Introduction
Cancer cell migration and invasion account for the
majority of cancer-associated mortalities (1). Increased motility of cancer cells
underlies the processes of migration and invasion (2,3) and is an
essential step in breast cancer metastasis (4). Targeting tumor cell motility is a
potential antitumor strategy (5,6). In
response to migratory and chemotactic stimuli, cancer cells form
membrane protrusions, which initiate the multi-step migration
process. Membrane protrusions result from localized polymerization
of sub-membrane actin and consequent formation of actin filaments
and the actin framework is widely accepted as the engine driving
cell motility; several actin-binding proteins regulate the assembly
and disassembly of actin filaments, and thus the dynamic behavior
of the actin cytoskeleton (7–9). Of these, the ubiquitous protein cofilin
is the most important effector of actin polymerization and
depolymerization, generating free barbed ends via pointed-end
depolymerization and filament severing (10,11).
The actin-depolymerizing factor (ADF)/cofilin family
includes ADF, cofilin and other proteins with similar biochemical
activities. Unicellular organisms such as yeasts usually express
only one ADF/cofilin isoform, whereas multicellular organisms
typically express several. In certain cultured mammalian cell lines
and invasive mammary tumor cells, cofilin-1 is the most abundant
isoform (12), whereas ADF is
expressed at much lower levels. In the present study, cofilin
refers to cofilin-1.
Previous studies have suggested that cofilin
activity correlates with cancer progression and cancer cell
migration and invasion; local activation of cofilin via uncaging
induces lamellipodia formation and determines the direction of cell
movement (13). siRNA-mediated
depletion of cofilin in carcinoma cells inhibits cell motility
(12) and the assembly and stability
of invadopodia and, consequently, cell invasion (14). Cofilin overexpression increases the
rate of cell migration in human glioblastoma cultures (15) and pancreatic cancer (16), and correlates with poor prognosis in
human pulmonary adenocarcinoma, gastric cancer, epithelial ovarian
cancer and gallbladder carcinoma (17–20).
Spontaneous overexpression of cofilin has been detected in invasive
subpopulations of mammary tumor cells (21), and is directly associated with the
invasion, intravasation and metastasis of mammary tumors (22). Tissue microarray analysis has
demonstrated that cofilin staining positively correlates with
breast tumor grade (23).
However, to the best of our knowledge there is no
direct evidence implicating deregulated cofilin expression in
breast cancer prognosis at present. The present study analyzed
cofilin expression in tissue microarrays of tumors from 310
patients with breast cancer via immunohistochemistry (IHC). These
data provide insight into the role of cofilin in invasive breast
cancer and establish correlations between cofilin expression and
clinical and pathological parameters.
Materials and methods
Patient material and immunostaining in
breast cancer tissue microarrays
Tissue arrays containing samples of invasive breast
tumors from 310 patients were purchased from the National
Engineering Center for BioChips in Shanghai, China. To prepare the
arrays, a 1.5 mm core of tumor tissue was removed from each tumor.
Tumors were formalin-fixed for at least 24 h and paraffin-embedded.
Cores were taken from the peripheral aspect of the tumor, and
necrotic tissue was avoided.
The expression of cofilin, estrogen receptor (ER),
progesterone receptor (PR), Ki-67, and human epidermal growth
factor receptor 2 (Her2) was determined in the arrays via
IHC, using the BenchMark ULTRA system (Ventana Medical Systems,
Inc., Tucson, AZ, USA) and Leica BOND-MAX system (Leica
Microsystems, Ltd., Milton Keynes, UK) according to the
manufacturer's protocol. Normal goat serum (10%; Boster Biological
Technology, Ltd., Wuhan, China) was used as blocking reagent, and
samples were blocked for 20 min at room temperature. UltraView
Universal HRP multimer in the DAB Detection Kit (cat. no. 760-500;
Ventana Medical Systems, Inc.) was used as the secondary antibody
at a ready-to-use dilution and incubated for 30 min at 37°C. For
cofilin expression, the cofilin-specific antibody from Abcam (cat.
no. ab42824; Cambridge, UK) was used at a 1:1,500 dilution and the
incubation time was 8 min at room temperature, while all other
primary antibodies required 20 min at 37°C. ER and PR were
demonstrated using SP1 (cat. no. 790-4325) and 1E2 (cat. no.
790-4296; both from Ventana Medical Systems, Inc.) antibodies,
respectively at a ready-to-use dilution according to the protocol
of the manufacturer. Negative expression was defined as <10%
positive nuclei (24). Ki-67 was
demonstrated using MM1 (cat. no. PA0410; Novocastra; Leica
Microsystems, Ltd.) at a ready-to-use dilution according to the
protocol of the manufacturer, and the expression was considered
positive (>14% immunostained nuclei) or negative (≤14%
immunostained nuclei). Her2 expression was assessed
semiquantitatively by using a standard protocol (HercepTest; Dako;
Agilent Technologies, Inc., Santa Clara, CA, USA) (25) and separated into 4 grades (from 0 to
3+).
Fluorescence in situ hybridization (FISH)
analysis was performed in Her2 2+ samples, using the PathVysion
HER-2 DNA Probe kit (Abbott Pharmaceutical Co., Ltd., Lake Bluff,
IL, USA) according to the manufacturer's protocol. Her2 expression
was designated as weak (IHC grade 0–1+ or FISH−) or
strong (IHC grade 3+ or FISH+). Lymph node metastasis
was staged according to the American Joint Committee on Cancer TNM
system (26). Ethical approval for
the present study was granted by the Human Research Ethics
Committee of the Taizhou Hospital of Zhejiang Province.
Scoring, evaluation and statistical
analysis
IHC staining was evaluated by two experienced
pathologists blinded to the clinical information. Cofilin staining
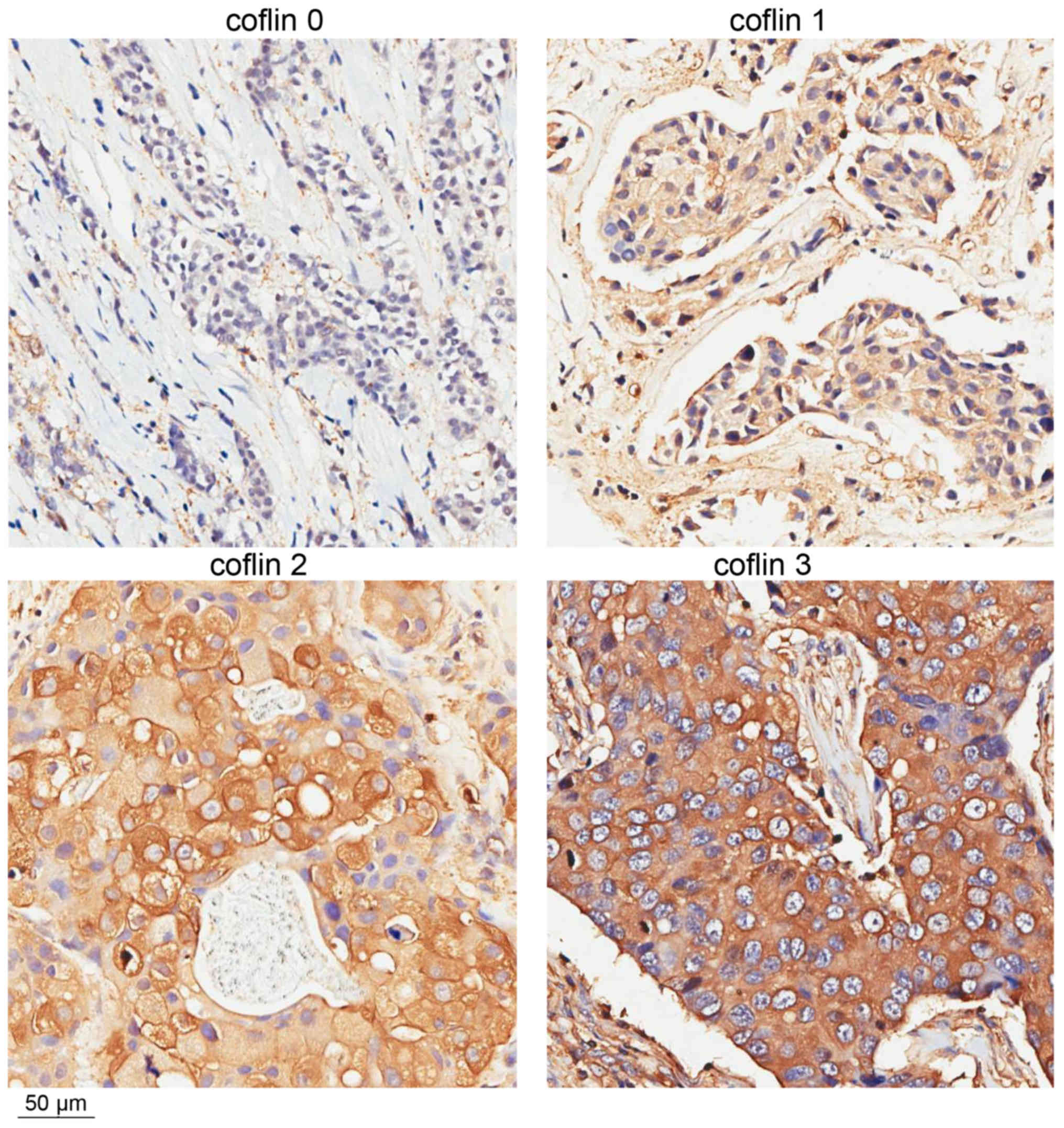
intensity in the cytoplasm of tumor cells was graded 0–3. The
percentage of cofilin-positive cells was scored 0–4 (0–5, 6–25,
26–50, 51–75 and 76–100%, respectively). The final cofilin
expression score ranged between 0 and 3 and was based on sum of the
intensity and percent positive scores (0–1, 2–3, 4–5 and 6–7,
respectively; Fig. 1). The slides
were scanned using an Aperio ScanScope slide scanner, and images of
representative areas were captured using Image Scope software
version 9.0 (Aperio Technologies, Ltd, Oxford, UK) followed by
Adobe Illustrator version 16.0 (Adobe Systems, San Jose, CA, USA)
(27).
Distributions of pathological and clinical
parameters [age, tumor size, Nottingham histological grade
(NHG)](28), and lymph node, ER, PR,
Her2, and Ki-67 status) according to the final cofilin score were
calculated using a one-way ANOVA or the Pearson χ2 test,
as indicated in Table I. Multiple
comparisons between the groups was performed using the
Student-Newman-Keuls method. Kaplan-Meier analysis and the Breslow
test were used to estimate the effect of high cofilin expression on
overall survival. For the Kaplan-Meier analysis, final cofilin
scores were analyzed in terms of weak (scores of 0, 1, and 2) and
strong (a score of 3) expression. Cox regression proportional
hazards models were used to estimate hazard ratios (HRs) for
mortality from breast cancer according to cofilin expression in
univariate and multivariate analyses. The covariates with P<0.05
in the univariate analysis (lymph node, ER, and PR status) were
included in the multivariate analysis. All statistical tests were
two-sided, and P<0.05 were considered to indicate a
statistically significant difference. All calculations were
performed using SPSS Statistics version 19 software (IBM Corp.,
Armonk, NY, USA).
 | Table I.Associations between cofilin
expression and clinicopathological features in breast cancer. |
Table I.
Associations between cofilin
expression and clinicopathological features in breast cancer.
|
|
| Cofilin staining
intensity |
|
|---|
|
|
|
|
|
|---|
| Factor | Number | 0 | 1 | 2 | 3 | P-value |
|---|
| All, n (%) | 310 | 53 (17) | 114 (37) | 92 (30) | 51 (16) |
|
| Age,
yearsa | 54 (29–88) | 50.5 (29–83) | 54 (31–88) | 57 (31–87) | 56 (37–88) | 0.055b |
| Tumor size,
mma | 30 (10–150) | 30 (10–100) | 30 (14–130) | 30 (10–150) | 35 (10–100) | 0.294b |
| NHG, n (%) |
|
|
|
|
| 0.030d |
| I | 19 (6) | 6 (32) | 8 (42) | 5 (26) | 0 (0) |
|
| II | 210 (68) | 36 (17) | 83 (40) | 61 (29) | 30 (14) |
|
|
III | 70 (22) | 10 (14) | 21 (30) | 19 (27) | 20 (29) |
|
|
Missing | 11 (4) |
|
|
|
|
|
| Nodal status, n
(%) |
|
|
|
|
| 0.082c |
| N0 | 141 (45) | 21 (15) | 48 (34) | 49 (34) | 23 (16) |
|
| N1 | 86 (28) | 14 (16) | 33 (38) | 21 (24) | 18 (21) |
|
| N2 | 56 (18) | 11 (20) | 26 (46) | 13 (23) | 6 (11) |
|
| N3 | 21 (7) | 7 (33) | 2 (10) | 8 (38) | 4 (19) |
|
|
Missing | 6 (2) |
|
|
|
|
|
| ER status, n
(%) |
|
|
|
|
| 0.084c |
|
Positive | 191 (62) | 26 (14) | 76 (40) | 62 (32) | 27 (14) |
|
|
Negative | 114 (37) | 24 (21) | 36 (32) | 30 (26) | 24 (21) |
|
|
Missing | 5 (2) |
|
|
|
|
|
| PR status, n
(%) |
|
|
|
|
| 0.176c |
|
Positive | 139 (45) | 21 (15) | 56 (40) | 45 (32) | 17 (12) |
|
|
Negative | 168 (54) | 31 (18) | 56 (33) | 47 (28) | 34 (20) |
|
|
Missing | 3 (1) |
|
|
|
|
|
| Ki67 status, n
(%) |
|
|
|
|
| 0.001c |
|
>14% | 99 (32) | 11 (11) | 29 (29) | 32 (32) | 27 (27) |
|
|
≤14% | 211 (68) | 42 (20) | 85 (40) | 60 (28) | 24 (13) |
|
|
Missing | 0 (0) |
|
|
|
|
|
| HER2 status, n
(%)e |
|
|
|
|
| 0.000d |
|
Strong | 77 (25) | 2 (2) | 29 (38) | 28 (36) | 18 (23) |
|
|
Weak | 233 (75) | 51 (22) | 85 (36) | 64 (27) | 33 (14) |
|
|
Missing | 0 (0) |
|
|
|
|
|
Results
Cofilin expression is associated with
clinicopathological variables
The association between cofilin staining intensity
and several clinical parameters [age, tumor size, NHG, lymph node
metastasis, and ER, PR, Ki-67, and Her2 expression] was determined
(Table I). There was a significant
association between cofilin staining intensity and NHG (P=0.030). A
trend toward a higher NHG in tumors with higher cofilin scores was
observed. No tumors exhibited a cofilin score of 3 in combination
with the lowest NHG score.
Cofilin expression was also associated with Her2
expression (P<0.001). The distribution of Her2-positive tumors
paralleled the distribution of cofilin scores: The majority of
Her2-positive tumors exhibited cofilin scores of 2 or 3. A similar
association between Ki-67 expression and cofilin expression was
observed (P=0.001). As Ki67 positive tumors exhibited a larger
percentage of cells with high cofilin expression compared with low
cofilin expression, it was hypothesized that positive Ki67 status
was associated with high cofilin expression. Cofilin expression was
not significantly associated with age, tumor size, lymph node
metastasis or ER or PR expression (P=0.055, 0.294, 0.082, 0.084 and
0.176, respectively).
High expression of cofilin is
associated with poor survival
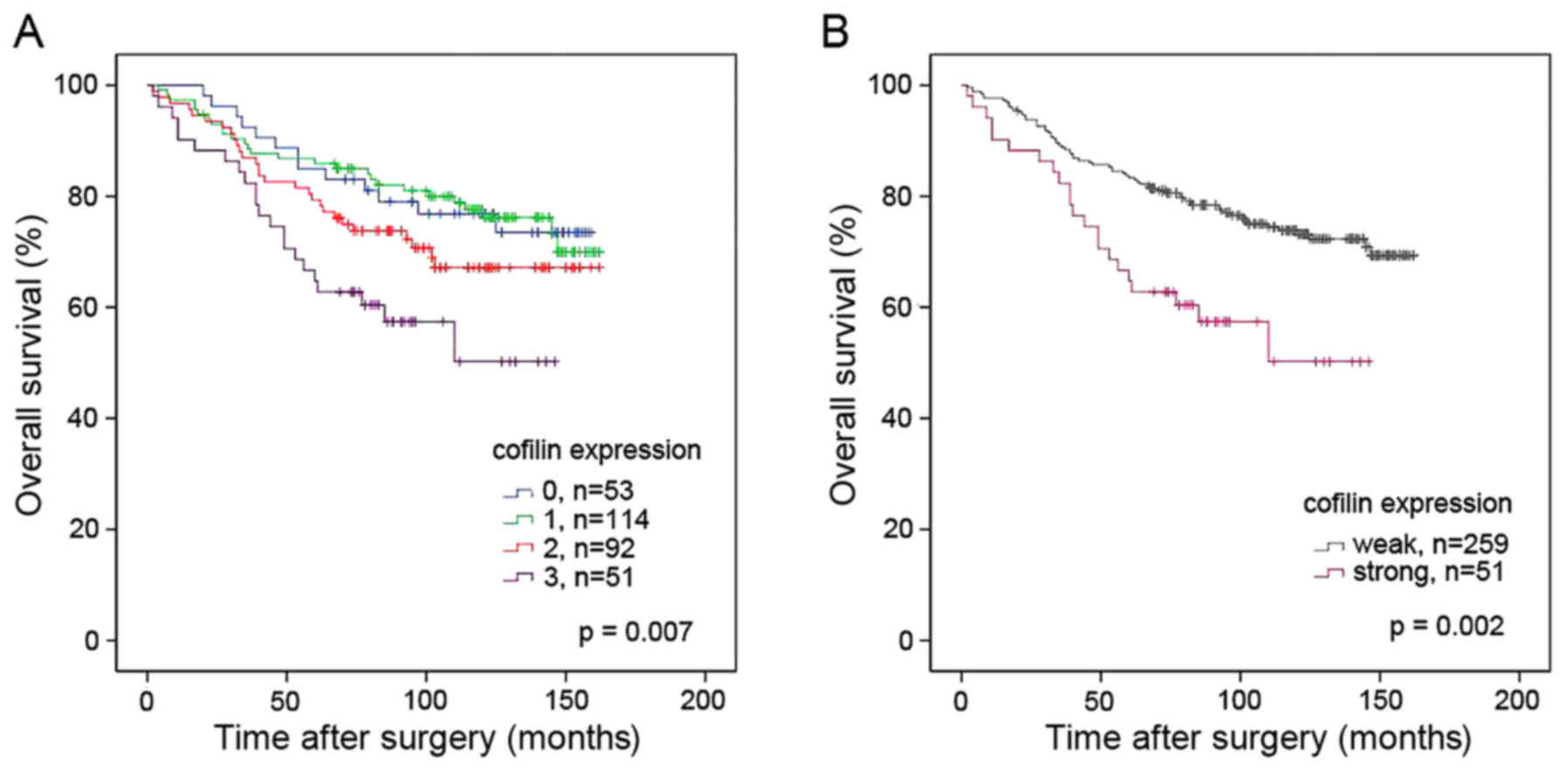
For survival analysis, the cofilin scores were
dichotomized: Scores of 0, 1 and 2 denoted weak expression, and a
score of 3 denoted strong expression. The rationale for this
grouping was the marked difference in cofilin staining intensity
between scores 2 and 3 (Fig. 1) and
the similarity of the survival curves for patients with scores of
0, 1, or 2 (Fig. 2). Kaplan-Meier
analysis demonstrated significant differences in overall survival
between patients bearing tumors with weak vs. strong cofilin
expression (P=0.002; Fig. 2).
Univariate and multivariate Cox regression analyses of survival in
association with cofilin expression were performed using the same
dichotomized variable as in the Kaplan-Meier analysis. The results
demonstrated that strong cofilin expression was an independent
indicator of reduced overall survival (P=0.002; HR, 2.22; 95%
confidence interval, 1.35–3.66) when the variables described in
Table II were included. Detailed
results of the Cox analyses are presented in Table II.
 | Table II.Univariate and multivariate COX
regression analyses of the effects on overall survival for
different patients and characteristics. |
Table II.
Univariate and multivariate COX
regression analyses of the effects on overall survival for
different patients and characteristics.
|
| Univariate |
|---|
|
|
|
|---|
| A, Variable | n | HR | 95% CI | P-value |
|---|
| Cofilin (strong vs.
weak)a | 310 | 2.15 | 1.32–3.49 | 0.002 |
| ER (positive vs.
negative) | 305 | 0.63 | 0.41–0.96 | 0.030 |
| PR (positive vs.
negative) | 307 | 0.59 | 0.39–0.92 | 0.019 |
| Her2 (strong vs.
weak) | 310 | 1.37 | 0.87–2.16 | 0.177 |
| Ki67 (positive vs.
negative) | 310 | 1.22 | 0.79–1.88 | 0.374 |
| Size (>20 mm vs.
≤20 mm) | 304 | 1.45 | 0.85–2.45 | 0.173 |
| Nottingham
histological grade (1–3) | 299 | 1.27 | 0.83–1.94 | 0.269 |
| Nodal status (N3,
N2, N1, N0) | 304 | 1.39 | 1.12–1.71 | 0.002 |
|
|
| Multivariate |
|
|
|
| B, Variable | n | HR | 95% CI | P-value |
|
| Cofilin (strong vs.
weak)a | 299 | 2.22 | 1.35–3.66 | 0.002 |
| ER (positive vs.
negative) | 299 | 0.80 | 0.47–1.36 | 0.400 |
| PR (positive vs.
negative) | 299 | 0.75 | 0.43–1.32 | 0.323 |
| Nodal status (N3,
N2, N1, N0) | 299 | 1.40 | 1.13–1.73 | 0.002 |
Discussion
Actin is the major component of the cytoskeleton,
which serves an important role in tumor cell migration, invasion
and mitosis. The actin-binding protein cofilin, a member of the
ADF/cofilin family, is a key regulator of actin polymerization and
depolymerization. The activity and output of the cofilin pathway
(cofilin and its regulatory proteins) are increased in cancer cells
(4,29,30).
Cofilin is thought to contribute to at least 3 cancer-associated
events: Initial cell transformation (31), increased cell motility during
metastasis and cell division (32).
Previous studies have demonstrated that tumors with
a higher NHG typically exhibited reduced tubule formation, nuclear
atypia and mitoses, and Her2 expression has been associated with
tumor cell proliferation and an aggressive phenotype (33–35). In
the present study, cofilin staining was associated with NHG, Her2
expression and Ki-67 expression, suggesting that cofilin may be a
marker of poor differentiation and high proliferation. In migrating
or invading cells, cofilin resides in cell membrane protrusions,
for example lamellipodia, invadopodia, and filopodia, which
initiate cell movement and determine cell polarity (36,37). This
localization is critical for cell movement, endocytosis and cell
division, all of which are important for normal cell proliferation,
differentiation and cancer development (38). This promotion may be responsible for
the positive association between cofilin expression and NHG, Her2
and Ki-67 status.
In agreement with previous studies, the present
study identified that cofilin expression did not correlate with ER
or PR status (23). There was also no
correlation observed between cofilin expression and tumor size. In
contrast, another study demonstrated a positive association between
cofilin expression and tumor stages T0, T1, and T2 (but not T3) in
breast cancer (39). Resolution of
this discrepancy requires additional study.
Owing to its effects on actin
polymerization/depolymerization, cofilin overexpression has been
associated with mammary tumor invasion, intravasation, metastasis,
lymph node metastasis and a higher nodal stage. Studies on other
human malignant tumor types support these associations (20,40,41).
However, in the present study, cofilin expression did not correlate
with the nodal stage. The present study demonstrated that cofilin
expression and the nodal stage are independent prognosis factors in
breast cancer. As the number of positive lymph nodes largely
depends on the completeness of axillary lymph node dissection, its
approximation may not always be accurate (42). Additionally, the time interval between
tumor diagnosis and surgery may affect the nodal stage (43). Consequently, the nodal stage may not
reflect a tendency for lymphatic metastasis. These considerations
may explain why cofilin expression does not necessarily correlate
with the clinical nodal stage. Active cofilin comprises only part
of the total level of cofilin in the cytoplasm. The present study
measured total cofilin abundance instead of cofilin activity, which
is difficult to estimate. Therefore, intensive studies are needed
to determine whether cofilin expression or activity is associated
with nodal metastasis in human breast cancer.
Notably, high cofilin expression was significantly
associated with shorter overall survival. This association remained
significant when other clinicopathological factors were included in
the COX regression analysis, suggesting that cofilin is a potential
independent prognostic factor in breast cancer.
In summary, the results of the present study suggest
that cofilin may promote the occurrence and development of breast
cancer, perhaps via its contribution to cell migration, invasion
and/or mitosis. How it does so is beyond the scope of the present
study, and requires additional study. The present study suggests
that cofilin is a potential independent prognostic factor in breast
cancer, and raises the possibility of targeting cofilin for more
effective treatment of breast cancer.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81001171) and the
Key Technologies R&D Program of Hubei Province (grant no.
2007AA302B07). The authors would like to thank Editage (https://www.editage.com/) for English language
editing.
Glossary
Abbreviations
Abbreviations:
|
ADF
|
actin-depolymerizing factor
|
|
ER
|
estrogen receptor
|
|
FISH
|
fluorescence in situ
hybridization
|
|
Her2
|
human epidermal growth factor receptor
2
|
|
HR
|
hazard ratio
|
|
IHC
|
immunohistochemistry
|
|
NHG
|
Nottingham histological grade
|
|
PR
|
progesterone receptor
|
References
|
1
|
Guan X: Cancer metastases: Challenges and
opportunities. Acta Pharm Sin B. 5:402–418. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Roussos ET, Condeelis JS and Patsialou A:
Chemotaxis in cancer. Nat Rev Cancer. 11:573–587. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sidani M, Wessels D, Mouneimne G, Ghosh M,
Goswami S, Sarmiento C, Wang W, Kuhl S, El-Sibai M, Backer JM, et
al: Cofilin determines the migration behavior and turning frequency
of metastatic cancer cells. J Cell Biol. 179:777–791. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang W, Eddy R and Condeelis J: The
cofilin pathway in breast cancer invasion and metastasis. Nat Rev
Cancer. 7:429–440. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Moretti RM, Marelli M Montagnani, Mai S
and Limonta P: Gonadotropin-releasing hormone agonists suppress
melanoma cell motility and invasiveness through the inhibition of
α3 integrin and MMP-2 expression and activity. Int J Oncol.
33:405–413. 2008.PubMed/NCBI
|
|
6
|
Limame R, de Beeck KO, Van Laere S, Croes
L, De Wilde A, Dirix L, Van Camp G, Peeters M, De Wever O, Lardon F
and Pauwels P: Expression profiling of migrated and invaded breast
cancer cells predicts early metastatic relapse and reveals
Krüppel-like factor 9 as a potential suppressor of invasive growth
in breast cancer. Oncoscience. 1:69–81. 2013.PubMed/NCBI
|
|
7
|
Carlier MF, Ressad F and Pantaloni D:
Control of actin dynamics in cell motility. Role of ADF/cofilin. J
Biol Chem. 274:33827–33830. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Achard V, Martiel JL, Michelot A, Guérin
C, Reymann AC, Blanchoin L and Boujemaa-Paterski R: A
‘primer’-based mechanism underlies branched actin filament network
formation and motility. Curr Biol. 20:423–428. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bugyi B and Carlier MF: Control of actin
filament treadmilling in cell motility. Annu Rev Biophys.
39:449–470. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bernstein BW and Bamburg JR: ADF/cofilin:
A functional node in cell biology. Trends Cell Biol. 20:187–195.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bravo-Cordero JJ, Magalhaes MA, Eddy RJ,
Hodgson L and Condeelis J: Functions of cofilin in cell locomotion
and invasion. Nat Rev Mol Cell Biol. 14:405–415. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hotulainen P, Paunola E, Vartiainen MK and
Lappalainen P: Actin-depolymerizing factor and cofilin-1 play
overlapping roles in promoting rapid F-actin depolymerization in
mammalian nonmuscle cells. Mol Biol Cell. 16:649–664. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ghosh M, Song X, Mouneimne G, Sidani M,
Lawrence DS and Condeelis JS: Cofilin promotes actin polymerization
and defines the direction of cell motility. Science. 304:743–746.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamaguchi H, Lorenz M, Kempiak S,
Sarmiento C, Coniglio S, Symons M, Segall J, Eddy R, Miki H,
Takenawa T and Condeelis J: Molecular mechanisms of invadopodium
formation: The role of the N-WASP-Arp2/3 complex pathway and
cofilin. J Cell Biol. 168:441–452. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yap CT, Simpson TI, Pratt T, Price DJ and
Maciver SK: The motility of glioblastoma tumour cells is modulated
by intracellular cofilin expression in a concentration-dependent
manner. Cell Motil Cytoskeleton. 60:153–165. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Y, Kuramitsu Y, Kitagawa T, Baron B,
Yoshino S, Maehara S, Maehara Y, Oka M and Nakamura K:
Cofilin-phosphatase slingshot-1L (SSH1L) is over-expressed in
pancreatic cancer (PC) and contributes to tumor cell migration.
Cancer Lett. 360:171–176. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Peng XC, Gong FM, Zhao YW, Zhou LX, Xie
YW, Liao HL, Lin HJ, Li ZY, Tang MH and Tong AP: Comparative
proteomic approach identifies PKM2 and cofilin-1 as potential
diagnostic, prognostic and therapeutic targets for pulmonary
adenocarcinoma. PLoS One. 6:e273092011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li D, Zhang Y, Li Z, Wang X, Qu X and Liu
Y: Activated Pak4 expression correlates with poor prognosis in
human gastric cancer patients. Tumour Biol. 36:9431–9436. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nishimura S, Tsuda H, Kataoka F, Arao T,
Nomura H, Chiyoda T, Susumu N, Nishio K and Aoki D: Overexpression
of cofilin 1 can predict progression-free survival in patients with
epithelial ovarian cancer receiving standard therapy. Hum Pathol.
42:516–521. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang ZL, Miao X, Xiong L, Zou Q, Yuan Y,
Li J, Liang L, Chen M and Chen S: CFL1 and Arp3 are biomarkers for
metastasis and poor prognosis of squamous cell/adenosquamous
carcinomas and adenocarcinomas of gallbladder. Cancer Invest.
31:132–139. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang W, Goswami S, Lapidus K, Wells AL,
Wyckoff JB, Sahai E, Singer RH, Segall JE and Condeelis JS:
Identification and testing of a gene expression signature of
invasive carcinoma cells within primary mammary tumors. Cancer Res.
64:8585–8594. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang W, Mouneimne G, Sidani M, Wyckoff J,
Chen X, Makris A, Goswami S, Bresnick AR and Condeelis JS: The
activity status of cofilin is directly related to invasion,
intravasation, and metastasis of mammary tumors. J Cell Biol.
173:395–404. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shaheed SU, Rustogi N, Scally A, Wilson J,
Thygesen H, Loizidou MA, Hadjisavvas A, Hanby A, Speirs V, Loadman
P, et al: Identification of stage-specific breast markers using
quantitative proteomics. J Proteome Res. 12:5696–5708. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Falck AK, Bendahl PO, Chebil G, Olsson H,
Fernö M and Rydén L: Biomarker expression and St Gallen molecular
subtype classification in primary tumours, synchronous lymph node
metastases and asynchronous relapses in primary breast cancer
patients with 10 years' follow-up. Breast Cancer Res Treat.
140:93–104. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rydén L, Jirström K, Bendahl PO, Fernö M,
Nordenskjöld B, Stål O, Thorstenson S, Jönsson PE and Landberg G:
Tumor-specific expression of vascular endothelial growth factor
receptor 2 but not vascular endothelial growth factor or human
epidermal growth factor receptor 2 is associated with impaired
response to adjuvant tamoxifen in premenopausal breast cancer. J
Clin Oncol. 23:4695–4704. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Edge S, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC Cancer Staging Handbook. 7th.
Springer; New York, NY: 2010
|
|
27
|
Krishnamurthy S, Mathews K, McClure S,
Murray M, Gilcrease M, Albarracin C, Spinosa J, Chang B, Ho J, Holt
J, et al: Multi-institutional comparison of whole slide digital
imaging and optical microscopy for interpretation of
hematoxylin-eosin-stained breast tissue sections. Arch Pathol Lab
Med. 137:1733–1739. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rakha EA, Reis-Filho JS, Baehner F, Dabbs
DJ, Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani
SR, et al: Breast cancer prognostic classification in the molecular
era: The role of histological grade. Breast Cancer Res. 12:2072010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ono S: Mechanism of depolymerization and
severing of actin filaments and its significance in cytoskeletal
dynamics. Int Rev Cytol. 258:1–82. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
van Rheenen J, Song X, van Roosmalen W,
Cammer M, Chen X, Desmarais V, Yip SC, Backer JM, Eddy RJ and
Condeelis JS: EGF-induced PIP2 hydrolysis releases and activates
cofilin locally in carcinoma cells. J Cell Biol. 179:1247–1259.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Garg P, Verma R, Cook L, Soofi A,
Venkatareddy M, George B, Mizuno K, Gurniak C, Witke W and Holzman
LB: Actin-depolymerizing factor cofilin-1 is necessary in
maintaining mature podocyte architecture. J Biol Chem.
285:22676–22688. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tammana TV, Sahasrabuddhe AA, Bajpai VK
and Gupta CM: ADF/cofilin-driven actin dynamics in early events of
Leishmania cell division. J Cell Sci. 123:1894–1901. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ellsworth RE, Hooke JA, Love B, Ellsworth
DL and Shriver CD: Molecular changes in primary breast tumors and
the nottingham histologic score. Pathol Oncol Res. 15:541–547.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Niikura N, Iwamoto T, Masuda S, Kumaki N,
Xiaoyan T, Shirane M, Mori K, Tsuda B, Okamura T, Saito Y, et al:
Immuno-histochemical Ki67 labeling index has similar proliferation
predictive power to various gene signatures in breast cancer.
Cancer Sci. 103:1508–1512. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Volpi A, Nanni O, De Paola F, Granato AM,
Mangia A, Monti F, Schittulli F, De Lena M, Scarpi E, Rosetti P, et
al: HER-2 expression and cell proliferation: Prognostic markers in
patients with node-negative breast cancer. J Clin Oncol.
21:2708–2712. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mouneimne G, DesMarais V, Sidani M, Scemes
E, Wang W, Song X, Eddy R and Condeelis J: Spatial and temporal
control of cofilin activity is required for directional sensing
during chemotaxis. Curr Biol. 16:2193–2205. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Oser M, Yamaguchi H, Mader CC,
Bravo-Cordero JJ, Arias M, Chen X, Desmarais V, van Rheenen J,
Koleske AJ and Condeelis J: Cortactin regulates cofilin and N-WASp
activities to control the stages of invadopodium assembly and
maturation. J Cell Biol. 186:571–587. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Huang X, Sun D, Pan Q, Wen W, Chen Y, Xin
X, Huang M, Ding J and Geng M: JG6, a novel marine-derived
oligosaccharide, suppresses breast cancer metastasis via binding to
cofilin. Oncotarget. 5:3568–3578. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang Y and Tong X: Expression of the
actin-binding proteins indicates that cofilin and fascin are
related to breast tumour size. J Int Med Res. 38:1042–1048. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Polachini GM, Sobral LM, Mercante AM,
Paes-Leme AF, Xavier FC, Henrique T, Guimarães DM, Vidotto A,
Fukuyama EE, Góis-Filho JF, et al: Proteomic approaches identify
members of cofilin pathway involved in oral tumorigenesis. PLoS
One. 7:e505172012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lu LI, Fu NI, Luo XU, Li XY and Li XP:
Overexpression of cofilin 1 in prostate cancer and the
corresponding clinical implications. Oncol Lett. 9:2757–2761.
2015.PubMed/NCBI
|
|
42
|
Naumann DN and Sintler M: The surgeon as
the most important factor in lymph node harvest during axillary
clearance. Anticancer Res. 33:3935–3939. 2013.PubMed/NCBI
|
|
43
|
Olivotto IA, Gomi A, Bancej C, Brisson J,
Tonita J, Kan L, Mah Z, Harrison M and Shumak R: Influence of delay
to diagnosis on prognostic indicators of screen-detected breast
carcinoma. Cancer. 94:2143–2150. 2002. View Article : Google Scholar : PubMed/NCBI
|