Introduction
Liver cancer is one of the most commonly occurring
malignant tumors; its global incidence and mortality rate are
ranked 5th and 3rd among malignant tumors, respectively (1,2). The
majority of cases of liver cancer are hepatocellular carcinoma
(HCC) (3). The global distribution of
HCC is disproportional, with the highest incidence reported in Asia
and Sub-Saharan Africa, particularly in China (3). Patients with HCC exhibit an overall
5-year survival rate of only 5% (4).
In total, ~70% of patients experience relapse within five years of
undergoing surgery and >80% of recurrences are within the
remaining liver tissue (5). Patients
with HCC often exhibit various outcomes, even when identical
clinicopathological features are observed; this suggests that the
development and rapid progression of HCC involves numerous complex
molecular and cellular events (6).
Therefore, developing effective methods for the prevention and
treatment of HCC requires an improved understanding of the
biological development of HCC.
As demonstrated in our previous analysis of the
tissue microarray data, the BTB domain-containing 3 (BTBD3) gene
was upregulated in HCC tissues, indicating that it may be a
cancer-associated gene and have a role in the occurrence and
development of HCC. The present study aimed to further analyze the
expression levels of BTBD3 in HCC cell lines and explore its role
in the occurrence, development and metastasis of HCC.
Materials and methods
Cell line and cell culture
The human immortalized normal hepatocyte LO2 cell
line, and HCC HepG2, Huh7, Bel7404 and Hep3B cell lines (Type
Culture Collection of the Chinese Academy of Sciences, Shanghai,
China), were maintained in Dulbecco's modified Eagle's medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fatal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The cells were
incubated at 37°C in 5% CO2.
Bioinformatics analysis
The HCC tissue microarray data of GSE14215 and
GSE29217 were downloaded from the Gene Expression Omnibus database
of the National Centre for Biotechnology Information (https://www.ncbi.nlm.nih.gov/geo/). The genes
with common various expressions were analyzed using Genespring
version 11.0 software (Agilent Technologies, Inc., Santa Clara, CA,
USA) and determined using meta-analysis.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA from all the cell lines was isolated using
miRNeasy Mini kit (Qiagen GmbH, Hilden, Germany) according to the
manufacturer's instructions. The RT-qPCR amplification for the
quantification of the BTBD3 and GAPDH mRNAs was performed using an
ABI PRISM 7500 Sequence Detection System (Applied Biosystems;
Thermo Fisher Scientific, Inc.) and a SYBR® Premix Ex Taq™ (Tli
RNaseH Plus) kit (Takara Bio, Inc., Otsu, Japan). The following
primers were used: BTBD3 sense, 5′-TGGCAGATGTACATTTTGTGG-3′ and
antisense, 5′-AACACAGAGCTCCCAACAGC-3′; GAP DH sense,
5′-GGGAAACTGTGGCGTGATG-3′ and antisense, 5′- GAG TGG GTG TCG CTG
TTGA-3′. The RT reaction was performed at 42°C for 30 min and then
70°C for 15 sec. qPCR was performed using the cDNA as template
under the following conditions: PCR initial activation at 95°C for
15 min, followed by 40 cycles at 94°C for 15 sec, 55°C for 30 sec
and 70°C for 30 sec. The expression level of BTBD3 was normalized
as relative expression to GAPDH. Relative expression was calculated
as 2−ΔΔCq (7). Each PCR
reaction was performed in triplicate.
Western blot analysis
Total proteins were extracted from LO2 and HepG2,
Huh7, Bel7404 and Hep3B cell lines. Equal amounts of protein (50
µg) were loaded for electrophoreses on 10% SDS-PAGE gels,
transferred to polyvinylidene fluoride membrane and incubated
overnight at 4°C with the appropriate primary antibodies as
follows: Monoclonal rabbit anti-human BTBD3 (dilution, 1:1,500;
catalog no. ABIN1398849; Abcam, Cambridge, MA, USA) and GAPDH
(dilution, 1:10,000; catalog no. G8140; US Biological, Swampscott,
MA, USA). Following incubation with the horseradish
peroxidase-conjugated bovine monoclonal anti-rabbit secondary
antibody (dilution, 1:5,000; catalog no. COL18A1; Boster Biological
Technology, Pleasanton, CA, USA) for 2 h at room temperature, the
immumoreactive proteins were visualized using the enhanced
chemiluminescence (GE Healthcare, Chicago, IL, USA) method. The
amounts of proteins were quantified by scanning densitometry using
Quantity One software (Bio-Rad Laboratories, Inc., Hercules, CA,
USA) and normalized to the relative internal standards by GAPDH
protein band density.
Short interfering (si)RNA
transfection
As presented in Table
I, two BTBD3 siRNAs, siRNA-79 and siRNA-81, and a negative
control siRNA, siRNA-negative control (NC), were designed and
synthesized by Sigma-Aldrich (Merck KGaA). Using Lipofectamine
RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.), siRNA-79,
siRNA-81 and siRNA-NC were transfected into Bel7404 cells,
according to the manufacturer's instructions. A total of 24-h after
transfection, the interference efficiency of siRNA was determined
by RT-qPCR assay.
 | Table I.BTB domain-containing 3 siRNA
sequences. |
Table I.
BTB domain-containing 3 siRNA
sequences.
| Strand | Sequence (5′-3′) |
|---|
| siRNA-79 |
|
Sense |
5′-CUUAGCUCAUCUGCAAAUAdTdT-3′ |
|
Antisense |
5′-UAUUUGCAGAUGAGCUAAGdTdT-3′ |
|
Target |
5′-AUAAACGUCUACUCGAUUC-3′ |
| siRNA-81 |
|
Sense |
5′-CCAGUUUGCAGUUGAUAAAdTdT-3′ |
|
Antisense |
5′-UUUAUCAACUGCAAACUGGdTdT-3′ |
|
Target |
5′-AAAUAGUUGACGUUUGACC-3′ |
| siRNA-NC |
|
Sense |
5′-UUCUCCGAACGUGUCACGUdTdT-3′ |
|
Antisense |
5′-ACGUGACACGUUCGGAGAAdTdT-3′ |
|
Target |
5′-UGCACUGUGCAAGCCUCUU-3′ |
MTS assay
Bel7407 cells were seeded into 96-well plates at a
density of 1×104 cells/well and cultivated at 37°C in 5%
CO2 for 24 h. Subsequently, Bel7407 cells were
transfected at 37°C for 6 h with BTBD3 siRNA-79 (25 nM) and
siRNA-NC (25 nM) using Lipofectamine RNAiMAX (Invitrogen; Thermo
Fisher Scientific, Inc.), respectively. MTS (1:10 dilution) was
added to the cells every 24 h (0, 24, 48 and 72 h), which were then
incubated for 4 h at 37°C. The optical density of the culture
medium at 490 nm was evaluated using an EnVision plate reader
(PerkinElmer, Inc., Waltham, MA, USA). Triplicate wells were
analyzed for each assay.
Flow cytometry
Bel7404 cells seeded at a density of
5×106 per well into 6-well plates were transfected with
siRNA-79 (25 nM) or siRNA-NC (25 nM). A total of 48-h after
transfection, cells were harvested, fixed with 70% ethanol and then
resuspended in propidium iodide/RNase Staining Buffer (BD
Biosciences, Franklin Lakes, NJ, USA). The DNA content of cells was
analyzed using a MoFlo XDPCell Sorter (Beckman Coulter, Inc., Brea,
CA, USA). The cell numbers in each phase of the cell cycle was
determined using FlowJo software (FlowJo version 7.6.3, LLC,
Ashland, OR, USA).
Wound healing assays
Bel7404 cells were grown to 90% confluence in the
6-well flat-bottomed plates. The monolayer cells were scratched
with a sterile 200-µl pipette tip to create a denuded zone (gap) of
1 mm width. The remaining cells were washed twice with PBS to
remove cell debris and incubated at 37°C in DMEM/F12 (Gibco; Thermo
Fisher Scientific, Inc.). The scratched areas were imaged at 0, 6,
24 and 48 h after wounding using a phase-contrast microscope (Leica
Micro-systems GmbH, Wetzlar, Germany, magnification, ×200). Cell
motility was evaluated as percentages of cell coverage to the
initial cell-free zone using ImageJ software (ImageJ version
2.1.4.7, National Institutes of Health, Bethesda, MA, USA). The
cell mobility rate was evaluated as follows: 1-distance at various
time points/distance at 0 h)x100% Three randomly selected wound
areas were analyzed.
Transwell invasion assay
Following siRNA transfection, 1×104
Bel7404 cells resuspended in serum-free DMEM/F12 (Gibco; Thermo
Fisher Scientific, Inc.) were seeded in the upper chambers, and 600
µl complete DMEM medium (Gibco; Thermo Fisher Scientific, Inc.) was
added to the lower chambers. Following incubation at 37°C for 18 h,
the non-migrated cells on the upper layer were removed using cotton
swabs and the penetrating cells in the lower layer were fixed in 4%
paraformaldehyde for 15 min and stained with 0.1% crystal violet
for 10–20 min at room temperature. The cells in five random
microscopic fields were counted and imaged using a light microscope
with a DP70CCD system (Olympus Corp., Tokyo, Japan). All
experiments were performed in triplicate.
Statistical analysis
All statistical analyses were performed using SPSS
(version 17.0; SPSS Inc., Chicago, IL, USA). Data are presented as
the mean ± standard deviation from three independent experiments
and statistical analyses were performed using Student's t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Analysis of the expression level of
the BTBD3 in HCC tissues using microarray data
Gene microarray data of GSE14215 and GSE29217 were
downloaded from NCBI and analyzed using Genespring software.
Compared with normal tissues, the analysis results demonstrated
that 227 and 281 genes were upregulated and downregulated,
respectively, in the HCC tissues. Among the upregulated genes,
BTBD3 ranked 60th, with 2.45-fold upregulation, indicating its
potential role in the occurrence and development of HCC.
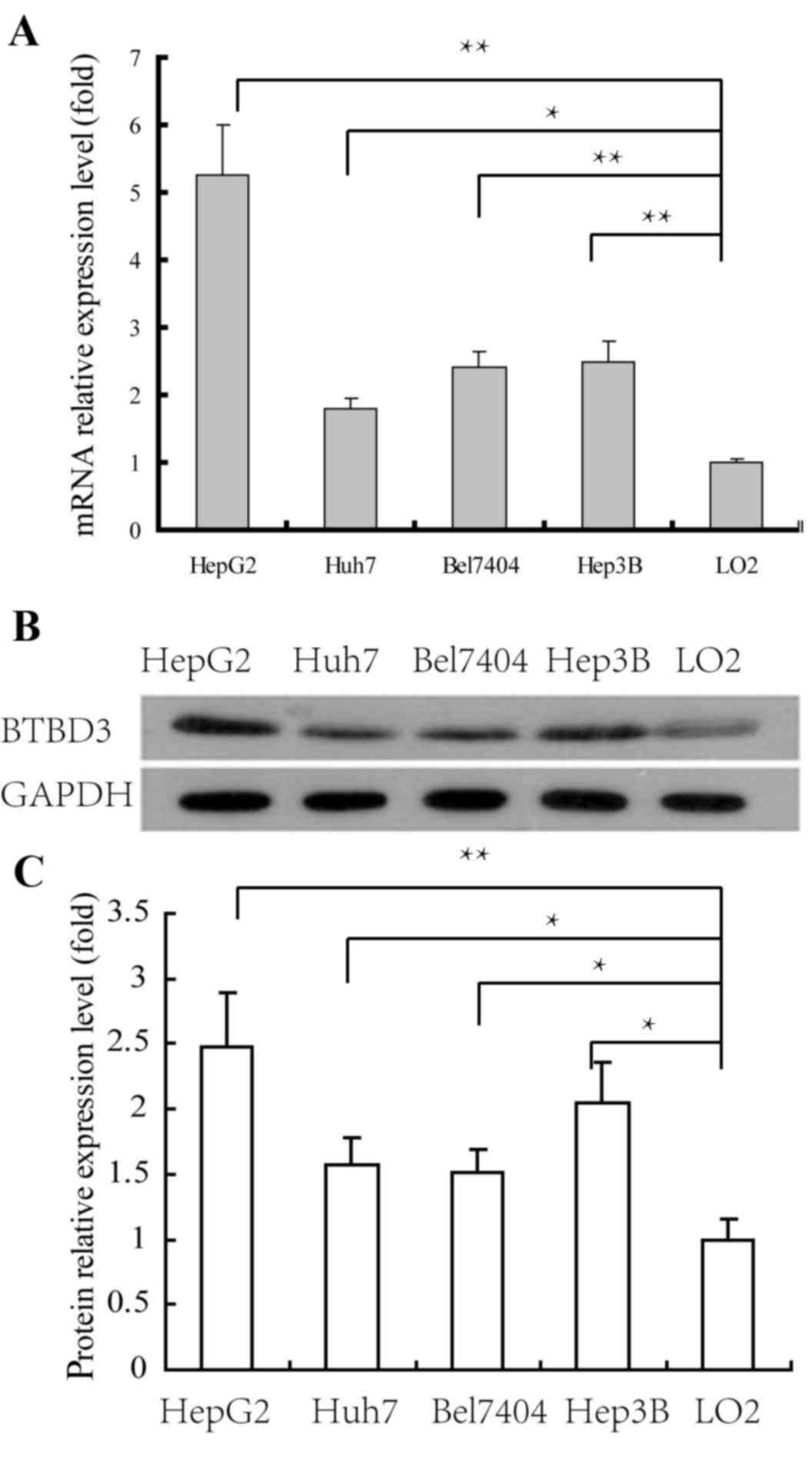
Expression levels of BTBD3 in HCC cell
lines
The expression levels of BTBD3 in HCC HepG2, Huh7,
Bel7407 and Hep3B cell lines and liver normal LO2 cell line were
determined by real-time RT-qPCR and western blotting. The
expression levels of BTBD3 mRNA in the HCC cell lines were
significantly increased compared with LO2 cells. As presented in
Fig. 1A, compared with LO2 cells,
expression was upregulated 5.3, 1.8, 2.5 and 2.6-fold in HepG2,
Huh7, Bel7404 and Hep3B cells, respectively. The results of the
western blot analysis confirmed that the expression levels of BTBD3
protein in HCC cell lines was significantly increased compared with
LO2 cells (Fig. 1B and C), which was
consistent with the findings of RT-qPCR. Therefore, it was
suggested that overexpression of BTBD3 gene was associated with the
proliferation or/and metastasis of HCC.
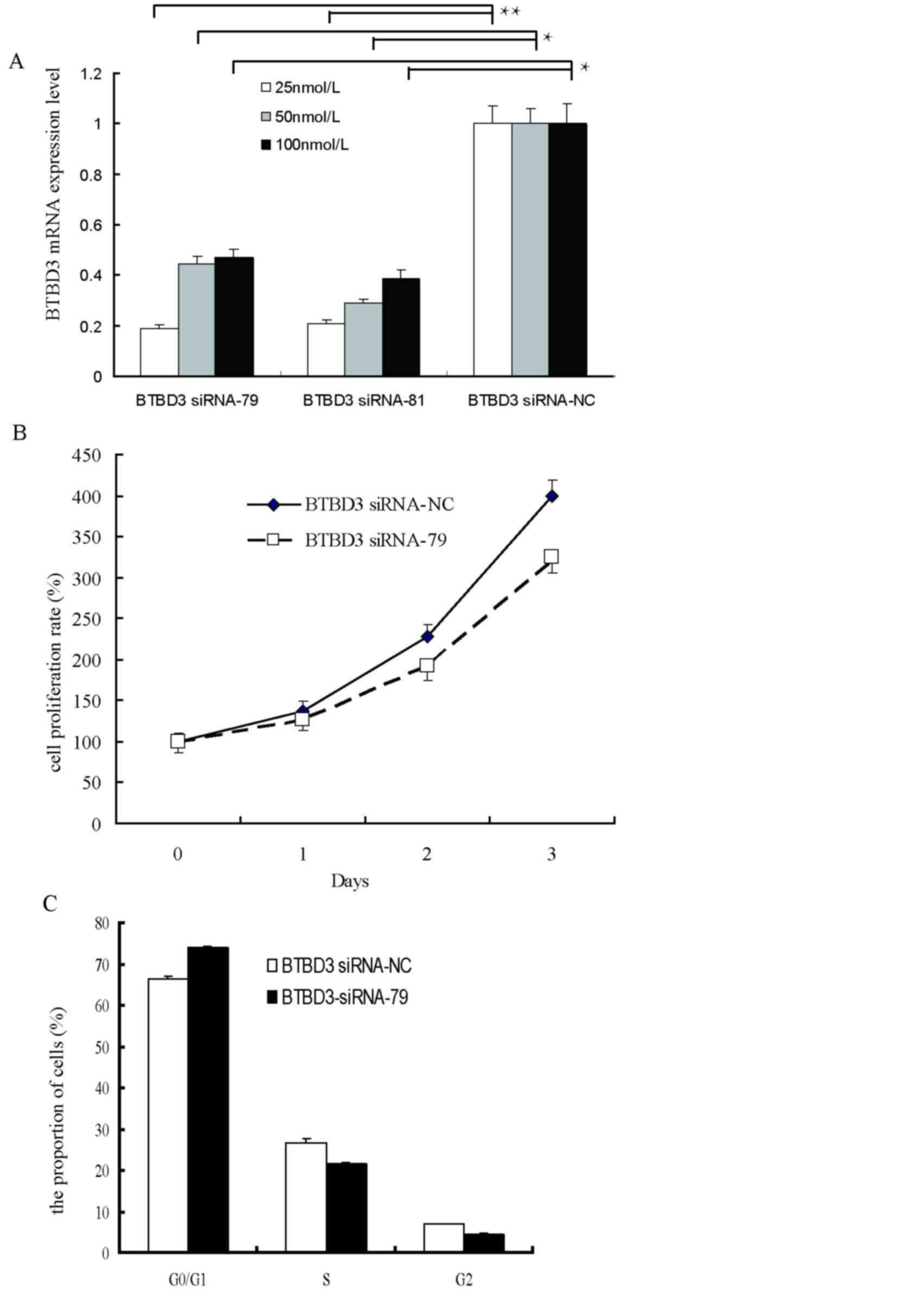
Effect of BTBD3 siRNA interference on
cell proliferation
To determine the effects of BTBD3 expression on cell
growth and migration, two BTBD3 siRNA sequences (siRNA-79 and
siRNA-81) and a negative control siRNA-NC were designed,
synthesized and transfected into Bel7407 cells. As demonstrated in
by RT-qPCR, 24 h after siRNA transfection, significant
interference, with 70% downregulation of the BTBD3 gene, was
observed at 25 nM of siRNA-79 and siRNA-81, and therefore 25 nM
siRNA-79 was selected for use in the following experiments
(Fig. 2A).
The present study analyzed cell proliferation of
Bel7407 cells upon silencing of BTBD3 by MTS assay (Fig. 2B). Compared with at 0 h, a 136, 227
and 398% increase in cell growth was observed in the negative
control group at 24, 48 and 72 h after transfection, respectively,
whereas a 127, 192 and 320% increase was observed at 24, 48 and 72
h after transfection, respectively, in the BTBD3 siRNA-79 group. No
significant difference was observed between the BTBD3 siRNA-79
group and the negative control (P>0.05), indicating a lack of
association between the BTBD3 gene and cell proliferation.
Flow cytometry was performed to determine the change
in cell cycle of Bel7407 cells at 48 h post-transfection (Fig. 2C). No significant difference in the
percentage of cells in the G0/G1,
G2 and S stages of the cell cycle were observed between
the BTBD3 siRNA-79 group and negative control, which further
confirmed that BTBD3 did not promote cell proliferation.
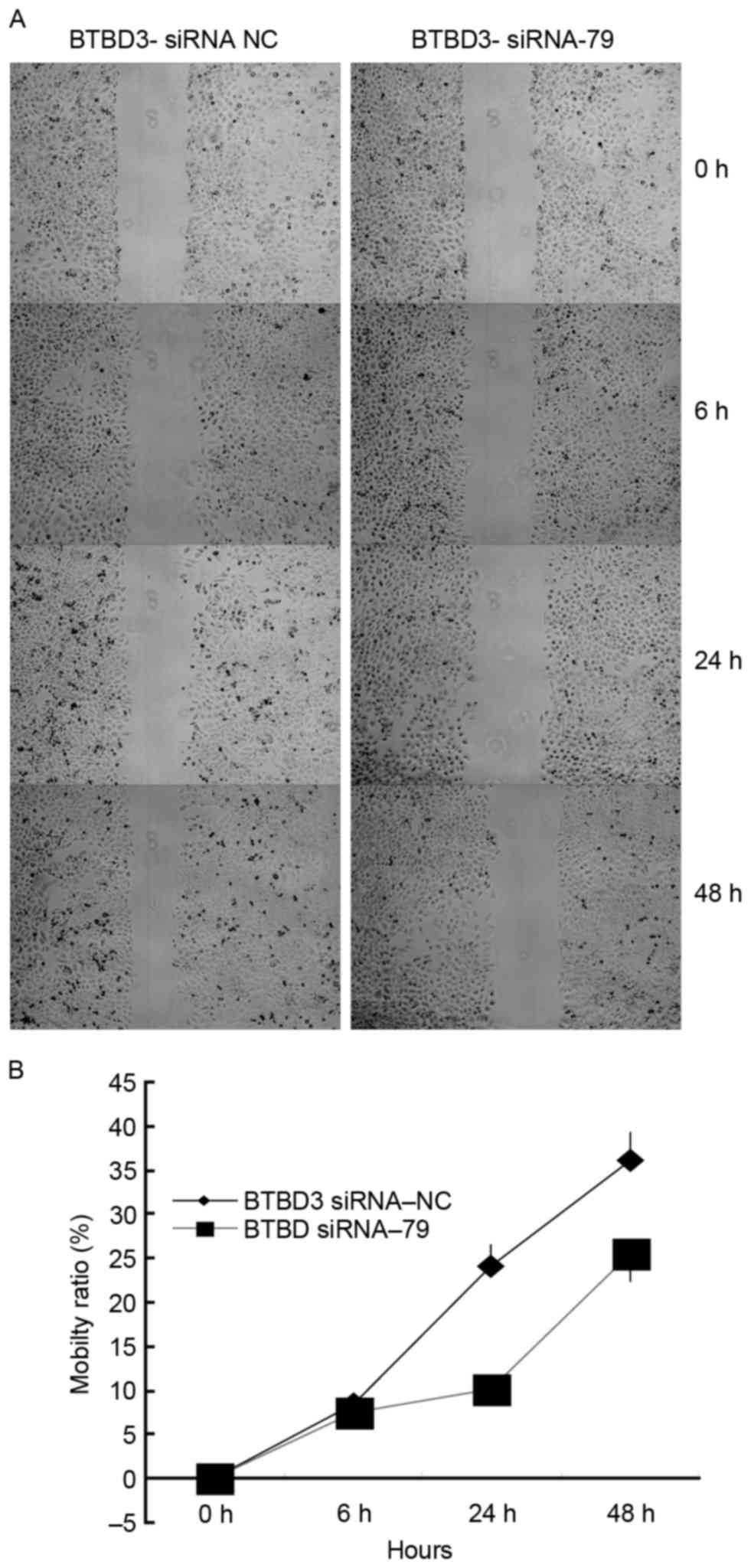
Effect of BTBD3 siRNA interference on
cell migration and invasion
In order to investigate of the role of BTBD3 gene in
the metastasis of HCC, a cell scratch test and a Transwell assay
were performed to determine the migration and invasion of Bel7407
cells following siRNA transfection.
As demonstrated by the cell scratch test (Fig. 3A), 6, 24 and 48 h after initiation,
the relative widths of the scratches in the control group were 91,
76 and 63% of the original width, respectively. The relative widths
in the siRNA-79 group were 93, 90 and 76%, respectively, which were
significantly wider than those of the control group (Fig. 3B; P=0.0361). These results indicated
that BTBD3 gene silencing resulted in a decreased capacity for cell
migration in Bel7404 cells.
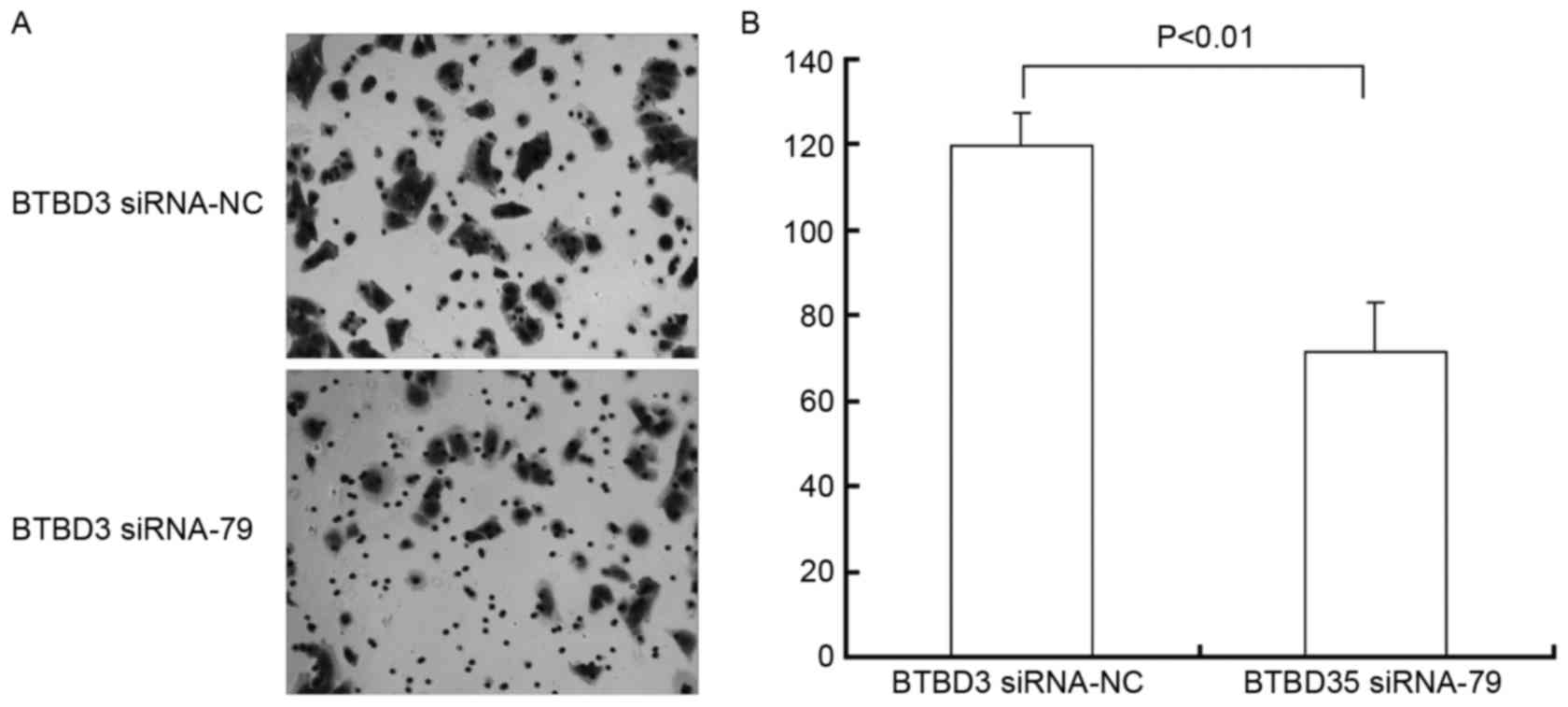
As demonstrated in the Transwell chamber assay, the
migration of Bel7407 cells was significantly inhibited (Fig. 4A) following siRNA transfection. As
presented in Fig. 4B, the number of
invasive and metastatic cells in the siRNA-NC control group was
119.5±7.31, which was significantly higher (P=0.007) than the BTBD3
siRNA-79 group (71.33±11.27).
Discussion
HCC is extremely malignant and highly invasive, with
a high incidence of both intra-hepatic and extra-hepatic
metastases. Metastatic recurrence is the most important reason for
the unsatisfactory prognosis following surgery (4–6,8,9). Owing to
its high incidence, metastasis and mortality rates, HCC has
attracted attention for the investigation of the mechanism
underlying its occurrence and development. With the development of
molecular biology techniques, including high-throughput detection
technology (microarray) and detailed sequencing techniques, there
has been great progress in the elucidation of the mechanisms
underlying HCC (10–12). Approximately 200 genes have been
associated with the proliferation, metastasis and recurrence of HCC
(11).
BTBD3 is located at 20p12.2, with two splice
variants coding for 482 and 385 amino acids, respectively. Within
this gene, there is a BTB (broad-complex, tramtrack and
bric-à-brac) structural domain (amino acids 113–219), a nuclear
localization signal region (amino acids 55–70), and a structural
domain containing BACK (amino acids 226–335) and PHR (amino acids
226–335) regions. At present, there have been limited studies
investigating the function of the BTBD3 gene (13–20). A
previous study by Zhang et al (13), investigating the function of
hsa-let-7i in colon cancer metastasis, is the only study to have
suggested the BTBD3 gene may be the target of hsa-let-7i. The
analysis of the microarray data of gene expression levels in HCC
tissues in the present study revealed that the BTBD3 gene was
upregulated by 2.45-fold in cancer tissues, which indicated the
potential role of the BTBD3 gene in the occurrence and development
of HCC. However, to the best of our knowledge, no previous studies
have investigated the association between the BTBD3 gene and
HCC.
In order to investigate the role of the BTBD3 gene
in the occurrence and development of HCC, RT-qPCR and western
blotting were performed to analyze the expression levels of BTBD3
in HCC cell lines. The overexpression of the BTBD3 gene in various
HCC cell lines indicated that BTBD3 may be a cancer-associated
gene. The expression level of BTBD3 was highest in HepG2 cells;
however, it was not in Bel7404 cells, as previously demonstrated by
a nude mouse transplantation tumor with the Bel7404 cell line
(21). To facilitate this study, the
present study used Bel7404 cells for the in vitro
experiments, including MTS and wound healing assays. Subsequently,
the siRNA interference technique was used to investigate the effect
of the BTBD3 gene on the proliferation and metastasis of Bel7407
cells. The results revealed there was a minimal impact on cell
proliferation following silencing of the BTBD3 gene. However,
significant inhibition of cell invasion (≤50%) was demonstrated in
the wound healing assay and the Transwell model. Based on the
aforementioned findings, it may be concluded that the BTBD3 gene
was overexpressed in HCC tissues and cell lines, which promoted the
invasion and metastasis of cancer cells without affecting cell
proliferation.
As there are limited previous studies regarding the
function of the BTBD3 gene, this is the first study to demonstrate
the promoting effect of the BTBD3 gene on HCC cell invasion;
however, further confirmation by performing in vivo
experiments, and investigation into the underlying mechanisms of
BTBD3 in HCC cell migration and invasion, are required.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81272483) and
Guangdong Natural Science Foundation (grant no.
S2013010016728).
Glossary
Abbreviations
Abbreviations:
|
BTBD3
|
BTB domain-containing 3
|
|
HCC
|
hepatocellular carcinoma
|
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yan T, Zhao J, Bi X, Zhao H, Huang Z, Li
ZY, Zhou JG, Li Y, Li C, Cai JQ and Zhao P: Prognosis of
hepatocenular carcinoma: A study of 832 cases. Zhong Hua Zhong Liu
Za Zhi. 35:54–58. 2013.(In Chinese).
|
|
4
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sherman M: Recurrence of hepatocellular
carcinoma. N Engl J Med. 359:2045–2047. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu M, Jiang L and Guan XY: The genetic
and epigenetic alterations in human hepatocellular carcinoma: A
recent update. Protein Cell. 5:673–691. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kishi Y, Hasegawa K, Sugawara Y and Kokudo
N: Hepatocellular carcinoma: Current management and future
development-improved outcomes with surgical resection. Int J
Hepatol. 2011:7281032011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shah SA, Cleary SP, Wei AC, Yang I, Taylor
BR, Hemming AW, Langer B, Grant DR, Greig PD and Gallinger S:
Recurrence after liver resection for hepatocellular carcinoma: Risk
factors, treatment, and outcomes. Surgery. 141:330–339. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Roessler S, Jia HL, Budhu A, Forgues M, Ye
QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX and Wang XW: A
unique metastasis gene signature enables prediction of tumor
relapse in early-stage hepatocellular carcinoma patients. Cancer
Res. 70:10202–10212. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fang Y, Xue JL, Shen Q, Chen J and Tian L:
MicroRNA-7 inhibits tumor growth and metastasis by targeting the
phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma.
Hepatology. 55:1852–1862. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mizuguchi Y, Mishima T, Yokomuro S, Arima
Y, Kawahigashi Y, Shigehara K, Kanda T, Yoshida H, Uchida E, Tajiri
T and Takizawa T: Sequencing and bioinformatics-based analyses of
the microRNA transcriptome in hepatitis B-related hepatocellular
carcinoma. PLoS One. 6:e153042011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang P, Ma Y, Wang F, Yang J, Liu Z, Peng
J and Qin H: Comprehensive gene and microRNA expression profiling
reveals the crucial role of hsa-let-7i and its target genes in
colorectal cancer metastasis. Mol Biol Rep. 39:1471–1478. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stewart SE, Yu D, Scharf JM, Neale BM,
Fagerness JA, Mathews CA, Arnold PD, Evans PD, Gamazon ER, Davis
LK, et al: Genome-wide association study of obsessive-compulsive
disorder. Mol Psychiatry. 18:788–798. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schonrock N, Humphreys DT, Preiss T and
Götz J: Target gene repression mediated by miRNAs miR-181c and
miR-9 both of which are down-regulated by amyloid-β. J Mol
Neurosci. 46:324–335. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu C, Xu B, Yuan P, Ott J, Guan Y, Liu Y,
Liu Z, Shen Y, Yu D and Lin D: Genome-wide examination of genetic
variants associated with response to platinum-based chemotherapy in
patients with small-cell lung cancer. Pharmacogenet Genomics.
20:389–395. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Damgaard T, Knudsen LM, Dahl IM, Gimsing
P, Lodahl M and Rasmussen T: Regulation of the CD56 promoter and
its association with proliferation, anti-apoptosis and clinical
factors in multiple myeloma. Leuk Lymphoma. 50:236–246. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sud A, Del Bono EA, Haines JL and Wiggs
JL: Fine mapping of the GLC1K juvenile primary open-angle glaucoma
locus and exclusion of candidate genes. Mol Vis. 14:1319–1326.
2008.PubMed/NCBI
|
|
19
|
Matsui A, Tran M, Yoshida AC, Kikuchi
SSUM, Ogawa M and Shimogori T: BTBD3 controls dendrite orientation
toward active axons in mammalian neocortex. Science. 342:1114–1118.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Q, Zhao Z, Shang J and Xia W: Targets
and candidate agents for type 2 diabetes treatment with
computational bioinformatics approach. J Diabetes Res.
2014:7639362014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen Y, Lin MC, Yao H, Wang H, Zhang AQ,
Yu J, Hui CK, Lau GK, He ML, Sung J and Kung HF:
Lentivirus-mediated RNA interference targeting enhancer of zeste
homolog 2 inhibits hepatocellular carcinoma growth through
down-regulation of stathmin. Hepatology. 46:200–208. 2007.
View Article : Google Scholar : PubMed/NCBI
|