Introduction
Esophageal squamous cell carcinoma (ESCC) is the
most common histological type of esophageal cancer worldwide
(2). ESCC tends to have a poor
prognosis due to high rates of lymph node metastasis and invasion
into surrounding organs (2). ESCC has
been identified to harbor frequent somatic mutations in tumor
protein 53 (3), Notch homolog 1,
tumor protein P53 (3) and
phosphatidylinositol-4,5-biphosphate 3-kinase catalytic subunit α
(4). However, previous cohorts used
to investigate the genomic abnormalities of ESCC were too small to
encompass the full range of genomic abnormalities (3,4). The
present authors have recently performed whole-exome sequence (WES)
analysis on 144 Japanese patients with ESCC, and identified a
previously uncharacterized mutant gene, zinc finger protein 750
(ZNF750) (1), which is an
essential regulator of epithelial proliferation and differentiation
(5–7).
In general, in order to identify a sporadic tumor
suppressor gene, studies should focus on the high frequency
aberrant genomic loci in familial cancer syndromes. As such, it was
identified that ZNF750 is located at chromosome 17q25, the
same location for the gene responsible for hereditary tylosis
esophageal cancer, a familial esophageal cancer (8,9).
In a previous study by the present authors,
mutations in ZNF750 were identified in 16.7% of ESCC tumors
and ZNF750 was identified to be the fifth most frequent
mutated genes. Additionally, 87.5% of mutations in ZNF750
led to decreased mRNA expression (1).
Similarly, Lin et al (10) and
Zhang et al (11) also
revealed inactivating mutations (70.0 and 64.0% of mutations in
each study, respectively) in ZNF750 by WES analysis of ESCC
and demonstrated that the depletion of ZNF750 in ESCC cell
lines promoted proliferation and invasiveness. These results
suggested that ZNF750 serves a role as a tumor suppressor in
ESCC. However to the best our knowledge, no study has investigated
whether aberrant ZNF750 expression affected clinical
outcomes in ESCC. Elucidation of the clinical significance of
ZNF750 expression may clarify whether ZNF750 may be
targeted in clinical therapy. Therefore, the present study aimed to
clarify the clinicopathological and prognostic significance of
ZNF750 expression in ESCC using two independent ESCC
datasets.
Materials and methods
ESCC patients and sample
collection
A total of 76 primary ESCC samples and 76 paired
normal tissues were obtained from 76 patients who underwent surgery
at Kyushu University Beppu Hospital (Beppu, Japan) and affiliated
hospitals [Iwate Medical University (Iwate, Japan), Kurume
University School of Medicine (Kurume, Japan), and Kagoshima
University Graduate School of Medical and Dental Sciences
(Kagoshima, Japan)] from January, 1998 to December, 2012 (Kyushu
dataset). The samples were taken as a part of routine examination.
The patient group comprised 67 males and 8 females (the gender of
one patient was not available). The mean age of the patients was
64.7 years (range, 40.0–79.0). All patients had a histological
diagnosis of ESCC and were closely followed at 3-month intervals.
The median follow-up period was 3.9 years. All patients were
treated in accordance with the Japan Esophageal Society Guidelines
for the treatment of esophageal cancer (12). Written informed consent was obtained
from all patients, and the Institutional Review Board of Kyushu
University (Fukuoka, Japan) approved the present study. Sample
collection was performed as previously described (13). Data on patient age, sex, histology,
tumor depth of invasion, lymph node metastasis, lymphatic invasion,
venous invasion and distant metastasis were obtained from the
clinical and pathological records. A total of two patients who
exhibited distant metastasis were included in the present study. Of
these patients, one exhibited distant lymph node metastasis and the
other liver metastasis.
ZNF750 mutation
ZNF750 mutation status data were obtained
from WES analysis that the authors of the present study previously
performed (1). RNA from 28 patients
of the 144 used in the previous study was available for
quantitative reverse transcription polymerase chain reaction
(RT-qPCR) analysis.
Total RNA extraction and RT
Total RNA from tissues was extracted by a modified
AGPC method using ISOGEN (Nippon Gene Co., Ltd., Tokyo, Japan), in
which the aqueous phase extraction step by chloroform is unrequired
(14). RT was performed using 8 µg of
total RNA with Moloney Murine leukemia virus reverse transcriptase
according to the manufacturer's protocol (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA).
Quantitative (q)PCR
qPCR reactions were performed using a LightCycler
480 probes master kit (Roche Diagnostics, Indianapolis, IN, USA)
and repeated three times as previously described (15). The following primers were used:
ZNF750; sense primer, 5′-GAACAGGTACTGCTTCCTGAGC-3′ and
antisense primer, 5′GAG AGC CTC CGT CAT CTGG-3′; small proline rich
protein 1A (SPRR1A); sense primer, 5′-TCCACCCCAGGAACCATGC-3′
and antisense primer, 5′-CTTGGTCTTCTGCTGGGCTG-3′;
glyceraldehyde-3-phosphate dehydrogenase (GAPDH); sense
primer, 5′-TTGGTATCGTGGAAGGACTCTA-3′ and antisense primer,
5′-TGTCATATTTGGCAGGTT-3. The expression levels of ZNF750 and
SPRR1A were normalized by GAPDH as an internal
control. The expression levels were calculated as the values
relative to the expression level of the cDNA using Human Universal
Reference Total RNA with the calibration curve method (Clontech
Laboratories, Inc., Mountainview, CA, USA) (16).
Immunohistochemical analysis
Immunohistochemistry of ZNF750 in 5 randomly
selected representative ESCC cases was performed on formalin-fixed,
paraffin-embedded tissues as previously described (15). A polyclonal rabbit anti-ZNF750
antibody (cat. no., HPA023012, Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany; dilution, 1:100) was used as the primary
antibody. Tumor histology was independently reviewed by an
experienced pathologist.
The Cancer Genome Atlas (TCGA) data
analysis
Paired RNA sequencing and survival data of 86
available patients with ESCC were obtained from TCGA (http://cancergenome.nih.gov/). ZNF750 mRNA
expression and survival data were extracted from this
reference.
Gene set enrichment analysis
(GSEA)
The correlations between ZNF750 expression
and previously annotated gene expression signatures were analyzed
by applying GSEA (17). ESCC
expression profiles from the National Center for Biotechnology
Information (NCBI) gene expression omnibus database (NCBI accession
no. GSE2533) were acquired, and analyzed using GSEA, as previously
described (18). Gene sets of
ZNF750 targets were extracted from C2 curated gene sets in
the Broad Institute database (http://www.broadinstitute.org/gsea/msigdb/collections.jsp).
Cancer Cell Line Encyclopedia (CCLE)
data analysis
Normalized mRNA expression data of 22 available ESCC
cell lines were obtained from the CCLE dataset (http://www.broadinstitute.org/ccle/home).
ZNF750 and SPRR1A mRNA expression data were extracted
from this reference.
Statistical analysis
For continuous variables, statistical analyses were
performed using Student's t-test for comparisons of ZNF750
mRNA expression according to the DNA mutation status (un-paired
t-test) and between corresponding tumor and normal tissues (paired
t-test). The degree of linearity was estimated by Pearson's
correlation coefficient. The association between ZNF750 mRNA
expression and clinicopathological factors was estimated by the
χ2 test with Yates' correction. Overall survival was
estimated using the Kaplan-Meier method, and survival curves were
compared using log-rank tests. Univariate and multivariate analyses
were used to determine patient prognosis and were performed by Cox
regression analysis with a backward stepwise model. Based on the
levels of ZNF750 mRNA expression in the Kyushu and the TCGA
datasets, patients were divided into two groups using the minimum
P-value approach, a comprehensive method that determines the
optimal risk separation cutoff point in continuous gene expression
measurement (19). The cutoff values
for high and low ZNF750 expression groups were 8.34
(ZNF750/GAPDH expression) in the Kyushu dataset and
2.75 (log2 expression) in the TCGA dataset. All tests were analyzed
by using the JMP v.11 software (JMP, Cary, NC, USA). Depth of tumor
invasion was classified according to the 7th edition of the
International Union against Cancer TNM classification system
(20). Histological grade of the
tumor was classified according to the 11th edition of the Japanese
Classification of Esophageal Cancer (21).
Results
Downregulation of ZNF750 expression in
ESCC
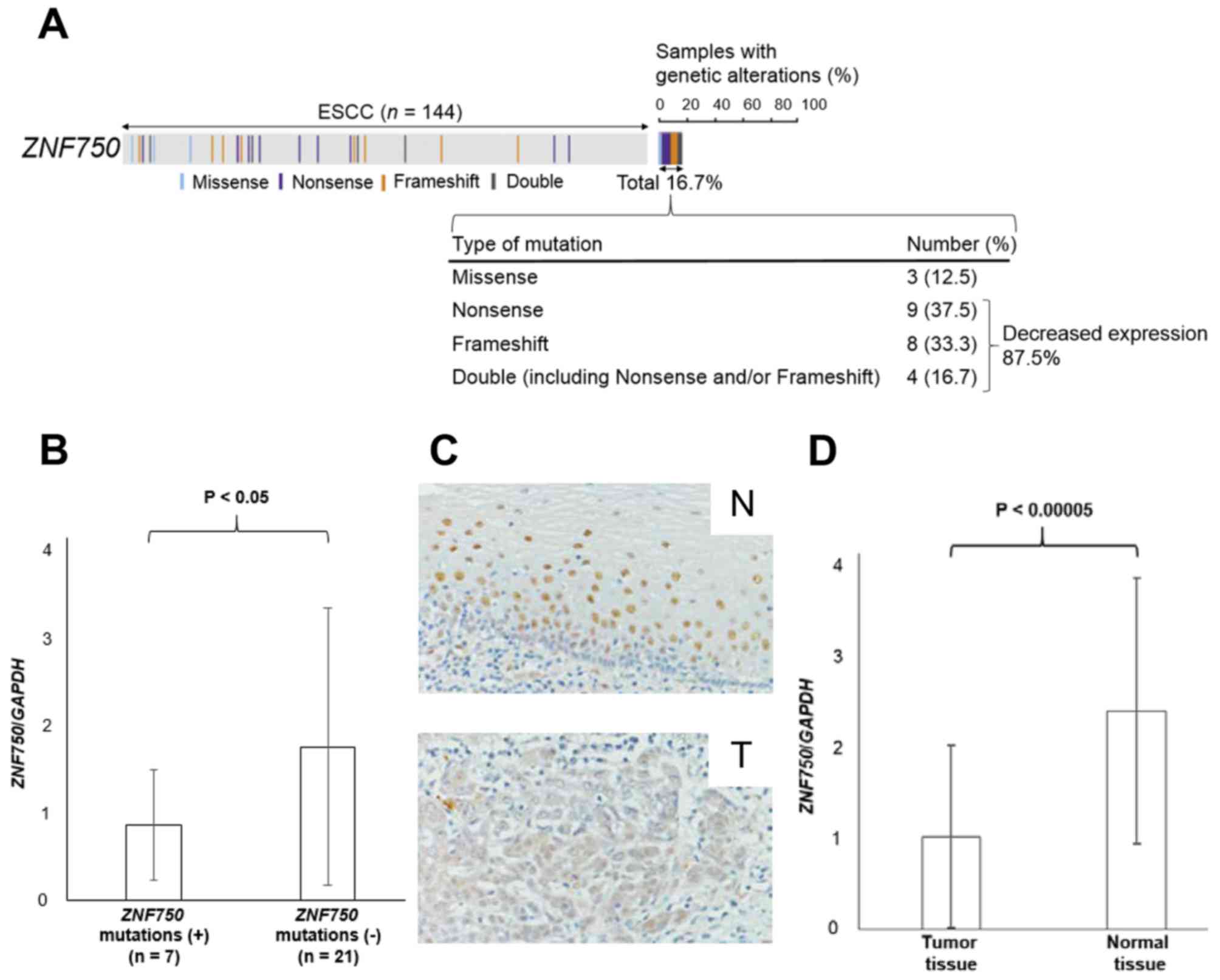
It was revealed that 16.7% of patients with ESCC
exhibited mutations in the ZNF750 gene (4) (Fig. 1A).
The majority of the mutations (87.5%) were nonsense and/or
frameshift mutations that caused reduced expression of
ZNF750 (Fig. 1A). As expected,
ZNF750 mRNA expression was significantly lower in patients
with ZNF750 nonsense and/or frameshift mutations (n=7)
compared with those without mutations (n=21) (P<0.05; Fig. 1B). Consistent with these data, the
immunohistochemical analysis revealed that ZNF750 was stained less
intensely in the nuclei of ESCC tumor cells compared with those of
normal esophageal epithelial cells in 4 of 5 (80.0%) patients
(Fig. 1C). Additionally, RT-qPCR
analysis also demonstrated that ZNF750 mRNA expression in
tumor tissues was significantly lower compared with paired normal
tissues (P<0.00005) (Fig. 1D). In
fact, ZNF750 mRNA expression levels in tumor tissues were
lower compared with normal tissues in 78.9% of 76 patients with
ESCC.
Clinicopathological significance of
ZNF750 mRNA expression in ESCC
The association between ZNF750 mRNA
expression levels in tumor tissues and clinicopathological factors
in ESCC is summarized in Table I. The
low ZNF750 expression group (n=37) exhibited a higher
frequency of tissues showing poorly differentiated histology
(P<0.05) compared with the high expression group (n=39). No
significant associations between ZNF750 mRNA expression and
age, sex, depth of invasion, lymph node metastasis, lymphatic
invasion, venous invasion or distant metastasis were observed.
 | Table I.Association between ZNF750
mRNA expression of tumor tissues and clinicopathological factors in
ESCC. |
Table I.
Association between ZNF750
mRNA expression of tumor tissues and clinicopathological factors in
ESCC.
| Factors | High ZNF750
expression (n=39) n, (%) | Low ZNF750
expression (n=37) n, (%) | P-value |
|---|
| <65 | 20 (51.3) | 18 (48.6) | NS |
| ≥65 | 19 (48.7) | 19 (51.4) |
|
| Male | 35 (89.7) | 32 (86.5) | NS |
| Female | 4 (10.3) | 4
(10.8) |
|
| NA | 0 (0) | 1
(2.7) |
|
| Well | 17 (43.6) | 7
(18.9) | <0.05 |
| Moderate/poor | 17 (43.6) | 22 (59.5) |
|
| NA | 5 (12.8) | 8 (21.6) |
|
| ≤submucosa
(T1) | 8 (20.5) | 2 (5.4) | NS |
| ≥muscularis propria
(T2-T4) | 31 (79.5) | 35 (94.6) |
|
| Absent | 12 (30.8) | 10 (27.0) | NS |
| Present | 27 (69.2) | 27 (73.0) |
|
| Absent | 8 (20.5) | 3 (8.1) | NS |
| Present | 31 (79.5) | 33 (89.2) |
|
| NA | 0 (0) | 1 (2.7) |
|
| Absent | 8 (20.5) | 2 (5.4) | NS |
| Present | 31 (79.5) | 34 (91.9) |
|
| NA | 0 (0) | 1 (2.7) |
|
| Absent | 38 (97.4) | 36 (97.3) | NS |
| Present | 1 (2.6) | 1 (2.7) |
|
Prognostic significance of ZNF750 mRNA
expression in ESCC
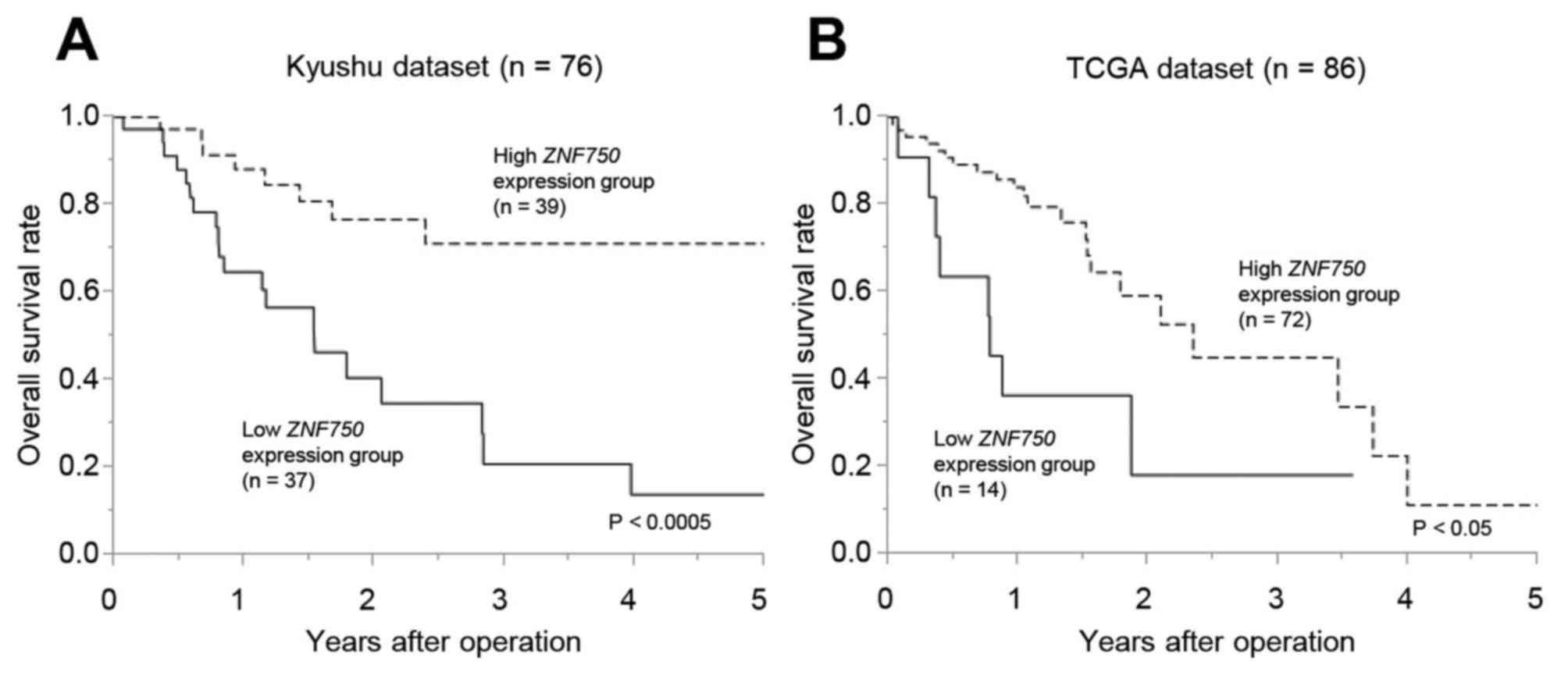
The overall survival rate of patients in the low
ZNF750 expression group was significantly lower compared
with the high expression group in the Kyushu dataset (P<0.0005;
Fig. 2A) and the TCGA dataset
(P<0.05) (Fig. 2B). In the
univariate analysis, higher T factor (T2-4), lymph node metastasis,
lymphovascular invasion and low ZNF750 mRNA expression were
significantly associated with a lower overall survival rate
(Table II). The multivariate
analysis demonstrated that lymphatic invasion (P<0.05) and low
ZNF750 mRNA expression (P<0.05) were independent poor
prognostic factors in ESCC (Table
II).
 | Table II.Univariate and multivariate analysis
of clinicopathological factors affecting overall survival rate. |
Table II.
Univariate and multivariate analysis
of clinicopathological factors affecting overall survival rate.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Factors | RR | 95% CI | P-value | RR | 95% CI | P-value |
|---|
| Age
(≥65/<65) | 0.93 | 0.46–1.91 | NS | 1.68 | 0.58–5.15 | NS |
| Sex
(male/female) | 1.19 | 0.45–4.13 | NS | 1.21 | 0.26–7.54 | NS |
| Histology grade
(well/moderate and poor) | 0.62 | 0.24–1.51 | NS | 1.89 | 0.59–5.66 | NS |
| T factor
(T1/T2-4) | 0.31 | 0.07–0.94 | <0.05 | 0.22 | 0.03–1.11 | NS |
| Lymph node
metastasis (negative/positive) | 0.43 | 0.17–0.98 | <0.05 | 0.53 | 0.11–1.92 | NS |
| Lymphatic invasion
(negative/positive) |
5.42×10−10 | 0.34–2.96 | <0.005 |
3.37×10−10 | 0.47–2.11 | <0.05 |
| Venous invasion
(negative/positive) |
6.02×10−10 | 0.48–2.08 | <0.05 |
1.67×10−9 | 0.37–2.68 | NS |
| Distant metastasis
(negative/positive) | 5.84 | 0.92–20.79 | NS | 11.01 | 0.51–23.34 | NS |
| ZNF750 mRNA
expression (high/low) | 3.83 | 1.82–8.59 | <0.0005 | 3.83 | 1.22–13.44 | <0.05 |
Correlation between ZNF750 mRNA
expression levels and epithelial differentiation genes in ESCC
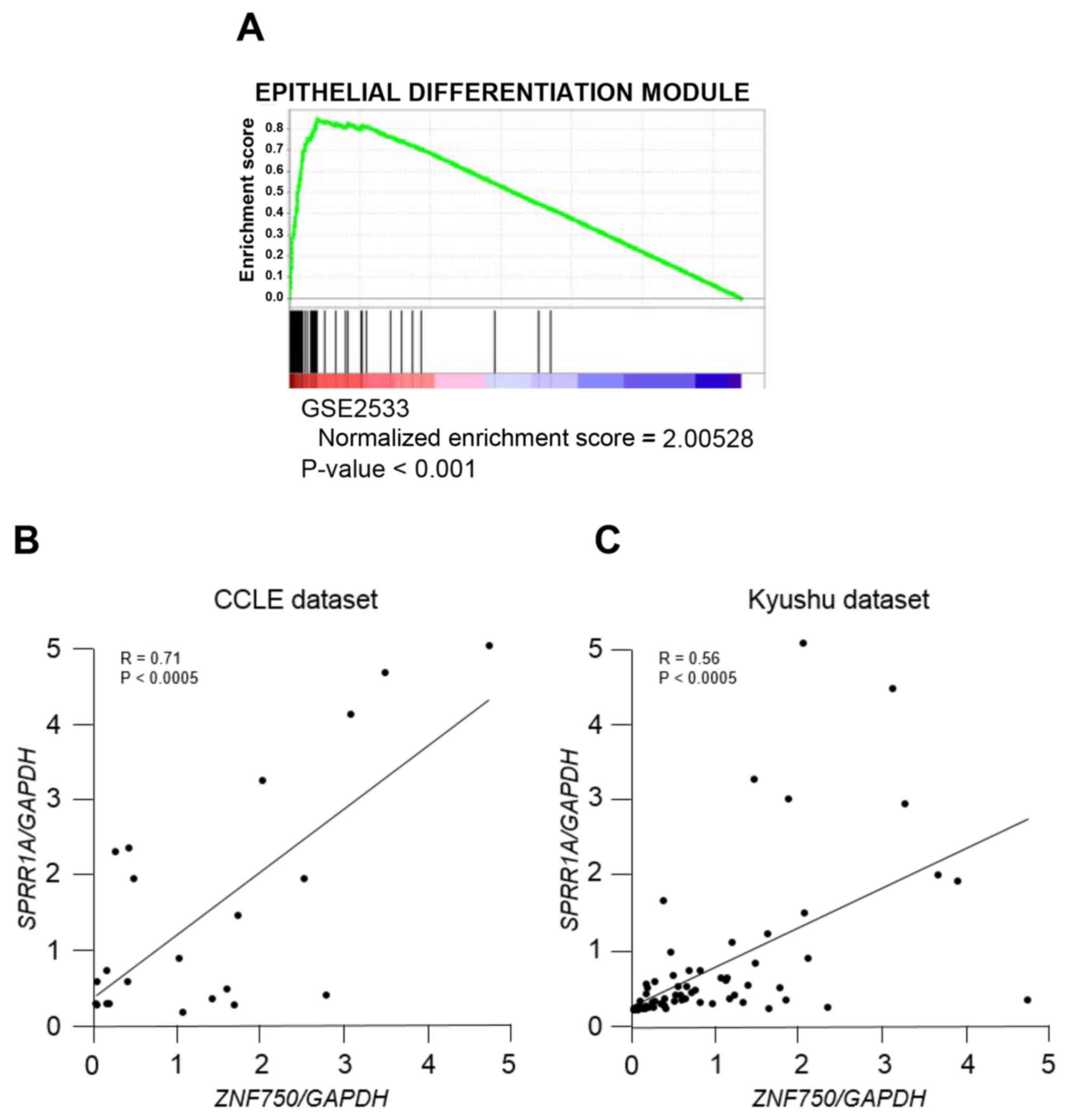
In order to validate the clinicopathological
analysis of ZNF750 in ESCC, GSEA was applied to patients
with ESCC (NCBI accession no. GSE2533). GSEA demonstrated that
ZNF750 expression was significantly correlated with
epithelial differentiation genes (Fig.
3A), which suggests that ZNF750 serves a role in
epithelial differentiation in the development of ESCC. Among those
genes, SPRR1A, an early epithelial differentiation marker
(22), was ranked at the upper level
of epithelial differentiation gene signature. The analysis
confirmed that ZNF750 mRNA expression was positively
correlated with SPRR1A mRNA expression using the CCLE
dataset (R=0.71; Fig. 3B) and the
Kyushu dataset (R=0.56; Fig. 3C).
Discussion
In the present study, the clinicopathological and
prognostic significance of ZNF750 expression, a novel
candidate tumor suppressor gene previously described by the present
authors (4), was assessed. In order
to assess the prognostic significance of ZNF750 expression,
two independent ESCC datasets were used. In addition, an
association between ZNF750 expression and epithelial
differentiation in ESCC was identified. The data of the present
study suggested that the downregulation of ZNF750 expression
due to inactivating mutations (nonsense/frameshift mutations)
induced the development or aggressiveness of ESCC, potentially via
dysregulation of esophageal epithelial differentiation. Although
other factors such as hyper-methylation or regulation by microRNA
may also downregulate the expression of ZNF750, to the best of our
knowledge, this is the first study to explore the clinical
significance of ZNF750 expression in ESCC.
The present clinicopathological study revealed that
the tissues from the low ZNF750 expression group exhibited a
higher frequency of poorly differentiated histology compared with
the high expression group. Previously, it was suggested that
ZNF750 function was associated with epithelial
differentiation (5–7,10,11). These data suggested that a low
expression of ZNF750 is associated with tumor aggressiveness
in ESCC.
Additionally, it was demonstrated that ZNF750
expression was reduced in tumor tissues, and that low expression of
ZNF750 was an independent poor prognostic factor in ESCC. In
a previous study by the present authors (4), it was also demonstrated that the
majority of the ZNF750 mutations were accompanied by loss of
heterozygosity. These data strongly support the hypothesis that
ZNF750 is a tumor suppressor gene, as previously identified
by WES analysis. In addition, low ZNF750 expression levels
in tumor tissues suggest that it is a promising biomarker capable
of predicting poor outcomes for patients with ESCC.
It has been reported that ZNF750 controls
epithelial homeostasis by repressing proliferation genes and
inducing differentiation genes (5–7). Previous
studies indicated that mutations in ZNF750 were pathogenic
and disrupt epithelial homeostasis, as observed in psoriasis
(23). The present study suggested
that ZNF750 expression was significantly correlated with
epithelial differentiation genes such as SPRR1A, an early
epithelial differentiation marker, in ESCC. The downregulation of
ZNF750 expression in ESCC may also disrupt epithelial
homeostasis, followed by the development of ESCC, according to
Tetreault et al (24), who
demonstrated that the downregulation of Kruppel like factor
4, an important epithelial differentiation gene, induced
malignant transformation of esophageal epithelium. However, the
function of the ZNF750 gene in ESCC has not been fully
elucidated. Additional large clinical studies and biological
experiments for ZNF750 in ESCC are required to clarify
this.
In conclusion, ZNF750 expression was low in
ESCC tumor tissues, and its reduction was associated with poor
differentiation. In addition, low ZNF750 expression was an
independent poor prognostic factor in ESCC. These data provide
evidence that ZNF750 is a novel tumor suppressor gene, and
may be a therapeutic target and useful biomarker of poor clinical
outcome in patients with ESCC.
Acknowledgements
The authors would like to thank Ms. M. Oshiumi, Mr.
M. Utou, Mrs. K. Oda, Ms. M. Kasagi, Mrs. S. Sakuma, Mrs. N.
Mishima and Ms. T. Kawano (Departments of Surgery and Pathology,
Kyushu University Beppu Hospital, Beppu, Japan) for their excellent
technical assistance. The present study was supported by the
following grants and foundations: Grants-in-Aid for Scientific
Research of MEXT (grant nos. 26461980, 26293303, 26670608, 26861003
and 26271401); Japan Agency for Medical Research and development,
AMED (grant no. P14009); The OITA Cancer Research Foundation 2015;
Daiwa Securities Health Foundation.
References
|
1
|
Sawada G, Niida A, Uchi R, Hirata H,
Shimamura T, Suzuki Y, Shiraishi Y, Chiba K, Imoto S, Takahashi Y,
et al: Genomic landscape of esophageal squamous cell carcinoma in a
Japanese population. Gastroenterology. 150:1171–1182. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Agrawal N, Jiao Y, Bettegowda C, Hutfless
SM, Wang Y, David S, Cheng Y, Twaddell WS, Latt NL, Shin EJ, et al:
Comparative genomic analysis of esophageal adenocarcinoma and
squamous cell carcinoma. Cancer Discov. 2:899–905. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shigaki H, Baba Y, Watanabe M, Murata A,
Ishimoto T, Iwatsuki M, Iwagami S, Nosho K and Baba H: PIK3CA
mutation is associated with a favorable prognosis among patients
with curatively resected esophageal squamous cell carcinoma. Clin
Cancer Res. 19:2451–2459. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cohen I, Birnbaum RY, Leibson K, Taube R,
Sivan S and Birk OS: ZNF750 is expressed in differentiated
keratinocytes and regulates epidermal late differentiation genes.
PLoS One. 7:e426282012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sen GL, Boxer LD, Webster DE, Bussat RT,
Qu K, Zarnegar BJ, Johnston D, Siprashvili Z and Khavari PA: ZNF750
is a p63 target gene that induces KLF4 to drive terminal epidermal
differentiation. Dev Cell. 22:669–677. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Boxer LD, Barajas B, Tao S, Zhang J and
Khavari PA: ZNF750 interacts with KLF4 and RCOR1, KDM1A, and
CTBP1/2 chromatin regulators to repress epidermal progenitor genes
and induce differentiation genes. Genes Dev. 28:2013–2026. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kelsell DP, Risk JM, Leigh IM, Stevens HP,
Ellis A, Hennies HC, Reis A, Weissenbach J, Bishop DT, Spurr NK and
Field JK: Close mapping of the focal non-epidermolytic palmoplantar
keratoderma (PPK) locus associated with oesophageal cancer (TOC).
Hum Mol Genet. 5:857–860. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Blaydon DC, Etheridge SL, Risk JM, Hennies
HC, Gay LJ, Carroll R, Plagnol V, McRonald FE, Stevens HP, Spurr
NK, et al: RHBDF2 mutations are associated with tylosis, a familial
esophageal cancer syndrome. Am J Hum Genet. 90:340–346. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin DC, Hao JJ, Nagata Y, Xu L, Shang L,
Meng X, Sato Y, Okuno Y, Varela AM, Ding LW, et al: Genomic and
molecular characterization of esophageal squamous cell carcinoma.
Nat Genet. 46:467–473. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang L, Zhou Y, Cheng C, Cui H, Cheng L,
Kong P, Wang J, Li Y, Chen W, Song B, et al: Genomic analyses
reveal mutational signatures and frequently altered genes in
esophageal squamous cell carcinoma. Am J Hum Genet. 96:597–611.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kuwano H, Nishimura Y, Oyama T, Kato H,
Kitagawa Y, Kusano M, Shimada H, Takiuchi H, Toh Y, Doki Y, et al:
Guidelines for diagnosis and treatment of carcinoma of the
esophagus april 2012 edited by the Japan esophageal society.
Esophagus. 12:1–30. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hirata H, Sugimachi K, Komatsu H, Ueda M,
Masuda T, Uchi R, Sakimura S, Nambara S, Saito T, Shinden Y, et al:
Decreased expression of fructose-1,6-bisphosphatase associates with
glucose metabolism and tumor progression in hepatocellular
carcinoma. Cancer Res. 76:3265–3276. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chomczynski P and Mackey K: Short
technical reports. Modification of the TRI reagent procedure for
isolation of RNA from polysaccharide- and proteoglycan-rich
sources. Biotechniques. 19:942–945. 1995.PubMed/NCBI
|
|
15
|
Ueda M, Iguchi T, Nambara S, Saito T,
Komatsu H, Sakimura S, Hirata H, Uchi R, Takano Y, Shinden Y, et
al: Overexpression of transcription termination factor 1 is
associated with a poor prognosis in patients with colorectal
cancer. Ann Surg Oncol. 22 Suppl 3:S1490–S1498. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sørensen BS, Schmidt H, von der Maase H,
Straten PT and Nexø E: Quantification of melanoma cell-specific
MART-1 mRNA in peripheral blood by a calibrated competitive reverse
transcription-PCR. Clin Chem. 46:1923–1928. 2000.PubMed/NCBI
|
|
17
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:pp. 15545–15550. 2005;
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei G, Luo H, Sun Y, Li J, Tian L, Liu W,
Liu L, Luo J, He J and Chen R: Transcriptome profiling of
esophageal squamous cell carcinoma reveals a long noncoding RNA
acting as a tumor suppressor. Oncotarget. 6:17065–17080. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mizuno H, Kitada K, Nakai K and Sarai A:
PrognoScan: A new database for meta-analysis of the prognostic
value of genes. BMC Med Genomics. 2:182009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yamasaki M, Miyata H, Miyazaki Y,
Takahashi T, Kurokawa Y, Nakajima K, Takiguchi S, Mori M and Doki
Y: Evaluation of the nodal status in the 7th edition of the
UICC-TNM classification for esophageal squamous cell carcinoma:
Proposed modifications for improved survival stratification: Impact
of lymph node metastases on overall survival after esophagectomy.
Ann Surg Oncol. 21:2850–2856. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Japan Esophageal Society, . Japanese
classification of esophageal cancer, 11th edition: Part I.
Esophagus. 14:1–36. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sark MW, Fischer DF, de Meijer E, van de
Putte P and Backendorf C: AP-1 and ets transcription factors
regulate the expression of the human SPRR1A keratinocyte terminal
differentiation marker. J Biol Chem. 273:24683–24692. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang CF, Hwu WL, Yang LC, Chung WH, Chien
YH, Hung CF, Chen HC, Tsai PJ, Fann CS, Liao F and Chen YT: A
promoter sequence variant of ZNF750 is linked with familial
psoriasis. J Invest Dermatol. 128:1662–1668. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tetreault MP, Yang Y, Travis J, Yu QC,
Klein-Szanto A, Tobias JW and Katz JP: Esophageal squamous cell
dysplasia and delayed differentiation with deletion of Krüppel-like
factor 4 in murine esophagus. Gastroenterology. 139:171–181.e9.
2010. View Article : Google Scholar : PubMed/NCBI
|