
is not typically over-expressed in normal or benign breast lesions
(46). However, a significantly lower
level of HER2/neu expression in invasive carcinoma has been
previously reported (47). The
results of the current study imply that the molecular phenotype
defined by the presence of ERa, ERβ and the absence of HER2 may be
a precursor for the development of a more aggressive and malignant
invasive ductal carcinoma. Furthermore, the coexpression of ERα and
ERβ may be a marker of hormonal sensitivity in association with
downregulation of HER2 expression.
In conclusion, the results of the current study
suggest an important role for ERβ in breast cancer development,
proliferation and metastasis, particularly when coexpressed with
ERα. The ERα+, ERβ+ subgroup is associated with high tumor grade,
metastasis and a high proliferation index. Therefore, the
co-expression of ERα and ERβ may be an indicator of aggressive
tumors. In addition, the ERα+, ERβ+ subgroup is significantly
associated with an HER2-negative status, and this may indicate a
sensitivity towards hormonal therapy due to the downregulation of
HER2 expression. Given that the ERα-ERβ balance is influenced by
the tumor microenvironment, including cytokines and growth factors,
decrypting signaling pathways elicited by those components and
their crosstalk with ER signaling may be investigated further in
future studies.
Acknowledgements
The authors would like to thank the Histology and
Immunocytology lab members at the Saleh Azeiz Cancer Institute
(Tunis, Tunisia). The present study was funded by The IMEC unit,
The Sciences Faculty of Bizerte, The University of Carthage and The
Tunisian ministry of higher education.
References
|
1
|
Couse JF and Korach KS: Estrogen receptor
null mice: What have we learned and where will they lead us? Endocr
Rev. 20:358–417. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nilsson S, Mäkelä S, Treuter E, Tujague M,
Thomsen J, Andersson G, Enmark E, Pettersson K, Warner M and
Gustafsson JA: Mechanisms of estrogen action. Physiol Rev.
81:1535–1565. 2001.PubMed/NCBI
|
|
3
|
Pettersson K and Gustafsson J: Role of
estrogen receptor beta in estrogen action. Annu Rev Physiol.
63:165–192. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hall JM and McDonnell DP: The estrogen
receptor beta-isoform (ERbeta) of the human estrogen receptor
modulates ERalpha transcriptional activity and is a key regulator
of the cellular response to estrogens and antiestrogens.
Endocrinology. 140:5566–5578. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tsai MJ and O'Malley BW: Molecular
mechanisms of action of steroid/thyroid receptor superfamily
members. Annu Rev Biochem. 63:451–486. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mangelsdorf DJ, Thummel C, Beato M,
Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M,
Chambon P and Evans RM: The nuclear receptor superfamily: The
second decade. Cell. 83:835–839. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
McKenna NJ and O'Malley BW: Combinatorial
control of gene expression by nuclear receptors and coregulators.
Cell. 108:465–474. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Walter P, Green S, Greene G, Krust A,
Bornert JM, Jeltsch JM, Staub A, Jensen E, Scrace G, Waterfield M,
et al: Cloning of the human estrogen receptor cDNA. Proc Natl Acad
Sci USA. 82:pp. 7889–7893. 1985; View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Green S, Walter P, Kumar V, Krust A,
Bornert JM, Argos P and Chambon P: Human estrogen receptor cDNA:
Sequence, expression and homology to v-erb-A. Nature. 320:134–139.
1986. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin Z, Reierstad S, Huang CC and Bulun SE:
Novel estrogen receptor-alpha binding sites and estradiol target
genes identified by chromatin immunoprecipitation cloning in breast
cancer. Cancer Res. 67:5017–5024. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Finn R, Dering J, Conklin D, Kalous O,
Cohen D, Desai A, Ginther C, Atefi M, Chen I, Fowst C, et al: PD
0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially
inhibits proliferation of luminal estrogen receptor-positive human
breast cancer cell lines in vitro. Breast Cancer Res. 11:R772009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kuiper GG, Enmark E, Pelto-Huikko M,
Nilsson S and Gustafsson JA: Cloning of a novel receptor expressed
in rat prostate and ovary. Proc Natl Acad Sci USA. 93:pp.
5925–5930. 1996; View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mosselman S, Polman J and Dijkema R: ER
beta: Identification and characterization of a novel human estrogen
receptor. FEBS Lett. 392:49–53. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Leung YK, Lee MT, Lam HM, Tarapore P and
Ho SM: Estrogen receptor-beta and breast cancer: Translating
biology into clinical practice. Steroids. 77:727–737. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fox EM, Davis RJ and Shupnik MA: ERbeta in
breast cancer-onlooker, passive player, or active protector?
Steroids. 73:1039–1051. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pettersson K, Delaunay F and Gustafsson
JA: Estrogen receptor beta acts as a dominant regulator of estrogen
signaling. Oncogene. 19:4970–4978. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lazennec G: Estrogen receptor beta, a
possible tumor suppressor involved in ovarian carcinogenesis.
Cancer Lett. 231:151–157. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jensen EV, Cheng G, Palmieri C, Saji S,
Mäkelä S, van Noorden S, Wahlström T, Warner M, Coombes RC and
Gustafsson JA: Estrogen receptors and proliferation markers in
primary and recurrent breast cancer. Proc Natl Acad Sci USA. 98:pp.
15197–16202. 2001; View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Skliris GP, Leygue E, Curtis-Snell L,
Watson PH and Murphy LC: Expression of oestrogen receptor-beta in
oestrogen receptor-alpha negative human breast tumours. Br J
Cancer. 95:616–626. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Speirs V, Parkes AT, Kerin MJ, Walton DS,
Carleton PJ, Fox JN and Atkin SL: Coexpression of estrogen receptor
alpha and beta: Poor prognostic factors in human breast cancer?
Cancer Res. 59:525–528. 1999.PubMed/NCBI
|
|
21
|
Grober OM, Mutarelli M, Giurato G, Ravo M,
Cicatiello L, De Filippo MR, Ferraro L, Nassa G, Papa MF, Paris O,
et al: Global analysis of estrogen receptor beta binding to breast
cancer cell genome reveals an extensive interplay with estrogen
receptor alpha for target gene regulation. BMC Genomics. 12:362011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real time quantitative PCR and
the 2(−Delta Delta C(T)). Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yan M, Rayoo M and Takano EA; kConFab
Investigators and Fox SB, : Nuclear and cytoplasmic expressions of
ERβ1 and ERβ2 are predictive of response to therapy and alters
prognosis in familial breast cancers. Breast Cancer Res Treat.
126:395–405. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Arafah M: Correlation of hormone receptors
with Her-2 Neu protein expression and the histological grade in
invasive breast cancers in a cohort of Saudi Arabia. Turk Patoloji
Dergisi. 28:38–43. 2012.PubMed/NCBI
|
|
25
|
Isola J and Tanner M: Chromogenic in situ
hybridization in tumor pathology. Methods Mol Med. 97:133–144.
2004.PubMed/NCBI
|
|
26
|
Jonsson P, Katchy A and Williams C:
Support of a bi-faceted role of estrogen receptor β (ERβ) in
ERα-positive breast cancer cells. Endocr Relat Cancer. 21:143–160.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang B, Warner M and Gustafsson JÅ:
Estrogen receptors in breast carcinogenesis and endocrine therapy.
Mol Cell Endocrinol 418 Pt. 3:240–244. 2015. View Article : Google Scholar
|
|
28
|
Bièche I, Parfait B, Laurendeau I, Girault
I, Vidaud M and Lidereau R: Quantification of estrogen receptor
alpha and beta expression in sporadic breast Cancer. Oncogene.
20:8109–8115. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Miller WR: Aromatase and the breast:
Regulation and clinical aspects. Maturitas. 54:335–341. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Qui WS, Yue L, Ding AP, Sun J, Yao Y, Shen
Z and Fan LH: Co-expression of ER-beta and HER2 associated with
poorer prognosis in primary breast cancer. Clin Invest Med.
32:E250–E260. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hou YF, Yuan ST, Li HC, Wu J, Lu JS, Liu
G, Lu LJ, Shen ZZ, Ding J and Shao ZM: ERbeta exerts multiple
simulative effects on human breast carcinoma cells. Oncogene.
23:5799–5806. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Oladimeji P, Skerl R, Rusch C and
Diakonova M: Synergistic activation of ERα by estrogen and
prolactin in breast cancer cells requires tyrosyl phosphorylation
of PAK1. Cancer Res. 76:2600–2611. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Morrison G, Fu X, Shea M, Nanda S,
Giuliano M, Wang T, Klinowska T, Osborne CK, Rimawi MF and Schiff
R: Therapeutic potential of the dual EGFR/HER2 inhibitor AZD8931 in
circumventing endocrine resistance. Breast Cancer Res Treat.
144:263–272. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Marotti JD, Collins LC, Hu R and Tamimi
RM: Estrogen receptor-beta expression in invasive breast cancer in
relation to molecular phenotype: Results from the Nurses' Health
Study. Mod Pathol. 23:197–204. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lindberg K, Helguero LA, Omoto Y,
Gustafsson JÅ and Haldosén LA: Estrogen receptorb β represses Akt
signaling in breast cancer cells via downregulation of HER2/ HER3
and upregulation of PTEN: Implications for tamoxifen sensitivity.
Breast Cancer Res. 13:R432011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Knoop AS, Lænkholm AV, Jensen MB, Nielsen
KV, Andersen J, Nielsen D and Ejlertsen B; Danish Breast Cancer
Cooperative Group, : Estrogen receptor, Progesterone receptor, HER2
status and Ki67 index and responsiveness to adjuvant tamoxifen in
postmenopausal high-risk breast cancer patients enrolled in the
DBCG 77C trial. Eur J Cancer. 50:1412–1421. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li W, Jia M, Qin X, Hu J, Zhang X and Zhou
G: Harmful effect of ERβ on BCRP-mediated drug resistance and cell
proliferation in ERα/PR-negative breast cancer. FEBS J.
280:6128–6140. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Järvinen TA, Pelto-Huikko M, Holli K and
Isola J: Estrogen receptor beta is coexpressed with ERalpha and PR
and associated with nodal status, grade and proliferation rate in
breast cancer. Am J Pathol. 156:29–35. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Leung YK, Lee MT, Lam HM, Tarapore P and
Ho SM: Estrogen receptor-beta and breast cancer: Translating
biology into clinical practice. Steroids. 77:727–737. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li X, Huang J, Yi P, Bambara RA, Hilf R
and Muyan M: Single-chain estrogen receptors (ERs) reveal that the
ERalpha/beta heterodimer emulates functions of the ERalpha dimer in
genomic estrogen signaling pathways. Mol Cell Biol. 24:7681–7694.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
McNamara KM and Sasano H: The
intracrinology of breast cancer. J Steroid Biochem Mol Biol.
145:172–178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Inwald EC, Klinkhammer-Schalke M,
Hofstädter F, Zeman F, Koller M, Gerstenhauer M and Ortmann O:
Ki-67 is a prognostic parameter in breast cancer patients: Results
of a large population-based cohort of a cancerregistry. Breast
Cancer Res Treat. 139:539–552. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Stope MB, Popp SL, Knabbe C and Buck MB:
Estrogen receptor alpha attenuates transforming growth factor-beta
signaling in breast cancer cells independent from agonistic and
antagonistic ligands. Breast Cancer Res Treat. 120:357–367. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yarden RI, Wilson MA and Chrysogelos SA:
Estrogen suppression of EGFR expression in breast cancer cells: A
possible mechanism to modulate growth. J Cell Biochem Suppl. 36
Suppl:S232–S246. 2001. View Article : Google Scholar
|
|
45
|
Subik K, Lee JF, Baxter L, Strzepek T,
Costello D, Crowley P, Xing L, Hung MC, Bonfiglio T, Hicks DG and
Tang P: The expression patterns of ER PR, HER2, CK5/6, EGFR, Ki-67
and AR by immunohistochemical analysis in-breast cancer cell lines.
Breast Cancer (Auckl). 4:35–41. 2010.PubMed/NCBI
|
|
46
|
Stark A, Hulka BS, Joens S, Novotny D,
Thor AD, Wold LE, Schell MJ, Melton LJ III, Liu ET and Conway K:
HER-2/neu amplification in benign breast disease and the risk of
subsequent breast cancer. J Clin Oncol. 18:267–274. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Allred DC, Clark GM, Molina R, Tandon AK,
Schnitt SJ, Gilchrist KW, Osborne CK, Tormey DC and McGuire WL:
Overexpression of HER-2/neu and its relationship with other
prognostic factors change during the progression of in situ to
invasive breast cancer. Hum Pathol. 23:974–979. 1992. View Article : Google Scholar : PubMed/NCBI
|