Introduction
Non-small cell lung cancer (NSCLC), the main type of
lung cancer, is one of the most common malignant tumors in males
worldwide (1). The incidence and
mortality of lung cancer is currently increasing in China, and the
most common risk factors are involved environmental, occupational
and genetic (2–4). Environmental factors, including tobacco
smoking, air pollution, contamination in drinking water and food,
are passing through the natural barriers such as shin and metabolic
elimination; the environmental carcinogens enter cells, damage DNA
and destroy the balance among growth, differentiation and apoptosis
of the cells, which cause carcinogenesis (3,4).
It has been well established that lung cancer is a
complex disease that is highly correlated with environmental
pollutants in the general population; in addition, genetic factors
are known to plays an important role in this disease (2–4). Studies
on the association between gene variants and lung cancer will
contribute to clarify the underlying mechanisms of lung cancer,
including its development, and will potentially provide therapeutic
targets that are important in the diagnosis and prognosis of lung
cancer.
p73, one of the p53 family members, is located at
the human chromosome 1p36.33, and shares relatively high structural
and functional homology with p53 (5,6). As a
candidate tumor-suppressor gene, it encodes two different proteins,
namely the transcriptionally active full-length TAp73 and the NH2
terminally truncated dominant-negative ΔNp73 (6), which have remarkable similarity with p53
in their DNA-binding, transactivation and oligomerization domains
(5). G4C14 and A4T14, the two single
nucleotide polymorphisms (SNPs) of p73 at positions 4 (G>A) and
14 (C>T) (rs2273953 and rs1801173, respectively), are located
upstream of the initiating codon AUG in exon 2, and potentially
influence the gene expression of p73 by forming a stem-loop
structure (5,7). Various studies have investigated p73
polymorphisms and cancer risk in a variety of tumors (8–14),
including lung cancer (15–17), but the conclusions of those studies
were inconsistent.
The human homolog of mouse double minute 2 (MDM2) is
known to act as a major negative regulator of p73. MDM2 is a
nuclear phosphoprotein that binds to the N-terminal TA domain of
TAp73 and inhibits the transcription of p73 by interrupting its
contact with the transcription adaptors/cofactors p300 and
CREB-binding protein (18–20). An important polymorphism (T>G,
rs2279744), termed SNP309, has been identified to be located in the
MDM2 intrinsic p53-response promoter (21–23). Cells
carrying the SNP309 GG genotype were observed to exhibit an
increased affinity for the transcriptional activator specificity
protein 1 (Sp1), which resulted in higher messenger RNA and protein
expression levels of MDM2 compared with those in cells carrying the
SNP309 TT genotype (21,22). Those results demonstrated that SNP309
T/G was associated with a decreased response to DNA-damaging agents
and an accelerated tumorigenesis in both hereditary and sporadic
cancer patients (22,23), suggesting that SNP309 may contribute
to individual differences in cancer susceptibility.
Several studies have shown that disruption of the
p53-MDM2 interaction by the suppression of MDM2 expression can lead
to p53 activation and tumor growth inhibition (21,22). The
p73 and MDM2 genes are involved in the genetics of the p53
signaling pathway in various tumors; however, their role in the
tumorigenesis of NSCLC remains to be clarified. In the present
study, the interaction between the p73 G4C14-A4T14 and MDM2 SNP309
polymorphisms was found to effect susceptibility to NSCLC in a
Southern Chinese population.
Materials and methods
Study subjects and samples
The present case-control study consisted of 186
patients with NSCLC and 196 cancer-free controls. All subjects were
consecutively recruited at the Central Hospital of Zhuzhou City
(Zhuzhou, China) and Hunan Provincial Tumor Hospital (Changsha,
China) between March 2013 and December 2014. All patients were
histopathologically confirmed and had not received preoperative
chemotherapy or radiotherapy. The cancer-free controls were
randomly recruited from healthy individuals who underwent a cancer
screening program in the same region and during the same period
than the case patients. At recruitment, informed consent forms
about the study were signed by all subjects, and each participant
was interviewed to collect baseline data, including demographic
factors, medical history, current or past tobacco use (yes or no)
and alcohol consumption (yes or no). The research protocol was
approved by the Institutional Review Board of the Central Hospital
of Zhuzhou City and Hunan Provincial Tumor Hospital (Zhuzhou,
China).
DNA extraction
Venous blood (5 ml) from each subject was collected
into Vacutainer (Hunan Ping'an Medical Machinery Technology Co.,
Ltd., Hunan, China) tubes containing EDTA and stored at −70°C. The
extraction of genomic DNA was performed using a Dzup (Blood)
Genomic DNA Isolation Reagent (Sangon Biotech Co., Ltd., Shanghai,
China), in accordance with the manufacturer's protocol.
MDM2 SNP309
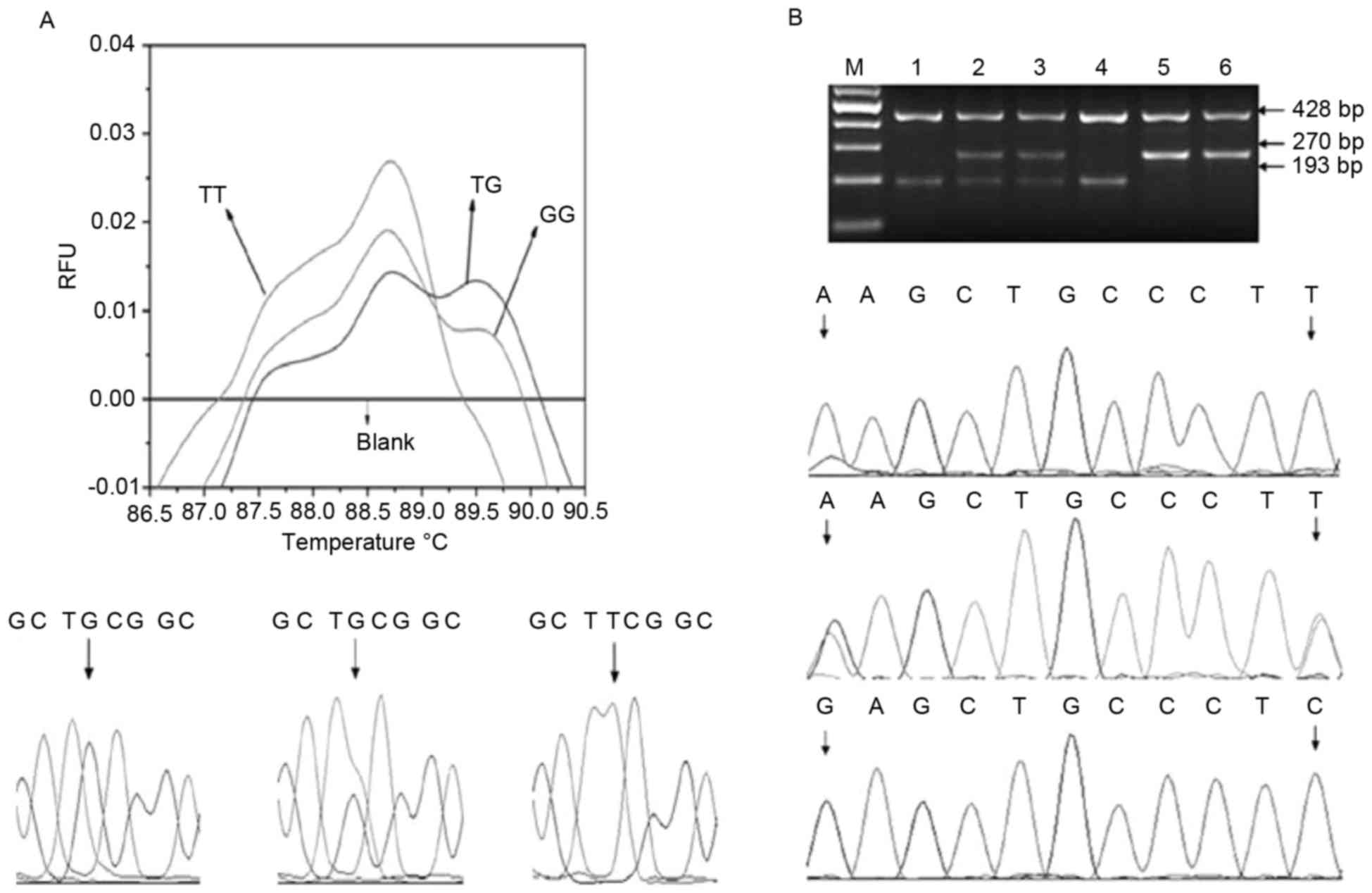
Polymorphism in MDM2 was determined by high
resolution melting (HRM) (24). The
primers used for quantitative polymerase chain reaction (PCR)
analysis were T, 5′-CTAGGGCTGCGGGGCCGATT-3′; G,
5′-GCGGGCTGGGCTAGGCTGCGGGGCCGTTG-3′; and common primer C,
5′-ACCCGACAGGCACCTGCGATC-3′ (all forward). The PCR volume was 20
µl, and included SYBR-Green PCR Master Mix (Shanghai GeneCore
BioTechnologies Co., Ltd., Shanghai, China; 10 µl), primer T (0.4
µl), primer G (0.1 µl), primer C (0.2 µl), double distilled (dd)
H2O (7.3 µl) and venous blood DNA (2 µl). The PCR conditions were
an initial denaturation step at 95°C for 10 min, followed by 40
cycles of 95°C for 20 sec, 61°C for 1 min and 72°C for 30 sec, and
a final extension step at 72°C for 10 min. Melting curves were
obtained following a denaturation period at 95°C for 60 sec and
holding for 30 sec at 60°C, and initial and final temperatures of
65 and 90°C, respectively, with a temperature gradient of
0.05°C/sec. PCR and HRM analyses were conducted with the
LightCyclerNano System (Roche Diagnostics, Basel, Switzerland), and
the melting curve and the accuracy of genotyping data were
validated by direct sequencing (Fig.
1A).
p73 G4C14-A4T14 polymorphism
Samples for the p73 G4C14-A4T14 genotype were
analyzed using PCR with confronting two-pair primers (CTPP)
(11,25). The two-pair primers use were forward
(F) 1, 5′-CCACGGATGGGTCTGATCC-3′; reverse (R) 1,
5′-GGCCTCCAAGGGCAGCTT-3′; F2, 5′-CCTTCCTTCCTGCAGAGCG-3′; and R2,
5′-TTAGCCCAGCGAAGGTGG-3′ (11). The
PCR volume was 15 µl, and contained 7.5 µl 2X Taq PCR Mastermix
(Tiangen Biotech Beijing Co., Ltd., Beijing, China), 2.5 µl dd H2O,
1 µl each of the above four primers and 1 µl venous blood genomic
DNA template. The PCR assay was performed in a GeneAmp PCR System
9700 (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham,
MA, USA), and the PCR conditions included an initial denaturation
step for 5 min at 95°C, followed by 35 cycles of 40 sec at 95°C, 40
sec at 60°C and 40 sec at 72°C, and a final elongation for 10 min
at 72°C (11). An aliquot (8 µl) of
the PCR product was analyzed on 2% agarose gel with ethidium
bromide (50 bp DNA Ladder Marker; Sinopharm Chemical Reagent Co.,
Ltd., Shanghai, China). The GC homozygote is expected to produce
two bands of 428 and 193 bp, respectively, while the AT homozygote
is expected to produce two bands of 428 and 270 bp, respectively,
and GC/AT heterozygosity is expected to produce three bands of 428,
270 and 193 bp, respectively (11,24). The
results of PCR-CTPP genotyping and sequencing analysis were
consistent (Fig. 1B).
Statistical analysis
Statistical analyses were performed using the
statistical software SPSS 13.0 (SPSS, Inc., Chicago, IL, USA). The
frequency of polymorphisms in the p73 and MDM2 genes among cases
and clinical outcome was statistically analyzed using the χ2 test.
The association between the polymorphisms of these two genes and
lung cancer was determined using an unconditional logistic
regression model to assess odds ratios (ORs) and 95% confidence
intervals (CIs). P<0.05 was considered to indicate a
statistically significant difference.
Results
Characteristics of the study
subjects
In the present case-control study, the frequency
distribution of selected characteristics of the subjects is
presented in Table I. There were no
significant differences in the distributions of age or sex between
cases and controls (P>0.05). However, compared with the control
subjects, the cases were more likely to be tobacco smokers
(P<0.05), which indicated that tobacco smoking was a high-risk
factor for NSCLC in the present study population.
 | Table I.Demographic characteristics of
non-small cell lung cancer cases and controls. |
Table I.
Demographic characteristics of
non-small cell lung cancer cases and controls.
| Characteristics | Cases, n (%) | Controls, n (%) | P-valuea |
|---|
| Total | 186 | 196 |
|
| Age, years |
|
| 0.99 |
| ≤45 | 90 (48.39) | 95 (48.47) |
|
|
>45 | 96 (51.61) | 101 (51.53) |
|
| Sex |
|
| 0.49 |
| Male | 136 (73.12) | 137 (69.90) |
|
|
Female | 50 (26.88) | 59 (30.10) |
|
| Cigarette
smoking |
|
| <0.01 |
| No | 54 (29.03) | 115 (58.67) |
|
| Yes | 132 (70.97) | 81 (41.33) |
|
| Alcohol
consumption |
|
| 0.95 |
| No | 122 (65.59) | 128 (65.31) |
|
| Yes | 64 (34.41) | 68 (34.69) |
|
| Histology |
|
Adenocarcinoma | 108 (58.06) | 0 |
|
|
Squamous cell | 60 (32.26) | 0 |
|
| Carcinoma |
|
Others | 18 (9.68) | 0 |
|
Distribution of p73 and MDM2
polymorphisms
The genotype and polymorphic allele frequencies of
the three polymorphisms (p73 G4C14-A4T14 and MDM2 309T/G SNPs)
between the cases and controls are shown in Table II.
 | Table II.Genotype and allele frequencies of
p73 and MDM2 among cases and controls, and their association with
the risk of developing non-small cell lung cancer. |
Table II.
Genotype and allele frequencies of
p73 and MDM2 among cases and controls, and their association with
the risk of developing non-small cell lung cancer.
| Genotype | Cases, n (%)
(n=186) | Controls, n (%)
(n=196) | P-value | OR (95%
CI)a |
|---|
| p73
G4C14-A4T14 |
|
GC/GC | 94 (50.54) | 98 (50.00) | – | 1.00
(Reference) |
|
GC/AT | 80 (43.01) | 71 (36.22) | 0.46 | 1.18
(0.77–1.80) |
|
AT/AT | 12 (6.45) | 27 (13.78) | 0.04 | 0.46
(0.22–0.97)b |
| AT
allele frequency | 104 (27.96) | 125 (31.89) | – | 1.00
(Reference) |
| GC
allele frequency | 268 (72.04) | 267 (68.11) | 0.24 | 1.21
(0.88–1.65) |
| MDM2 309 T/G |
| GG | 58 (31.18) | 44 (22.45) | – | 1.00
(Reference) |
| TG | 96 (51.61) | 101 (51.53) | 0.18 | 0.72
(0.45–1.17) |
| TT | 32 (17.21) | 51 (26.02) | 0.01 | 0.48
(0.26–0.86)b |
| G
allele frequency | 212 (56.99) | 189 (48.21) | – | 1.00
(Reference) |
| T
allele frequency | 160 (23.01) | 203 (51.79) | 0.02 | 0.70
(53–0.94)b |
The frequencies of the three genotypes (GC/GC, GC/AT
and AT/AT) of p73 G4C14-A4T14 polymorphism in NSCLC cases and
controls were 50.54, 43.01 and 6.45%, and 50.00, 36.22 and 13.78%,
respectively. There were significant differences in the genotype
distributions of p73 G4C14-A4T14 polymorphism between cases and
controls (P<0.05). The frequencies of the three genotypes (GG,
TG and TT) of MDM2 309T/G SNPs in NSCLC cases and controls were
31.18, 51.61 and 17.21%, and 22.45, 51.53 and 26.02%, respectively.
A similar result was obtained with the MDM2 309T/G SNPs
(P<0.05). Furthermore, the frequencies of the p73 AT and MDM2 G
alleles were 27.96 and 56.99%, respectively, among cases, and 31.89
and 48.21%, respectively, among controls. The frequencies the G
allele of MDM2 309T/G SNPs were significantly different between
cases and controls (P<0.05). By contrast, there was no
difference in the frequencies the AT allele of p73 G4C14-A4T14
(P>0.05).
Association between p73 and MDM2
polymorphisms and NSCLC risk
An unconditional logistic regression model was used
to estimate the association between genotypes and the risk of
developing NSCLC (Table II). The p73
AT/AT genotype was associated with a decreased risk of developing
NSCLC (OR=0.46; 95% CI=0.22–0.97), compared with that of the GC/GC
genotype. Similarly, the MDM2 TT genotype was also associated with
a decreased risk of developing NSCLC (OR=0.48; 95% CI=0.26–0.86),
compared with that of the GG genotype. However, the heterozygous
genotypes for both polymorphisms (p73 GC/AT or MDM2 TG) were not
associated with such risk. p73 GC/AT and GC/GC, or MDM2 TG and GG,
were combined the into one group for subsequent analysis. The
present study examined whether there was a statistical combination
effect between the p73 and MDM2 polymorphisms (Table III). It was observed that controls
carrying the p73 AT/AT genotype were more likely to carry the MDM2
TT genotype than the cases (7.14 vs. 1.07%, respectively;
P<0.05). Furthermore, the OR significantly decreased to 0.13
(95% CI=0.03–0.59) among subjects carrying both the p73 AT/AT and
MDM2 TT genotypes.
 | Table III.Risk of lung cancer in association
with MDM2 SNP309 and p73 G4C14-to-A4T14 polymorphism. |
Table III.
Risk of lung cancer in association
with MDM2 SNP309 and p73 G4C14-to-A4T14 polymorphism.
| p73 genotype | MDM2 genotype | Cases, n (%)
(n=186) | Control, n (%)
(n=196) | P-value | OR (95%
CI)a |
|---|
| GC/AT + GC/GC | GG + TG | 144 (77.42) | 132 (67.35) | – | 1.00
(Reference) |
| GC/AT + GC/GC | TT | 30 (16.13) | 37 (18.88) | 0.28 | 0.74
(0.44–1.27) |
| AT/AT | GG + TG | 10 (5.38) | 13 (6.63) | 0.42 | 0.71
(0.30–1.66) |
| AT/AT | TT | 2 (1.07) | 14 (7.14) | <0.01 | 0.13
(0.03–0.59)b |
Discussion
The present study is a case-control study aimed to
detect the association between the p73 G4C14-A4T14 and MDM2 SNP309
polymorphisms, alone or under interaction, and the risk of
developing NSCLC in a Southern Chinese population. In the present
hospital-based analysis, it was demonstrated that both p73 and MDM2
polymorphisms were associated with a decreased risk of developing
NSCLC. Furthermore, the interact was detected between the p73 AT/AT
and MDM2 TT genotypes using a multiplicative manner.
p73 has structural and functional homology with the
p53 gene, and induces cell cycle arrest or apoptosis in response to
DNA damage (26,27). A dinucleotide polymorphism at
positions 4 and 14 (G4C14-A4T14) is located at exon 2 of the p73
gene, which possibly forms a stem-loop structure to influence p73
translation and gene expression (5).
Although the association between the p73 G4C14-A4T14 polymorphism
and the risk of developing cancer has been investigated in a
variety of tumors, the results were not consistent (8–17,28,29). Niwa
et al (12) and Zhang et
al (17) reported that the AT/AT
genotype was associated with a significantly increased risk of
developing endometrial cancer and lung in Japanese (12) and Northern Chinese (17) populations, respectively. However,
Hamajima et al (28) did not
identify any associations between this polymorphism and the risk of
developing digestive tract cancers, including colorectal cancer, in
a Japanese population. Similar results were obtained with lung
cancer in Korea (29). On the
contrary, Hu et al (16)
reported that the AT allele of p73 was associated with a
significantly decreased risk of developing lung cancer in a Chinese
population. In accordance with this result, the present study also
observed a significant association between the p73 G4C14-A4T14
polymorphism and the risk of developing NSCLC, and the frequency of
the p73 GC allele among the healthy Chinese was 0.707, which was
similar to that of the healthy Japanese (12). Besides, there was a significant
interaction between p73 G4C14-A4T14 and MDM2 309T/G SNPs in NSCLC
in a Southern Chinese population.
The 309T/G SNP is a common SNP in the promoter
region of the MDM2 gene, which has been demonstrated to increase
MDM2 expression by increasing the binding affinity of Sp1 to the
promoter of MDM2 (22,30), and represses the transcriptional
activity of p73, thus attenuating its activity in G1 cell-cycle
control and apoptosis (18–20). It has been reported that individuals
carrying the G allele of MDM2 SNP309 were more sensitized to
develop both hereditary and sporadic cancers (22). Various studies have reported an
increased risk of developing lung cancer for the carriers of the G
allele in Korean and Chinese populations (31,32).
Previous studies confirmed that the G allele of MDM2 SNP309 was
associated with higher expression levels of MDM2 protein (21). In addition, various studies failed to
show a significant association between the G allele of MDM2 SNP309
and the risk of developing breast (33,34) or
lung cancer (35). In the present
case-control study, it was investigated whether the MDM2 309T/G
SNPs were associated with the risk of developing NSCLC in a
Southern Chinese population. The results revealed that the
frequencies of the three genotypes and the allele distributions
were significantly different between the NSCLC patients and the
healthy controls. In addition, there was a significant difference
between the GG and TT genotypes, and subjects carrying the T allele
had a significantly decreased risk of developing NSCLC in the
current Southern Chinese population compared with that of patients
carrying the GG genotype, which was consistent with the results of
Jun et al (31). Therefore,
these findings further supported the present result that the MDM2
SNP309 T allele may reduce the risk of developing NSCLC. It could
be hypothesized that different molecular pathogeneses in different
tumors may cause this discrepancy, which would require further
study.
The present study examined whether the p73 gene
G4C14-A4T14 polymorphism and the MDM2 gene SNP309 were associated
with the risk of developing NSCLC in a Chinese population. Compared
with the p73 GC/AT + GC/GC and MDM2 GG + TG genotypes, there was a
significant difference in the p73 AT/AT and MDM2 TT genotypes with
NSCLC in a Chinese population, and the OR was significantly
decreased to 0.13 (95% CI=0.03–0.59). Therefore, the present
results revealed that there was a significant interaction between
p73 G4C14-to-A4T14 and MDM2 309T/G SNPs in NSCLC patients within
the present Southern Chinese population. However, the small sample
size used in the present study caused broad CIs in the regression
model. Thus, further larger population-based studies are required
to confirm the association of these two polymorphisms with the risk
of developing NSCLC in other populations.
In conclusion, the present study demonstrated a
significant association in the decreased risk of developing NSCLC
between p73 AT/AT and MDM2 TT genotypes. Furthermore, the p73
G4C14-A4T14 and MDM2 309 T/G SNPs possibly had a interaction on the
lower risk of developing NSCLC, and these findings may be a
protective factor for NSCLC development in Chinese populations.
Acknowledgements
The present study was supported by funds from the
National Natural Science Foundation of China (grant nos. 61171061
and 31501538), the key program of Hunan Provincial Department of
Science and Technology (grant no. 2016NK2096), the Natural Science
Foundation of Hunan Province (grant no. 2015JJ2049), Changsha Key
Science & Technology Program of Hunan Province of China (grant
no. kh1601106), the Scientific Research Fund of Hunan Provincial
Education Department (grant nos. 15C0412 and 15C0411) and the
Postdoctoral Science Foundation of China (grant nos. 2016T90769,
2015M580707 and 2016M592456). The authors sincerely thank the
patients and healthy volunteers for their participation in the
present study, as well as the Hunan Provincial Tumor Hospital.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhou BB and Elledge SJ: The DNA damage
response: Putting checkouts in perspective. Nature. 408:433–439.
2000. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hu ZB, Ma HX, Tan W, et al: The research
between genetic susceptibility to lung cancer and p73 gene
polymorphism. Chin J Epidemiol. 26:106–109. 2005.
|
|
4
|
Cheng Z, Wang W, Song YN, Kang Y and Xia
J: hoGG1, p53 genes and smoking interactions are associated with
the development of lung cancer. Asian Pac J Cancer Prev.
13:1803–1808. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kaghad M, Bonnet H, Yang A, Creancier L,
Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, et al:
Monoallelically expressed gene related to p53 at lp36, a region
frequently deleted in neuroblastoma and other human cancers. Cell.
90:809–819. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Melino G, De Laurenzi V and Vousden KH:
p73: Friend or foe in tumorigenesis. Nat Rev Cancer. 2:605–615.
2002. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peters MA, Janer M, Kolb S, Jarvik GP,
Ostrander EA and Stanford JL: Germline mutations in the p73 gene do
not predispose to familial prostate-brain cancer. Prostate.
48:292–296. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ryan BM, Mcmanus R, Daly JS, Carton E,
Keeling PW, Reynolds JV and Kelleher D: A common p73 polymorphism
is associated with a reduced incidence of oesophageal carcinoma. Br
J Cancer. 85:1499–1503. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang XE, Hamajima N, Katsuda N, Matsuo K,
Hirose K, Mizutani M, Iwata H, Miura S, Xiang J, Tokudome S and
Tajima K: Association of p53 codon Arg72pro and p73 G4C14-to-A4T14
at exon 2 genetic polymorphisms with the risk of Japanese breast
cancer. Breast Cancer. 10:307–311. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li G, Sturgis EM, Wang LE, Chamberlain RM,
Amos CI, Spitz MR, El-Naggar AK, Hong WK and Wei Q: Association of
a p73 exon 2 G4C14-to-A4T14 polymorphism with risk of squamous cell
carcinoma of the head and neck. Carcinogenesis. 25:1911–1916. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee KE, Hong YS, Kim BG, Kim NY, Lee KM,
Kwak JY and Roh MS: p73 G4C14 to A4T14 polymorphism is associated
with colorectal cancer risk and survival. World J Gastroenterol.
16:4448–4454. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Niwa Y, Hamajima N, Atsuta Y, Yamamoto K,
Tamakoshi A, Saito T, Hirose K, Nakanishi T, Nawa A, Kuzuya K and
Tajima K: Genetic polymorphisms of p73 G4C14-to-A4T14 at exon 2 and
p53 Arg72Pro and the risk of cervical cancer in Japanese. Cancer
Lett. 205:55–60. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pfeifer D, Arbman G and Sun XF:
Polymorphism of the p73 gene in relation to colorectal cancer risk
and survival. Carcinogenesis. 26:103–107. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Niwa Y, Hirose K, Matsuo K, Tajima K,
Ikoma Y, Nakanishi T, Nawa A, Kuzuya K, Tamakoshi A and Hamajima N:
Association of p73 G4C14-to-A4T14 polymorphism at exon 2 and p53
Arg72Pro polymorphism with the risk of endometrial cancer in
Japanese subjects. Cancer Lett. 219:183–190. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li G, Wang LE, Chamberlain RM, Amos CI,
Spitz MR and Wei Q: p73 G4C14-to-A4T14 polymorphism and risk of
lung cancer. Cancer Res. 64:6863–6866. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu Z, Miao X, Ma H, Tan W, Wang X, Lu D,
Wei Q, Lin D and Shen H: Dinucleotide polymorphism of p73 gene is
associated with a reduced risk of lung cancer in a Chinese
population. Int J Cancer. 114:455–460. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang X, Li X, Wu Z, Lin F and Zhou H: The
p73 G4C14-to-A4T14 polymorphism is associated with risk of lung
cancer in the Han Nationality of North China. Mol Carcinog.
52:387–391. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ozaki T and Nakagawara A: p73, a
sophisticated p53 family member in the cancer world. Cancer Sci.
96:729–737. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Levrero M, De Laurenzi V, Costanzo A, Gong
J, Melino G and Wang JY: Structure, function and regulation of p63
and p73. Cell Death Differ. 6:1146–1153. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zeng X, Chen L, Jost CA, Maya R, Keller D,
Wang X, Kaelin WG Jr, Oren M, Chen J and Lu H: MDM2 suppresses p73
function without promoting p73 degradation. Mol Cell Biol.
19:3257–3266. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lundgren K, de Oca Luna R Montes, McNeill
YB, Emerick EP, Spencer B, Barfield CR, Lozano G, Rosenberg MP and
Finlay CA: Targeted expression of MDM2 uncouples S phase from
mitosis and inhibits mammary gland development independent of p53.
Genes Dev. 11:714–725. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bond GL, Hu W, Bond EE, Robins H, Lutzker
SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, et al: A
single nucleotide polymorphism in the MDM2 promoter attenuates the
p53 tumor suppressor pathway and accelerates tumor formation in
humans. Cell. 119:591–602. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bougeard G, Baert-Desurmont S, Tournier I,
Vasseur S, Martin C, Brugieres L, Chompret A, Bressac-de Paillerets
B, Stoppa-Lyonnet D, Bonaiti-Pellie C and Frebourg T: Impact of the
MDM2 SNP309 and p53 Arg72Pro polymorphism on age tumor onset in
Li-Fraumeni syndrome. J Med Genet. 43:531–533. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cheng H, Xie L, Hu WJ, et al: Detection of
the MDM2 SNP309 by QRT-PCR. J Clin Oncol. 16:577–581. 2011.
|
|
25
|
Wang SS, Guo HY, Dong LL, Zhu XQ, Ma L, Li
W and Tang JX: Association between a p73 gene polymorphism and
genetic susceptibility to non-small cell lung cancer in the South
of China. Asian Pac J Cancer Prev. 15:10387–19391. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu J, Jiang J, Zhou W and Chen X: The
potential tumor suppressor p73 differently regulates cellular p53
target genes. Cancer Res. 58:5061–5065. 1998.PubMed/NCBI
|
|
27
|
Jost CA, Marin MC and Kaelin WG Jr: p73 is
a simian [correction of human] p53-related protein that can induce
apoptosis. Nature. 389:191–194. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hamajima N, Matsuo K, Suzuki T, Nakamura
T, Matsuura A, Hatooka S, Shinoda M, Kodera Y, Yamamura Y, Hirai T,
et al: No association of p73 G4C14-to-A4T14 at exon 2 and p53
Arg72Pro polymorphism with the risk of digestive tract cancers in
Japanese. Cancer Lett. 181:81–85. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Choi JE, Kang HG, Chae MH, Kim EJ, Lee WK,
Cha SI, Kim CH, Jung TH and Park JY: No association between p73
G4C14-to-A4T14 polymorphism and the risk of lung cancer in a Korean
population. Biochem Genet. 44:543–550. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bond GL, Hirshfield KM, Kirchhoff T, Alexe
G, Bond EE, Robins H, Bartel F, Taubert H, Wuerl P, Hait W, et al:
MDM2 SNP309 accelerates tumor formation in a gender-specific and
hormone-dependent manner. Cancer Res. 66:5104–5110. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jun HJ, Park SH, Lee WK, Choi JE, Jang JS,
Kim EJ, Cha SI, Kim DS, Kam S, Kim CH, et al: Combined effects off
p73 and MDM2 polymorphisms on the risk of lung cancer. Mol
Carcinog. 46:100–105. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang X, Miao X, Guo Y, Tan W, Zhou Y, Sun
T, Wang Y and Lin D: Genetic polymorphisms in cell cycle regulatory
genes MDM2 and TP53 are associated with susceptibility to lung
cancer. Hum Mutat. 27:110–117. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ma H, Hu Z, Zhai X, Wang S, Wang X, Qin J,
Jin G, Liu J, Wang X, Wei Q and Shen H: Polymorphisms in the MDM2
promoter and risk of breast cancer: A case-control analysis in a
Chinese population. Cancer Lett. 240:261–267. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Campbell IG, Eccles DM and Choong DY: No
association of the MDM2 SNP309 polymorphism with risk of breast or
ovarian cancer. Cancer Lett. 240:195–197. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hu Z, Ma H, Lu D, Qian J, Zhou J, Chen Y,
Xu L, Wang X, Wei Q and Shen H: Genetic variants in the MDM2
promoter and lung cancer risk in a Chinese population. Int J
Cancer. 118:1275–1278. 2006. View Article : Google Scholar : PubMed/NCBI
|