Introduction
Each year around 250,000 new cases of kidney cancer
(KC) are diagnosed in the world, accounting for about 2% of all
cancers (1). It is one of the highest
incidences of cancer in the urinary system tumors. The morbidity
and mortality of KC is continuously growing. The clear cell renal
cell carcinoma (ccRCC), the most common subtype of KC, is estimated
as 80% of all patients (2). The
comprehensive biological process of the molecular mechanism of
ccRCC may involve in stepwise accumulation of genomic instability,
epigenetic changes, gene mutations and deviant expression of
protein-coding genes and noncodings (3,4).
The genome-wide association researches have found
several mutations conferring risk of ccRCC, including von
Hippel-Lindau gene (VHL), which codes for a tumor suppressor
gene. Various causes, such as mutation, loss of heterozygosity,
allelic deletion and promoter hypermethylation, makes VHL
inactivation on chromosome 3p as a high frequent genetic event in
ccRCC (5). Carcinogenesis and
cancer-associated angiogenesis is well-characterized by the
alteration in the pathway of VHL gene/hypoxia inducible
factor/vascular endothelial growth factor (6). Moreover, some genes, such as
KDM5C [lysine (K)-specific demethylase 5C], PBRM1
(polybromo-1), BAP1 (BRCA1 associated protein-1),
KDM6A [lysine (K)-specific demethylase 6A] and SETD2
(SET domain containing 2), are involved in chromatin modification
in primary human renal cell carcinoma (RCC) and their mutations
will profoundly change their chromatin organization and their
transcriptional program (7).
On the other side, the non-coding RNA transcripts
also showed significant difference in ccRCC, such as overexpression
of miRNA-210 (8) and downregulation
of miR-141 and miR-200c (9).
miRNA-210 is one of the HIF-1 targets, reported to correlate with
poor patient survival (8).
Long non-coding RNAs (lncRNAs), mainly locating
within nucleus or cytosolic compartment (10), many identified of which are
transcribed by RNA polymerase II, belong to a new class of
non-coding RNA transcripts more than 200 nucleotides in length.
Based on their location regarding protein-coding genes, LncRNAs are
classified into five locus biotypes: Sense, antisense,
bidirectional, intronic and intergenic (11). lncRNAs can regulate gene expression by
the ways of transcriptional regulation, splicing, imprinting,
epigenetic regulation and subcellular transport (12). Therefore, lncRNAs are important in
gene expression and chromatin remodeling and exhibit previously
unrecognized functions, such as promotion of the occurrence,
development, proliferation and differentiation of the malignant
disease including cancers.
In this study, some lncRNAs were identified to be
differentially expressed in cancer while comparing with normal
tissue, providing some potential candidates for discovery of new
cancer biomarkers and therapeutic targets to improve diagnosis and
therapy in ccRCC.
Materials and methods
Source of data sets
Three data sets of human exon arrays, GSE53757,
GSE46699 and GSE47352, were downloaded from GEO. GSE53757 contained
72 paired ccRCC and normal adjacent tissues (13). Data processing and methodology were as
previously described (14). GSE46699
contained 65 paired ccRCC and normal adjacent tissues, including
stages 1–3 (15). GSE47352 contained
4 primary metastatic and 5 non-metastatic tumor samples (16).
Probe re-annotation pipeline
The following steps as described by Zhang et
al (17) were used to assess the
lncRNA expressions in the ccRCC gene expression data that a
pipeline uniquely mapped to lncRNAs was used to determine the probe
sets from the Affymetrix array. First of all, Affymetrix HG-U133
Plus 2.0 (Affymetrix, Santa Clara, CA, USA) probe set ID was mapped
to the NetAffx Annotation Files (HG-U133 Plus 2.0 Annotations, CSV
format, release 33, 10/30/12), which the Ensembl gene ID, probe set
ID, Refseq transcript ID gene symbol, gene symbol, gene title and
other informative items for the specific probe set were included in
the annotations. Then, we used the following criteria to filter the
probe sets: i) ‘NR’ (NR indicates non-coding RNA in the Refseq
database) was labeled and retained for the probe sets which were
only assigned with Refseq IDs; ii) ‘antisense’, ‘sense intronic’,
‘processed transcripts’, ‘sense overlapping’, ‘lncRNA’ or ‘non
sense mediated decay’ in Ensembl annotations were annotated and
retained for the probe sets which were only assigned with Ensembl
gene IDs; and iii) those annotated with Ensembl gene titles above
and labeled as ‘NR’ in Refseq database were retained at the same
time for the probe sets with both Ensembl gene IDs and Refseq IDs.
Secondly, rRNAs, pseudogenes, microRNAs and other short RNAs
including snRNAs, tRNAs and snoRNAs were filtered out and 2,935
Affymetrix probe sets with corresponding annotated lncRNA
transcripts were obtained.
Data analysis
After quality control assessment was done, each
dataset was normalized by applying a Bioconductor package Robust
multi-array average (RMA) through R 3.0.2. Then the linear models
for microarray data (LIMMA), a modified t-test method incorporating
the Benjamini-Hochberg multiple hypotheses correction technique,
were used to analyze the normalized data. The probe sets of which
the adjusted P-value was below 0.01 between two groups were defined
as significantly different lncRNAs.
Results
LncRNA expression profiles on
Affymetrix HG-U133 Plus 2.0 arrays
According to the Ensembl and Ref-seq annotations of
lncRNAs and the NetAffx annotation of probe sets, 2,935 probe sets
corresponding to lncRNA genes were identified on the Affymetrix
HG-U133 Plus 2.0 arrays. Among them, we annotated 1,002 probe sets
mapping to lncRNA genes by both the RefSeq and Ensemble database;
we annotated 500 probe sets only by the RefSeq database; we
annotated 1,423 probe sets only by the Ensemble database. Those
probe sets with controversial annotations were excluded in the two
databases.
LncRNAs differentially expressed in
ccRCC
We used R to process the CEL files for background
correction, normalization, and summarizations with RMA algorithm.
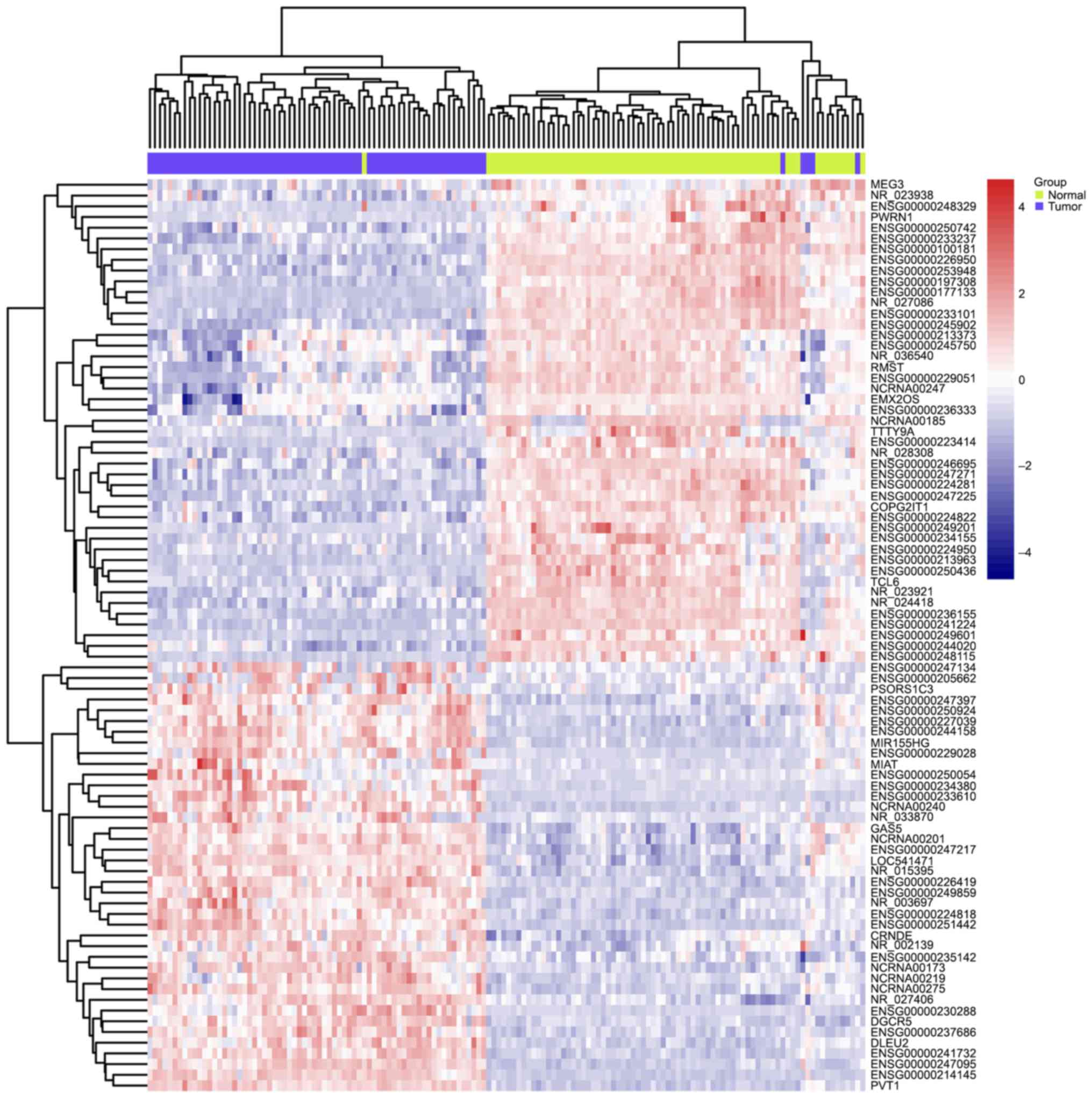
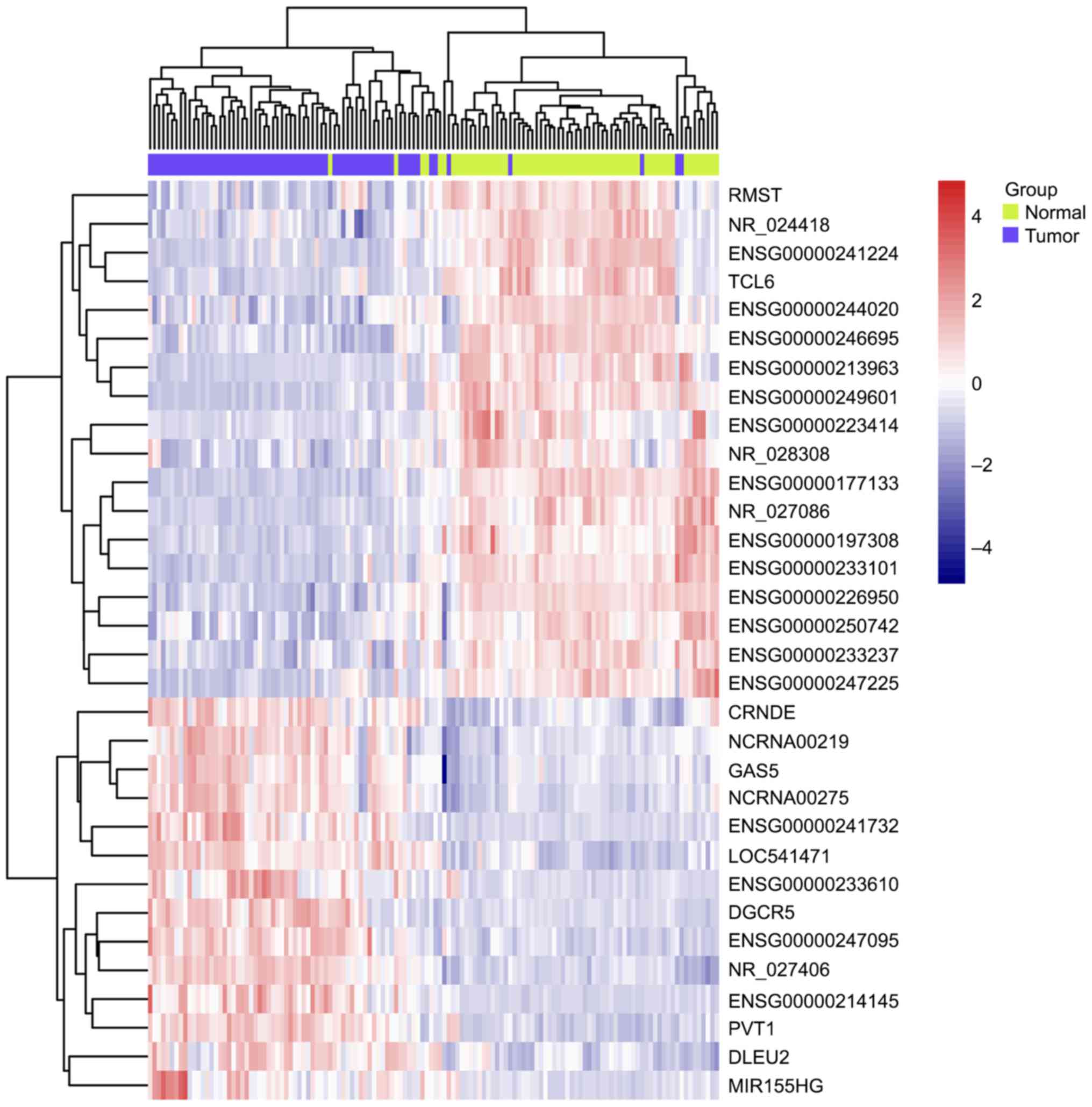
Comparing to normal tissue by using LIMMA at a threshold of an
adjusted P-value <0.01, 85 lncRNAs in GSE53737 (Fig. 1) and 32 lncRNAs in GSE46699 (Fig. 2) were identified to be differentially
expressed in ccRCC. Interestingly, the overlapping lncRNAs of both
arrays were exactly the differentially expressed lncRNAs in
GSE46699, listed in Table I. A total
of 18 downregulated and 14 upregulated lncRNAs was found among the
32 differentially expressed lncRNAs. Any official Human Genome
Nomenclature Committee symbols were not yet owned to most of these
lncRNAs and their functions are still unknown. Some lncRNAs, such
as ENSG00000177133 (18), NR_024418
(19), T-cell leukemia/lymphoma 6
(TCL6) (20,21), GAS5 (22–25), DLEU2
(26), colorectal neoplasia
differentially expressed (CRNDE) (27), and MIR155HG (28), have been reported to play a pro- or
anticancer role in various kind of cancers. Among them, NR_24418
and TCL6 have been reported to be associated with ccRCC (19,20)
(Table I).
 | Table I.Overlapping 32 long non-coding RNAs
differentially expressed in ccRCC in both GSE53757 and
GSE46699. |
Table I.
Overlapping 32 long non-coding RNAs
differentially expressed in ccRCC in both GSE53757 and
GSE46699.
|
| GSE53757 | GSE46699 |
|
|---|
|
|
|
|
|
|---|
| Symbol | logFC | adj.P.Val | logFC | adj.P.Val | Cancers link to
lncRNA |
|---|
|
ENSG00000214145 |
3.60594 | 2.69E-46 |
1.22867 | 1.99E-14 |
|
| PVT1 |
2.81551 | 8.93E-50 |
1.33005 | 3.33E-25 |
|
| MIR155HG |
2.78895 | 3.74E-31 |
1.03184 | 4.44E-08 | Lymphocytic
leukemia |
|
ENSG00000247095 |
2.47881 | 2.38E-45 |
1.05478 | 2.80E-17 |
|
| DGCR5 |
2.25397 | 2.79E-28 |
1.52029 | 5.50E-16 |
|
|
ENSG00000233610 |
2.21139 | 6.22E-25 |
1.07377 | 2.00E-10 |
|
|
ENSG00000241732 |
2.12166 | 6.49E-39 |
1.10549 | 1.88E-14 |
|
| LOC541471 |
2.02896 | 1.93E-33 |
1.62800 | 2.20E-20 |
|
| CRNDE |
1.55389 | 1.21E-18 |
1.45479 | 2.19E-12 | Colorectal adenomas
and adenocarcinomas |
| DLEU2 |
1.45483 | 3.76E-30 |
1.04097 | 4.62E-10 | Lymphocytic
leukemia |
| NCRNA00275 |
1.26093 | 7.08E-33 |
1.07722 | 7.33E-13 |
|
| NCRNA00219 |
1.22560 | 2.99E-20 |
1.14880 | 5.95E-13 |
|
| GAS5 |
1.09636 | 1.08E-25 |
1.02773 | 1.84E-12 | Kidney, cervical,
colorectal, hepatocellular, gastric, prostate, breast and bladder
cancer |
| NR_027406 |
1.01639 | 6.34E-24 |
1.22427 | 2.42E-23 |
|
|
ENSG00000197308 | −1.29187 | 3.71E-34 | −1.00448 | 4.44E-20 |
|
| NR_028308 | −1.43885 | 1.43E-15 | −1.29727 | 1.25E-12 |
|
|
ENSG00000250742 | −1.57523 | 7.05E-20 | −1.20294 | 6.30E-15 |
|
|
ENSG00000246695 | −1.62689 | 4.44E-32 | −1.35738 | 1.23E-21 |
|
|
ENSG00000226950 | −1.68462 | 1.53E-42 | −1.17068 | 7.12E-22 |
|
|
ENSG00000223414 | −1.75572 | 6.42E-23 | −1.24300 | 1.02E-13 |
|
|
ENSG00000233237 | −1.91752 | 1.23E-27 | −1.39309 | 1.25E-16 |
|
|
ENSG00000244020 | −1.99999 | 3.99E-27 | −1.46462 | 5.89E-14 |
|
| RMST | −2.15814 | 6.77E-26 | −1.17918 | 5.01E-14 |
|
| TCL6 | −2.20017 | 1.33E-32 | −1.19247 | 3.70E-15 | RCC, T-cell
leukemia |
|
ENSG00000213963 | −2.24474 | 1.47E-37 | −1.86640 | 1.69E-16 |
|
| NR_024418 | −2.50614 | 1.80E-37 | −1.28191 | 2.79E-15 | RCC |
|
ENSG00000247225 | −2.73504 | 6.39E-37 | −1.45770 | 4.41E-22 |
|
|
ENSG00000233101 | −2.92826 | 1.26E-38 | −1.82633 | 1.14E-22 |
|
|
ENSG00000177133 | −3.10089 | 2.87E-37 | −2.71809 | 9.17E-30 | Gastric cancer |
|
ENSG00000249601 | −3.19708 | 2.22E-43 | −2.14564 | 1.67E-24 |
|
| NR_027086 | −3.42128 | 9.26E-40 | −1.69737 | 4.08E-23 |
|
|
ENSG00000241224 | −4.12725 | 1.75E-43 | −1.83665 | 4.44E-20 |
|
Independent validation
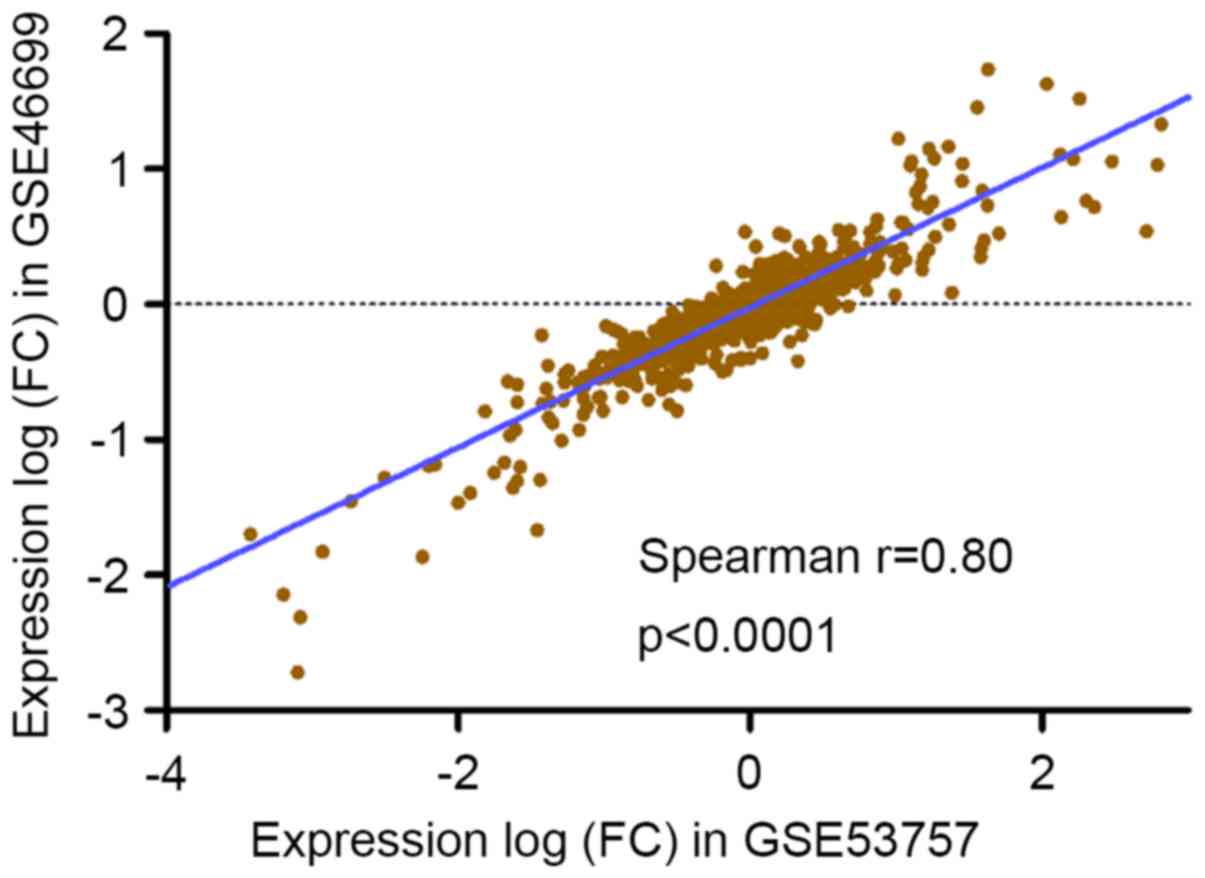
We analyzed GSE46699 and GSE53737 to independently
validate the results (Fig. 3),
finding that significant expression changes (adjusted P<0.01) at
the same direction were presented for the above differentially
expressed lncRNAs. A significant concordance shown in Fig. 3 was existed in the distribution of
expression differentials between the experiment and the validation
data sets, reflecting that the expression patterns of these genes
were highly consistent among different sample sets.
Differentially expressed lncRNAs
associated with RCC metastasis
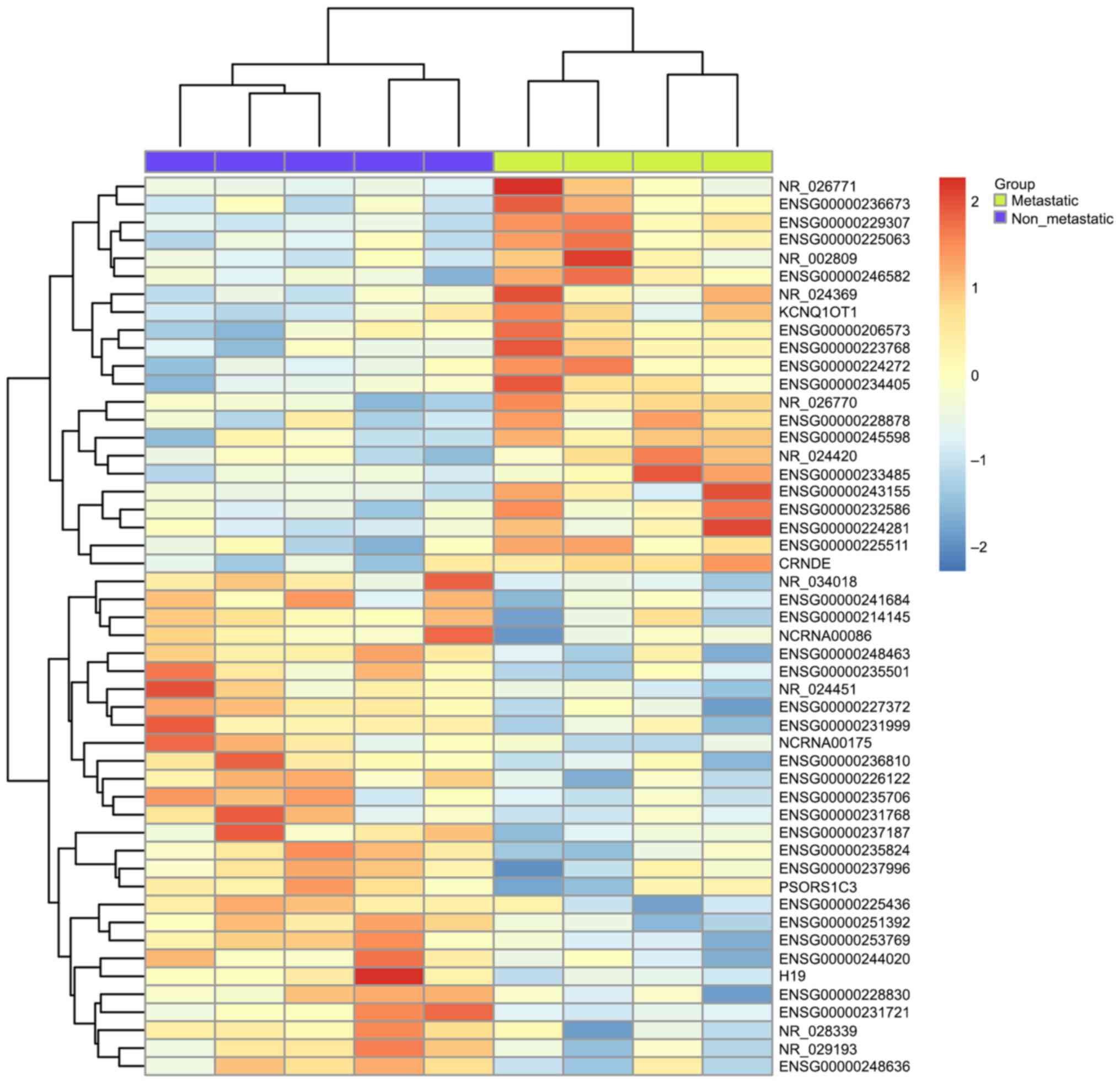
To validate the utility of exon array data in
combination with clinical annotation to identify cancer
metastasis-related lncRNA, we examined the expression pattern of
GSE47352 consisting of 4 primary metastatic and 5 non-metastatic
tumor samples (Fig. 4). Comparing to
non-metastatic samples, the top 50 differentially expressed lncRNAs
in metastatic ccRCC were identified and we found 22 lncRNAs were
upregulated and 28 lncRNAs were downregulated (Mann-Whitney U test,
P<0.05). By comparing with the results above, we found that
three out of these fifty lncRNAs were also present in the
overlapping genes shown in Table I,
which are ENSG00000214145, CRNDE and ENSG00000244020. Among them,
CRNDE showed higher expression in both ccRCC and metastatic samples
compared to normal tissue and non-metastatic ones, while
ENSG00000244020 is lower expressed. However, the lncRNA
ENS00000214145 showed an increased expression in ccRCC but
decreased in metastatic RCC.
Discussion
LncRNAs function in comprehensive biological
processes through miscellaneous mechanisms (29,30). In
recent years emerging evidence indicate that lncRNAs are disordered
and play important roles in tumorigenesis and tumor progression
(10,12,22,27).
Because the existing microarrays may fortuitously contain some
lncRNAs probes, some researchers (31,32)
recently suggested that we mine these array data and may achieve
lncRNA expression profiling.
In the present study, a set of lncRNAs
differentially expressed between ccRCC and normal renal tissue was
analyzed. Previous studies have reported some lncRNAs in human
cancers, including ENSG00000177133, NR_024418, TCL6, GAS5, DLEU2,
CRNDE and MIR155HG, while recent studies have reported NR_24418 and
TCL6 associated with ccRCC (19,20).
TCL6 was first identified in T-cell leukemia
(21), located 7 kb upstream of the
TML1 locus on chromosome 14q32.1. The TCL6 gene expressed at least
11 isoforms through very complex alternative-splicing, including
splicing with the TML1 gene. Since the TCL6 gene was expressed in
T-cell leukemia carrying a t(14;14) (q11;q32.1) chromosome
translocation and was not expressed in normal T-cells, it is
thought to be a candidate gene potentially involved in
leukemogenesis. Recently, a microarray experiment performed with
custom-designed arrays enriched with probes for lncRNAs mapping to
intronic genomic regions (20),
analyzing 18 primary RCC tumors and 11 non-tumor adjacent matched
tissues. Forty lncRNAs differentially expressed between RCC and
non-tumor samples was obtained (false discovery rate <5%),
including TCL6. However, the fold change (FC) of TCL6 in RCC showed
an increased expression (FC=1.96), which is opposite to our results
(logFC=−2.20 in GSE53757 and logFC=−1.19 in GSE46699, P<0.01).
As our study have integrated two microarray analysis, we stand more
about our results and it is also necessary to process more research
work to find out the role of TCL6 in RCC.
Another lncRNA gene, NR_024418, is a 723 bp
intragenic lncRNA transcribed from chromosome 5. We found NR_024418
is decreased in ccRCC in two arrays (logFC=−2.51 in GSE53757 and
logFC=−1.28 in GSE46699, P<0.01), which is consistent with the
results (logFC=−4.16) published in 2012 (19). Nevertheless, the characters and
biological function of NR_024418 were still unclear.
GAS5 was reported as tumor suppressor in various
cancers, including KC (33),
colorectal cancer (22), breast
cancer (34) and hepatocellular
carcinoma (23). RCC tissue and cell
line a498 displayed reduced GAS5 expression. Progressed cell death,
arrested cell cycle, retarded growth and devoid of invasion and
migration were displayed in the cells a498 ectopically expressing
GAS5 (35). These findings supported
the tumor suppressor role of GAS5 in RCC. GAS5 is critical to
control cell apoptosis and cell population growth in mammalian
(34). GAS5 transcripts are subject
to complex post-transcriptional processing and some, but not all,
GAS5 transcripts sensitize mammalian cells to apoptosis inducers.
Molecular study identified the GAS5 gene as a novel partner of BCL6
(encoding a POZ/zinc finger protein that acts as a transcriptional
repressor), which is frequently altered at its 5′non-coding region
by 3q27 chromosomal translocation in B-cell lymphoma (36). However, in our study, we found GAS5
upregulated in ccRCC in both arrays (logFC=1.10 in GSE53757 and
logFC=1.03 in GSE46699, P<0.01). More research work is needed to
find out the role of GAS5 in ccRCC.
By comparing the overlapping genes in ccRCC and
metastatic ccRCC, we found that the gene CRNDE was increased in
both ccRCC samples compared to normal tissues and in metastatic RCC
compared to non-metastatic RCC, which remind us that CRNDE probably
display a crucial role in ccRCC development. CRNDE is transcribed
from chromosome 16 on the strand opposite to the adjacent IRX5
gene, with which it may share a bidirectional promoter (37). CRNDE was initially identified highly
elevated in colorectal cancer (27),
but it is also upregulated in many other solid tumors, such as
hepatocellular carcinoma (38),
prostate cancer (39), KC (40,41) and
hematological malignancies (42,43). CRNDE
is associated with a ‘stemness’ signature, because CRNDE was
implicated as one of several lncRNAs required for the maintenance
of pluripotency in mouse embryonic stem cells, and thus,
potentially involved in tumorigenesis (44). Specifically, at the locus for mouse
CRNDE, the promoter was bound by select pluripotency-related
transcription factors, including c-Myc and n-Myc, while knockdown
of CRNDE was associated with decreased levels of a different suite
of critical pluripotency markers: Sox2, Klf4, Nanog, and Oct4.
Therefore, we believe that CRNDE is an important gene in ccRCC
progression, and more research work was demanded to declare the
biological mechanisms in ccRCC.
However, studies about the other genes, such as
ENSG00000177133, DLEU2, MIR155HG, ENSG00000214145 and
ENSG00000244020 are really rare. The only research that mentioned
ENS00000177133 is that it is low expressed in gastric cancer
(18). The genes DLEU2 and MIR155HG
have been reported to be related to myeloproliferative disorders
and leukemia (26,28,45). One
research published recently shows that the gene ENSG00000244020 is
decreased in pancreatic ductal adenocarcinoma (46). Unfortunately, no report was published
about the gene ENSG00000214145 in any other lncRNA study.
We mined three existing microarray data sets and
achieved global lncRNA expression profiles in ccRCC. A set of
lncRNAs differentially expressed in metastatic RCC were also
identified, providing potential candidates for discovery of new
cancer biomarkers and therapeutic targets to improve diagnosis and
therapy in RCC.
Acknowledgements
The present study was funded by JiangXi, Provincial
Department of Science and Technology (grant no. 20161BBG70202),
Jiangxi Provincial Health Commission (grant no. 20161071) and the
Second Affiliated Hospital of Nanchang University (grant no.
2014YNLC12014).
Glossary
Abbreviations
Abbreviations:
|
lncRNAs
|
long non-coding RNAs
|
|
ccRCC
|
clear cell renal cell carcinoma
|
|
VHL
|
von Hippel-Lindau gene
|
|
PBRM1
|
polybromo-1
|
|
SETD2
|
SET domain containing 2
|
|
KDM5C
|
lysine (K)-specific demethylase 5C
|
|
KDM6A
|
lysine (K)-specific demethylase 6A
|
|
BAP1
|
BRCA1 associated protein-1
|
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Patard JJ, Leray E, Rioux-Leclercq N,
Cindolo L, Ficarra V, Zisman A, De La Taille A, Tostain J, Artibani
W, Abbou CC, et al: Prognostic value of histologic subtypes in
renal cell carcinoma: A multicenter experience. J Clin Oncol.
23:2763–2771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maher ER: Genomics and epigenomics of
renal cell carcinoma. Semin Cancer Biol. 23:10–17. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rydzanicz M, Wrzesinski T, Bluyssen HA and
Wesoły J: Genomics and epigenomics of clear cell renal cell
carcinoma: Recent developments and potential applications. Cancer
Lett. 341:111–126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jonasch E, Futreal PA, Davis IJ, Bailey
ST, Kim WY, Brugarolas J, Giaccia AJ, Kurban G, Pause A, Frydman J,
et al: State of the science: An update on renal cell carcinoma. Mol
Cancer Res. 10:859–880. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shen C and Kaelin WG Jr: The VHL/HIF axis
in clear cell renal carcinoma. Semin Cancer Biol. 23:18–25. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Simon JM, Hacker KE, Singh D, Brannon AR,
Parker JS, Weiser M, Ho TH, Kuan PF, Jonasch E, Furey TS, et al:
Variation in chromatin accessibility in human kidney cancer links
H3K36 methyltransferase loss with widespread RNA processing
defects. Genome Res. 24:241–250. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nakada C, Tsukamoto Y, Matsuura K, Nguyen
TL, Hijiya N, Uchida T, Sato F, Mimata H, Seto M and Moriyama M:
Overexpression of miR-210, a downstream target of HIF1α, causes
centrosome amplification in renal carcinoma cells. J Pathol.
224:280–288. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nakada C, Matsuura K, Tsukamoto Y,
Tanigawa M, Yoshimoto T, Narimatsu T, Nguyen LT, Hijiya N, Uchida
T, Sato F, et al: Genome-wide microRNA expression profiling in
renal cell carcinoma: Significant down-regulation of miR-141 and
miR-200c. J Pathol. 216:418–427. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Derrien T, Johnson R, Bussotti G, Tanzer
A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG,
et al: The GENCODE v7 catalog of human long noncoding RNAs:
Analysis of their gene structure, evolution, and expression. Genome
Res. 22:1775–1789. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Enfield KS, Pikor LA, Martinez VD and Lam
WL: Mechanistic roles of noncoding RNAs in lung cancer biology and
their clinical implications. Genet Res Int.
2012:7374162012.PubMed/NCBI
|
|
13
|
von Roemeling CA, Radisky DC, Marlow LA,
Cooper SJ, Grebe SK, Anastasiadis PZ, Tun HW and Copland JA:
Neuronal pentraxin 2 supports clear cell renal cell carcinoma by
activating the AMPA-selective glutamate receptor-4. Cancer Res.
74:4796–4810. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tun HW, Marlow LA, von Roemeling CA,
Cooper SJ, Kreinest P, Wu K, Luxon BA, Sinha M, Anastasiadis PZ and
Copland JA: Pathway signature and cellular differentiation in clear
cell renal cell carcinoma. PLoS One. 5:e106962010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Eckel-Passow JE, Serie DJ, Bot BM, Joseph
RW, Cheville JC and Parker AS: ANKS1B is a smoking-related
molecular alteration in clear cell renal cell carcinoma. BMC Urol.
14:142014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ni D, Ma X, Li HZ, Gao Y, Li XT, Zhang Y,
Ai Q, Zhang P, Song EL, Huang QB, et al: Downregulation of FOXO3a
promotes tumor metastasis and is associated with metastasis-free
survival of patients with clear cell renal cell carcinoma. Clin
Cancer Res. 20:1779–1790. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang X, Sun S, Pu JK, Tsang AC, Lee D,
Man VO, Lui WM, Wong ST and Leung GK: Long non-coding RNA
expression profiles predict clinical phenotypes in glioma.
Neurobiol Dis. 48:1–8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cao WJ, Wu HL, He BS, Zhang YS and Zhang
ZY: Analysis of long non-coding RNA expression profiles in gastric
cancer. World J Gastroenterol. 19:3658–3664. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu G, Yao W, Wang J, Ma X, Xiao W, Li H,
Xia D, Yang Y, Deng K, Xiao H, et al: LncRNAs expression signatures
of renal clear cell carcinoma revealed by microarray. PLoS One.
7:e423772012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fachel AA, Tahira AC, Vilella-Arias SA,
Maracaja-Coutinho V, Gimba ER, Vignal GM, Campos FS, Reis EM and
Verjovski-Almeida S: Expression analysis and in silico
characterization of intronic long noncoding RNAs in renal cell
carcinoma: Emerging functional associations. Mol Cancer.
12:1402013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Saitou M, Sugimoto J, Hatakeyama T, Russo
G and Isobe M: Identification of the TCL6 genes within the
breakpoint cluster region on chromosome 14q32 in T-cell leukemia.
Oncogene. 19:2796–2802. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yin D, He X, Zhang E, Kong R, De W and
Zhang Z: Long noncoding RNA GAS5 affects cell proliferation and
predicts a poor prognosis in patients with colorectal cancer. Med
Oncol. 31:2532014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tu ZQ, Li RJ, Mei JZ and Li XH:
Down-regulation of long non-coding RNA GAS5 is associated with the
prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol.
7:4303–4309. 2014.PubMed/NCBI
|
|
24
|
Pickard MR and Williams GT: Regulation of
apoptosis by long non-coding RNA GAS5 in breast cancer cells:
Implications for chemotherapy. Breast Cancer Res Treat.
145:359–370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cao S, Liu W, Li F, Zhao W and Qin C:
Decreased expression of lncRNA GAS5 predicts a poor prognosis in
cervical cancer. Int J Clin Exp Pathol. 7:6776–6783.
2014.PubMed/NCBI
|
|
26
|
Morenos L, Chatterton Z, Ng JL, Halemba
MS, Parkinson-Bates M, Mechinaud F, Elwood N, Saffery R and Wong
NC: Hypermethylation and down-regulation of DLEU2 in paediatric
acute myeloid leukaemia independent of embedded tumour suppressor
miR-15a/16-1. Mol Cancer. 13:1232014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Graham LD, Pedersen SK, Brown GS, Ho T,
Kassir Z, Moynihan AT, Vizgoft EK, Dunne R, Pimlott L, Young GP, et
al: Colorectal neoplasia differentially expressed (CRNDE), a novel
gene with elevated expression in colorectal adenomas and
adenocarcinomas. Genes Cancer. 2:829–840. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cui B, Chen L, Zhang S, Mraz M, Fecteau
JF, Yu J, Ghia EM, Zhang L, Bao L, Rassenti LZ, et al: MicroRNA-155
influences B-cell receptor signaling and associates with aggressive
disease in chronic lymphocytic leukemia. Blood. 124:546–554. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guttman M and Rinn JL: Modular regulatory
principles of large non-coding RNAs. Nature. 482:339–346. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Michelhaugh SK, Lipovich L, Blythe J, Jia
H, Kapatos G and Bannon MJ: Mining Affymetrix microarray data for
long non-coding RNAs: Altered expression in the nucleus accumbens
of heroin abusers. J Neurochem. 116:459–466. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liao Q, Liu C, Yuan X, Kang S, Miao R,
Xiao H, Zhao G, Luo H, Bu D, Zhao H, et al: Large-scale prediction
of long non-coding RNA functions in a coding-non-coding gene
co-expression network. Nucleic Acids Res. 39:3864–3878. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou S, Wang J and Zhang Z: An emerging
understanding of long noncoding RNAs in kidney cancer. J Cancer Res
Clin Oncol. 140:1989–1995. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mourtada-Maarabouni M, Pickard MR, Hedge
VL, Farzaneh F and Williams GT: GAS5, a non-protein-coding RNA,
controls apoptosis and is downregulated in breast cancer. Oncogene.
28:195–208. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Qiao HP, Gao WS, Huo JX and Yang ZS: Long
non-coding RNA GAS5 functions as a tumor suppressor in renal cell
carcinoma. Asian Pac J Cancer Prev. 14:1077–1082. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ohno H: Pathogenetic role of BCL6
translocation in B-cell non-Hodgkin's lymphoma. Histol Histopathol.
19:637–650. 2004.PubMed/NCBI
|
|
37
|
Cabili MN, Trapnell C, Goff L, Koziol M,
Tazon-Vega B, Regev A and Rinn JL: Integrative annotation of human
large intergenic noncoding RNAs reveals global properties and
specific subclasses. Genes Dev. 25:1915–1927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chang Q, Chen J, Beezhold KJ, Castranova
V, Shi X and Chen F: JNK1 activation predicts the prognostic
outcome of the human hepatocellular carcinoma. Mol Cancer.
8:642009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pressinotti NC, Klocker H, Schäfer G, Luu
VD, Ruschhaupt M, Kuner R, Steiner E, Poustka A, Bartsch G and
Sültmann H: Differential expression of apoptotic genes PDIA3 and
MAP3K5 distinguishes between low- and high-risk prostate cancer.
Mol Cancer. 8:1302009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ooi A, Wong JC, Petillo D, Roossien D,
Perrier-Trudova V, Whitten D, Min BW, Tan MH, Zhang Z, Yang XJ, et
al: An antioxidant response phenotype shared between hereditary and
sporadic type 2 papillary renal cell carcinoma. Cancer Cell.
20:511–523. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cifola I, Spinelli R, Beltrame L, Peano C,
Fasoli E, Ferrero S, Bosari S, Signorini S, Rocco F, Perego R, et
al: Genome-wide screening of copy number alterations and LOH events
in renal cell carcinomas and integration with gene expression
profile. Mol Cancer. 7:62008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rager JE and Fry RC: The aryl hydrocarbon
receptor pathway: A key component of the microRNA-mediated AML
signalisome. Int J Environ Res Public Health. 9:1939–1953. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Broyl A, Hose D, Lokhorst H, de Knegt Y,
Peeters J, Jauch A, Bertsch U, Buijs A, Stevens-Kroef M, Beverloo
HB, et al: Gene expression profiling for molecular classification
of multiple myeloma in newly diagnosed patients. Blood.
116:2543–2553. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Guttman M, Donaghey J, Carey BW, Garber M,
Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al:
lincRNAs act in the circuitry controlling pluripotency and
differentiation. Nature. 477:295–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Iosue I, Quaranta R, Masciarelli S,
Fontemaggi G, Batassa EM, Bertolami C, Ottone T, Divona M,
Salvatori B, Padula F, et al: Argonaute 2 sustains the gene
expression program driving human monocytic differentiation of acute
myeloid leukemia cells. Cell Death Dis. 4:e9262013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fu XL, Liu DJ, Yan TT, Yang JY, Yang MW,
Li J, Huo YM, Liu W, Zhang JF, Hong J, et al: Analysis of long
non-coding RNA expression profiles in pancreatic ductal
adenocarcinoma. Sci Rep. 6:335352016. View Article : Google Scholar : PubMed/NCBI
|