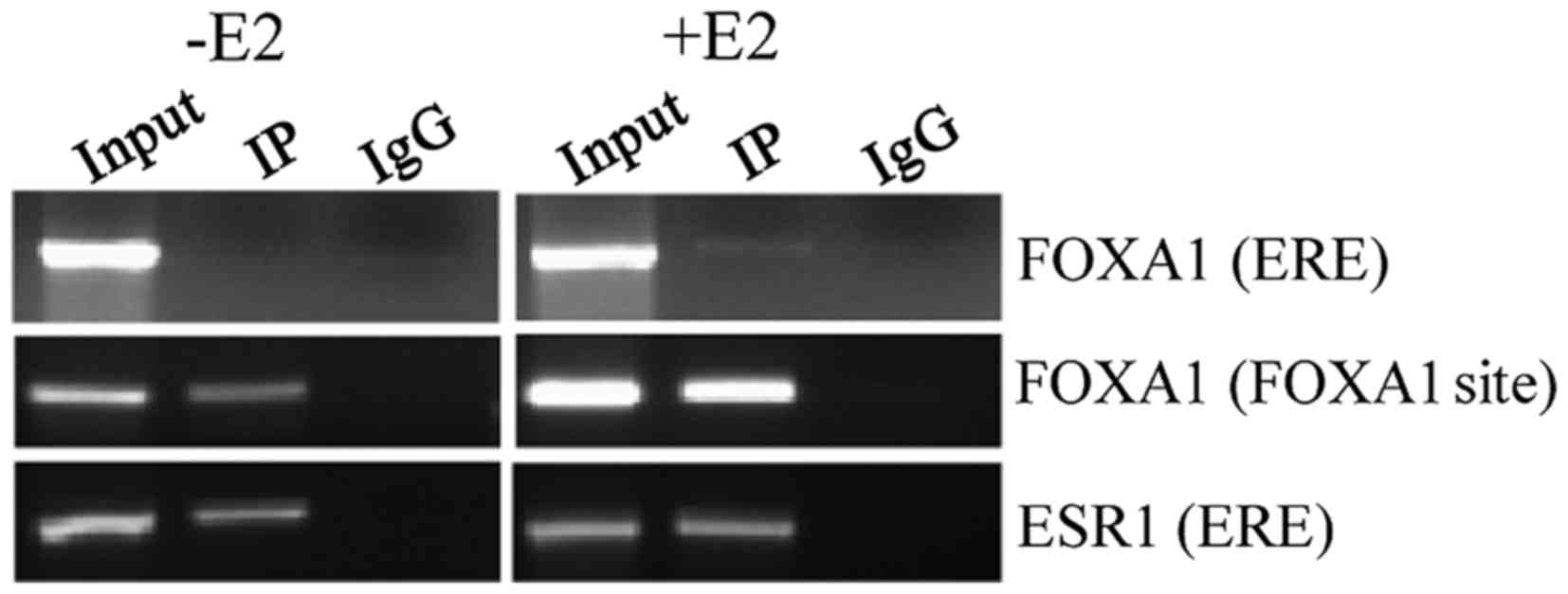
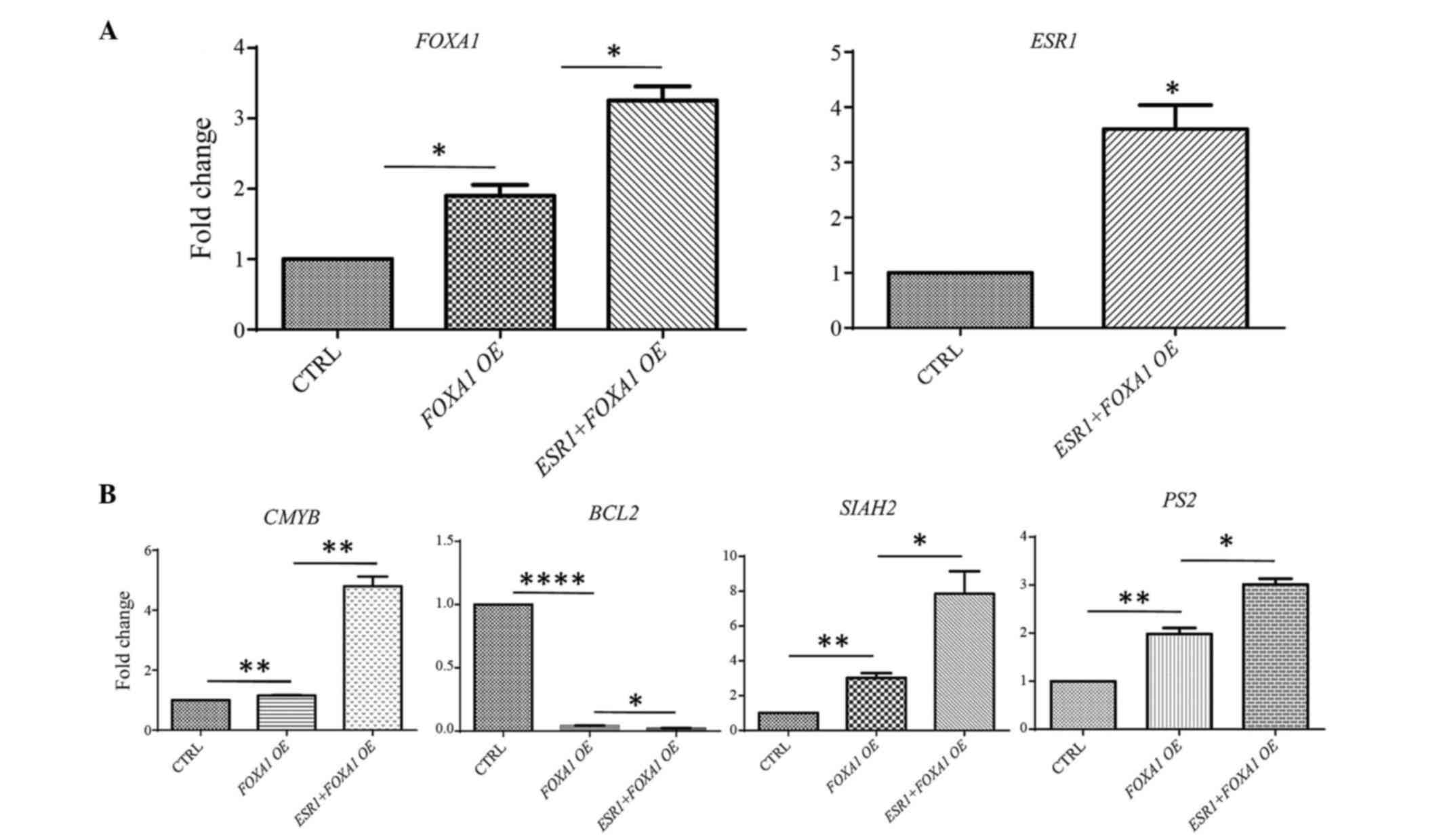
|
1
|
Colditz GA: Relationship between estrogen
levels, use of hormone replacement therapy, and breast cancer. J
Natl Cancer Inst. 90:814–823. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nomura Y, Tashiro H, Hamada Y and
Shigematsu T: Relationship between estrogen receptors and risk
factors of breast cancer in Japanese pre- and postmenopausal
patients. Breast Cancer Res Treat. 4:37–43. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Borellini F and Oka T: Growth control and
differentiation in mammary epithelial cells. Environ Health
Perspect. 80:85–99. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang HZ, Bennett JM, Smith KT, Sunil N
and Haslam SZ: Estrogen mediates mammary epithelial cell
proliferation in serum-free culture indirectly via mammary
stroma-derived hepatocyte growth factor. Endocrinology.
143:3427–3434. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Richert MM, Schwertfeger KL, Ryder JW and
Anderson SM: An atlas of mouse mammary gland development. J Mammary
Gland Biol Neoplasia. 5:227–241. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Clarke RB, Howell A, Potten CS and
Anderson E: Dissociation between steroid receptor expression and
cell proliferation in the human breast. Cancer Res. 57:4987–4991.
1997.PubMed/NCBI
|
|
7
|
Russo J, Ao X, Grill C and Russo IH:
Pattern of distribution of cells positive for estrogen receptor
alpha and progesterone receptor in relation to proliferating cells
in the mammary gland. Breast Cancer Res Treat. 53:217–227. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ali S and Coombes RC: Endocrine-responsive
breast cancer and strategies for combating resistance. Nat Rev
Cancer. 2:101–112. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang Y, Zou L, Lu WQ, Zhang Y and Shen
AG: Foxo3a expression is a prognostic marker in breast cancer. PloS
One. 8:e707462013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gross JM and Yee D: How does the estrogen
receptor work? Breast Cancer Res. 4:62–64. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Power KA and Thompson LU: Ligand-induced
regulation of ERalpha and ERbeta is indicative of human breast
cancer cell proliferation. Breast Cancer Res Treat. 81:209–221.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Akaogi K, Nakajima Y, Ito I, Kawasaki S,
Oie SH, Murayama A, Kimura K and Yanagisawa J: KLF4 suppresses
estrogen-dependent breast cancer growth by inhibiting the
transcriptional activity of ERalpha. Oncogene. 28:2894–2902. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lumachi F, Brunello A, Maruzzo M, Basso U
and Basso SM: Treatment of estrogen receptor-positive breast
cancer. Curr Med Chem. 20:596–604. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Breast Cancer Trialists' Collaborative
Group, . Systemic treatment of early breast cancer by hormonal,
cytotoxic, or immune therapy. 133 randomised trials involving
31,000 recurrences and 24,000 deaths among 75,000 women. Early
Breast Cancer Trialists' Collaborative Group. Lancet. 339:71–85.
1992.PubMed/NCBI
|
|
15
|
Osborne CK: Tamoxifen in the treatment of
breast cancer. N Engl J Med. 339:1609–1618. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Osborne CK and Schiff R: Mechanisms of
endocrine resistance in breast cancer. Annu Rev Med. 62:233–247.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Garcia-Becerra R, Santos N, Diaz L and
Camacho J: Mechanisms of resistance to endocrine therapy in breast
cancer: Focus on signaling pathways, miRNAs and genetically based
resistance. Int J Mol Sci. 14:108–145. 2013. View Article : Google Scholar
|
|
18
|
Kabel AM, Altalhi D, Alsharabi H, Qadi O
and Ad Khan M: Tamoxifen-resistant breast cancer: Causes of
resistance and possible management. Journal of Cancer Research and
Treatment. 4:37–40. 2016.
|
|
19
|
Lin L, Miller CT, Contreras JI, Prescott
MS, Dagenais SL, Wu R, Yee J, Orringer MB, Misek DE, Hanash SM, et
al: The hepatocyte nuclear factor 3 alpha gene, HNF3alpha (FOXA1),
on chromosome band 14q13 is amplified and overexpressed in
esophageal and lung adenocarcinomas. Cancer Res. 62:5273–5279.
2002.PubMed/NCBI
|
|
20
|
Sekiya T, Muthurajan UM, Luger K, Tulin AV
and Zaret KS: Nucleosome-binding affinity as a primary determinant
of the nuclear mobility of the pioneer transcription factor FoxA.
Genes Dev. 23:804–809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Costa RH, Grayson DR and Darnell JE Jr:
Multiple hepatocyte-enriched nuclear factors function in the
regulation of transthyretin and alpha 1-antitrypsin genes. Mol Cell
Biol. 9:1415–1425. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tilghman SM and Belayew A: Transcriptional
control of the murine albumin/alpha-fetoprotein locus during
development. Proc Natl Acad Sci USA. 79:pp. 5254–5257. 1982;
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kaestner KH: The hepatocyte nuclear factor
3 (HNF3 or FOXA) family in metabolism. Trends Endocrinol Metab.
11:281–285. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee CS, Friedman JR, Fulmer JT and
Kaestner KH: The initiation of liver development is dependent on
Foxa transcription factors. Nature. 435:944–947. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bernardo GM and Keri RA: FOXA1: A
transcription factor with parallel functions in development and
cancer. Biosci Rep. 32:113–130. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cirillo LA, McPherson CE, Bossard P,
Stevens K, Cherian S, Shim EY, Clark KL, Burley SK and Zaret KS:
Binding of the winged-helix transcription factor HNF3 to a linker
histone site on the nucleosome. EMBO J. 17:244–254. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lupien M, Eeckhoute J, Meyer CA, Wang Q,
Zhang Y, Li W, Carroll JS, Liu XS and Brown M: FoxA1 translates
epigenetic signatures into enhancer-driven lineage-specific
transcription. Cell. 132:958–970. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lacroix M and Leclercq G: About GATA3,
HNF3A, and XBP1, three genes co-expressed with the oestrogen
receptor-alpha gene (ESR1) in breast cancer. Mol Cell Endocrinol.
219:1–7. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mirosevich J, Gao N, Gupta A, Shappell SB,
Jove R and Matusik RJ: Expression and role of Foxa proteins in
prostate cancer. Prostate. 66:1013–1028. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Carroll JS and Brown M: Estrogen receptor
target gene: An evolving concept. Molecular Endocrinol.
20:1707–1714. 2006. View Article : Google Scholar
|
|
31
|
Laganiere J, Deblois G, Lefebvre C,
Bataille AR, Robert F and Giguère V: From the Cover: Location
analysis of estrogen receptor alpha target promoters reveals that
FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci
USA. 102:pp. 11651–11656. 2005; View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Thorat MA, Marchio C, Morimiya A, Savage
K, Nakshatri H, Reis-Filho JS and Badve S: Forkhead box A1
expression in breast cancer is associated with luminal subtype and
good prognosis. J Clin Pathol. 61:327–332. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Badve S, Turbin D, Thorat MA, Morimiya A,
Nielsen TO, Perou CM, Dunn S, Huntsman DG and Nakshatri H: FOXA1
expression in breast cancer - correlation with luminal subtype A
and survival. Clinical Cancer Res. 13:4415–4421. 2007. View Article : Google Scholar
|
|
34
|
Burch JB: Regulation of GATA gene
expression during vertebrate development. Semin Cell Dev Biol.
16:71–81. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zheng R and Blobel GA: GATA transcription
factors and cancer. Genes Cancer. 1:1178–1188. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yoon NK, Maresh EL, Shen D, Elshimali Y,
Apple S, Horvath S, Mah V, Bose S, Chia D, Chang HR and Goodglick
L: Higher levels of GATA3 predict better survival in women with
breast cancer. Hum Pathol. 41:1794–1801. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kouros-Mehr H, Slorach EM, Sternlicht MD
and Werb Z: GATA3 maintains the differentiation of the luminal cell
fate in the mammary gland. Cell. 127:1041–1055. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Asselin-Labat ML, Sutherland KD, Barker H,
Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG,
van der Wees J, et al: GATA3 is an essential regulator of
mammary-gland morphogenesis and luminal-cell differentiation. Nat
Cell Biol. 9:201–209. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Schaner ME, Ross DT, Ciaravino G, Sorlie
T, Troyanskaya O, Diehn M, Wang YC, Duran GE, Sikic TL, Caldeira S,
et al: Gene expression patterns in ovarian carcinomas. Mol Biol
Cell. 14:4376–4386. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Flouriot G, Griffin C, Kenealy M,
Sonntag-Buck V and Gannon F: Differentially expressed messenger RNA
isoforms of the human estrogen receptor-alpha gene are generated by
alternative splicing and promoter usage. Mol Endocrinol.
12:1939–1954. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Albergaria A, Paredes J, Sousa B, Milanezi
F, Carneiro V, Bastos J, Costa S, Vieira D, Lopes N, Lam EW, et al:
Expression of FOXA1 and GATA3 in breast cancer: The prognostic
significance in hormone receptor-negative tumours. Breast Cancer
Res. 11:R402009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wilson BJ and Giguère V: Identification of
novel pathway partners of p68 and p72 RNA helicases through
Oncomine™ meta-analysis. BMC Genomics. 8:4192007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wilson BJ and Giguère V: Meta-analysis of
human cancer microarrays reveals GATA3 is integral to the estrogen
receptor alpha pathway. Mol Cancer. 7:492008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
Oncomine™: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Higgins JPT, Wang L, Kambham N, Montgomery
K, Mason V, Vogelmann SU, Lemley KV, Brown PO, Brooks JD and van de
Rijn M: Gene expression in the normal adult human kidney assessed
by complementary DNA microarray. Mol Biol Cell. 15:1–656.
2004.PubMed/NCBI
|
|
46
|
Roth RB, Hevezi P, Lee J, Willhite D,
Lechner SM, Foster AC and Zlotnik A: Gene expression analyses
reveal molecular relationships among 20 regions of the human CNS.
Neurogenetics. 7:67–80. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shyamsundar R, Kim YH, Higgins JP,
Montgomery K, Jorden M, Sethuraman A, van de Rijn M, Botstein D,
Brown PO and Pollack JR: A DNA microarray survey of gene expression
in normal human tissues. Genome Biol. 6:R222005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tabchy A, Valero V, Vidaurre T, Lluch A,
Gomez H, Martin M, Qi Y, Barajas-Figueroa LJ, Souchon E, Coutant C,
et al: Evaluation of a 30-gene paclitaxel, fluorouracil,
doxorubicin and cyclophosphamide chemotherapy response predictor in
a multicenter randomized trial in breast cancer. Clinc Cancer Res.
16:5351–5361. 2010. View Article : Google Scholar
|
|
49
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Su AI, Cooke MP, Ching KA, Hakak Y, Walker
JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al:
Large-scale analysis of the human and mouse transcriptomes. Proc
Natl Acad Sci USA. 99:pp. 4465–4470. 2002; View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhao H, Langerød A, Ji Y, Nesland JM,
Tibshirani R, Bukholm IK, Kåresen R, Botstein D, Børresen-Dale A
and Jeffrey SS: Different gene expression patterns in invasive
Lobular and ductal carcinomas of the breast. Mol Biol Cell.
15:2523–2536. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yu K, Ganesan K, Miller LD and Tan P: A
modular analysis of breast cancer reveals a novel low-grade
molecular signature in estrogen receptor-positive tumors. Clin
Cancer Res. 12:3288–3296. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang Y, Klijn JG, Zhang Y, Sieuwerts AM,
Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu
J, et al: Gene-expression profiles to predict distant metastasis of
lymph-node-negative primary breast cancer. Lancet. 365:671–679.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Waddell N, Cocciardi S, Johnson J, Healey
S, Marsh A, Riley J, da Silva L, Vargas AC, Reid L; kConFab
Investigators, ; Simpson PT, Lakhani SR and Chenevix-Trench G: Gene
expression profiling of formalin-fixed, paraffin-embedded familial
breast tumours using the whole genome-DASL assay. J Pathol.
221:452–461. 2010.PubMed/NCBI
|
|
55
|
van 't Veer LJ1, Dai H, van de Vijver MJ,
He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ,
Witteveen AT, et al: Gene expression profiling predicts clinical
outcome of breast cancer. Nature. 415:530–536. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Schmidt M, Böhm D, von Törne C, Steiner E,
Puhl A, Pilch H, Lehr HA, Hengstler JG, Kölbl H and Gehrmann M: The
humoral immune system has a key prognostic impact in node-negative
breast cancer. Cancer Res. 68:5405–5413. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Pollack JR, Sørlie T, Perou CM, Rees CA,
Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Børresen-Dale AL
and Brown PO: Microarray analysis reveals a major direct role of
DNA copy number alteration in the transcriptional program of human
breast tumors. Proc Natl Acad Sci USA. 99:pp. 12963–12968. 2002;
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu
W, Giri DD, Viale A, Olshen AB, Gerald WL and Massagué J: Genes
that mediate breast cancer metastasis to lung. Nature. 436:518–524.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Lu X, Lu X, Wang ZC, Iglehart JD, Zhang X
and Richardson AL: Predicting features of breast cancer with gene
expression patterns. Breast Cancer Res Treat. 108:191–201. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Korde LA, Lusa L, McShane L, Lebowitz PF,
Lukes L, Camphausen K, Parker JS, Swain SM, Hunter K and Zujewski
JA: Gene expression pathway analysis to predict response to
neoadjuvant docetaxel and capecitabine for breast cancer. Breast
Cancer Res Treat. 119:685–699. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kao KJ, Chang KM, Hsu HC and Huang AT:
Correlation of microarray-based breast cancer molecular subtypes
and clinical outcomes: implications for treatment optimization. BMC
Cancer. 11:1432011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Julka PK, Chacko RT, Nag S, Parshad R,
Nair A, Oh DS, Hu Z, Koppiker CB, Nair S, Dawar R, et al: A phase
II study of sequential neoadjuvant gemcitabine plus doxorubicin
followed by gemcitabine plus cisplatin in patients with operable
breast cancer: prediction of response using molecular profiling. Br
J Cancer. 98:1327–1335. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Hatzis C, Pusztai L, Valero V, Booser DJ,
Esserman L, Lluch A, Vidaurre T, Holmes F, Souchon E, Wang H, et
al: A genomic predictor of response and survival following
taxane-anthracycline chemotherapy for invasive breast cancer. JAMA.
305:1873–1881. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Glück S, Ross JS, Royce M, McKenna EF Jr,
Perou CM, Avisar E and Wu L: TP53 genomics predict higher clinical
and pathologic tumor response in operable early-stage breast cancer
treated with docetaxel-capecitabine ± trastuzumab. Breast Cancer
Res Treat. 132:781–791. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Farmer P, Bonnefoi H, Becette V,
Tubiana-Hulin M, Fumoleau P, Larsimont D, Macgrogan G, Bergh J,
Cameron D, Goldstein D, et al: Identification of molecular apocrine
breast tumours by microarray analysis. Oncogene. 24:4660–4771.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Desmedt CI, Piette F, Loi S, Wang Y,
Lallemand F, Haibe-Kains B, Viale G, Delorenzi M, Zhang Y,
d'Assignies MS, et al: TRANSBIG Consortium: Strong time dependence
of the 76-gene prognostic signature for node-negative breast cancer
patients in the TRANSBIG multicenter independent validation series.
Clin Cancer Res. 13:3207–3214. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Bos PD, Zhang XH, Nadal C, Shu W, Gomis
RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA and
Massagué J: Genes that mediate breast cancer metastasis to the
brain. Nature. 459:1005–1009. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Bonnefoi H, Potti A, Delorenzi M, Mauriac
L, Campone M, Tubiana-Hulin M, Petit T, Rouanet P, Jassem J, Blot
E, et al: Validation of gene signatures that predict the response
of breast cancer to neoadjuvant chemotherapy: a substudy of the
EORTC 10994/BIG 00–01 clinical trial. Lancet Oncol. 8:1071–1078.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Livak and Schmittgen: Analysis of relative
gene expression data using real-time quantitative PCR and the
2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Jozwik KM and Carroll JS: Pioneer factors
in hormone-dependent cancers. Nat Rev Cancer. 12:381–385. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Tozlu S, Girault I, Vacher S, Vendrell J,
Andrieu C, Spyratos F, Cohen P, Lidereau R and Bieche I:
Identification of novel genes that co-cluster with estrogen
receptor alpha in breast tumor biopsy specimens, using a
large-scale real-time reverse transcription-PCR approach. Endocr
Relat Cancer. 13:1109–1120. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Cimino-Mathews A, Subhawong AP, Illei PB,
Sharma R, Halushka MK, Vang R, Fetting JH, Park BH and Argani P:
GATA3 expression in breast carcinoma: Utility in triple-negative,
sarcomatoid, and metastatic carcinomas. Hum Pathol. 44:1341–1349.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Voduc D, Cheang M and Nielsen T: GATA3
expression in breast cancer has a strong association with estrogen
receptor but lacks independent prognostic value. Cancer Epidemiol
Biomarkers Prev. 17:365–373. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Mehra R, Varambally S, Ding L, Shen R,
Sabel MS, Ghosh D, Chinnaiyan AM and Kleer CG: Identification of
GATA3 as a breast cancer prognostic marker by global gene
expression meta-analysis. Cancer Res. 65:11259–11264. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Lin CY, Ström A, Vega VB, Kong SL, Yeo AL,
Thomsen JS, Chan WC, Doray B, Bangarusamy DK, Ramasamy A, et al:
Discovery of estrogen receptor alpha target genes and response
elements in breast tumor cells. Genome Biol. 5:R662004. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Kim K, Barhoumi R, Burghardt R and Safe S:
Analysis of estrogen receptor alpha-Sp1 interactions in breast
cancer cells by fluorescence resonance energy transfer. Mol
Endocrinol. 19:843–854. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Lambertini E, Tavanti E, Torreggiani E,
Penolazzi L, Gambari R and Piva R: ERalpha and AP-1 interact in
vivo with a specific sequence of the F promoter of the human
ERalpha gene in osteoblasts. J Cell Physiol. 216:101–110. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Li L and Davie JR: Association of Sp3 and
estrogen receptor alpha with the transcriptionally active trefoil
factor 1 promoter in MCF-7 breast cancer cells. J Cell Biochem.
105:365–369. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Oh DS, Troester MA, Usary J, Hu Z, He X,
Fan C, Wu J, Carey LA and Perou CM: Estrogen-regulated genes
predict survival in hormone receptor-positive breast cancers. J
Clin Oncol. 24:1656–1664. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Buache E, Etique N, Alpy F, Stoll I,
Muckensturm M, Reina-San-Martin B, Chenard MP, Tomasetto C and Rio
MC: Deficiency in trefoil factor 1 (TFF1) increases tumorigenicity
of human breast cancer cells and mammary tumor development in
TFF1-knockout mice. Oncogene. 30:3261–3273. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Amiry N, Kong X, Muniraj N, Kannan N,
Grandison PM, Lin J, Yang Y, Vouyovitch CM, Borges S, Perry JK, et
al: Trefoil factor-1 (TFF1) enhances oncogenicity of mammary
carcinoma cells. Endocrinology. 150:4473–4483. 2009. View Article : Google Scholar : PubMed/NCBI
|