Introduction
Urothelial cell carcinoma (UCC) of the bladder is
the fifth most common type of tumor and the second most common
cause of mortality in patients with genitourinary tract
malignancies in developed countries (1). Bladder cancer (BC) comprises two
long-recognized disease entities, non-muscle invasive bladder
cancer (NMIBC) and muscle invasive bladder cancer (MIBC), which
have distinct molecular features and clinical outcomes (2). Although 70–80% of patients are diagnosed
with NMIBC at the time of initial presentation, high recurrence
rates (50–70%) have been observed in these patients (3). Furthermore, about one-third of recurrent
cases will progress to MIBC and eventually succumb to the disease
(4). The present study acknowledges
that the biology of tumors, particularly pT1 bladder tumors, of a
similar stage and grade can vary greatly. Thus, identifying the
patients that are at risk of developing MIBC and the patients that
are not is important for appropriate disease management.
Currently, pathological analyses (including clinical
stage and tumor grade) are key determinants for risk assessment and
therapeutic decision making in BC (5). However, none of the predictive values
derived from conventional histopathological parameters have
demonstrated sufficient sensitivity or specificity for detecting,
monitoring and determining the prognosis of BC (5,6). These
limitations have led to numerous previous studies that aimed to
identify molecular markers that enable clinicians to classify BCs
in more detail, thereby enabling appropriate selection of the
optimal treatment regimen (2). Recent
genome-wide expression and sequencing studies identified the genes
and signaling pathways that are key drivers of urothelial cancer
and revealed a more complex picture comprising multiple molecular
subclasses that traverse conventional grade and stage groupings
(7,8).
Numerous studies have revealed that low-grade noninvasive and
high-grade invasive BC are genetically and clinically disparate
entities (9,10). Low-grade noninvasive bladder tumors
are characterized by gain-of-function mutations, which mainly
affect classical oncogenes including fibroblast growth factor
receptor 3 (FGFR3) and Harvey rat sarcoma viral oncogene
homolog genes, whereas invasive tumors are characterized by
loss-of-function mutations resulting in inactivation of tumor
suppressors including p53, RB, and phosphate and
tensin homolog (11,12). FGFR3 belongs to a family of
structurally associated tyrosine kinase receptors that are involved
in numerous aspects of embryogenesis and tissue homeostasis, as
well as being implicated in the tumorigenesis of bladder and other
urothelial types of cancer, multiple myeloma and cervical cancer
(13–15). Mutated FGFR3 is constitutively
activated and induces a number of oncogenic signaling pathways,
including the RAS/mitogen activated protein kinases (MAPK),
phospholipase Cc1 (PLCc1), phosphoinositide 3-kinase (PI3K) and
signal transducer and activator of transcription (STAT) signaling
pathways (11,16–18).
Activating mutations in FGFR3 genes are associated with
genetically stable Ta and low-grade BC, which represent the
favorable BC pathway (19).
Activating mutations of FGFR3 are observed in ≤70% of NMIBC
cases, whereas overexpression of a wild-type receptor has been
revealed in ~40% of patients with invasive disease (20). Although numerous studies identified
associations between FGFR3 mutation status and pathological
phenotype, the prognostic implications of these activating
mutations has not been clearly established (20–23). To
the best of our knowledge, no previous studies have undertaken a
comprehensive analysis of FGFR3 mutation status and gene
expression as prognostic markers in primary pT1 BC.
In the present study, the association between
FGFR3 gene expression level, mutation status and
pathological phenotype in primary pT1 BC tissues was examined. Of
note, the present study also evaluated the implications of
FGFR3 as a prognostic indicator for pT1 BC.
Materials and methods
Study population and follow-up
protocols
Tissue samples were obtained from 151 consecutive
patients with primary pT1 BC who underwent transurethral resection
(TUR) for histologically diagnosed transitional cell carcinomas
between January 1996 and December 2008 at Chungbuk National
University Hospital (South Korea). The tissue samples for the
present study were provided by Chungbuk National University
Hospital, a member of the National Biobank of Korea, which is
supported by the Ministry of Health, Welfare and Family Affairs.
All tumors were macrodissected within 15 min of surgical resection,
fresh-frozen in liquid nitrogen and stored at −80°C until use. Each
patient was independently reviewed by a genitourinary pathologist
who was unaware of how the clinical data were to be used. All
patients received six cycles of induction Bacillus Calmette-Guerin
(BCG) therapy (12.5 mg of Tice strain BCG in 50 ml of physiological
bacteriostatic-free saline solution), according to European
Association of Urology guidelines, and were confirmed to be disease
free 3 months following transurethral resection of the bladder
tumor (TURB) following BCG induction therapy. In order to reduce
confounding factors affecting the analyses and to delineate a more
homogenous study population, patients undergoing immediate
postoperative therapy with single-dose mitomycin C (n=8) or BCG
maintenance therapy (n=12), or those diagnosed with a concomitant
carcinoma in situ (n=6), were excluded from the study. To
avoid the risk of under staging, cases where bladder muscle was not
clearly identifiable (n=5) were also excluded. Therefore, 120
primary pT1 BC cases were finally used for analysis. The study
cohort included 97 males and 23 females. The mean age of patients
was 65.93 years (range, 24–88 years).
Tumors were staged according to the 2002
tumor-node-metastasis classification system and the 1973 World
Health Organization grading system (5,24). When a
BC specimen did not include sufficient muscle or when a grade 3
tumor was detected, a second-look TURB was systematically conducted
2–4 weeks after the initial resection. Following initial TURB, each
patient was monitored according to standard guidelines (5). Standard follow-up included cystoscopy
and urinary cytology at 3-monthly intervals for 2 years, then
6-monthly intervals for 2 years and yearly intervals thereafter.
Radiographic evaluation including chest and abdominal computed
tomography was performed on an annual basis for evaluation of the
upper urinary tract and early detection of metastasis. Recurrence
was defined as the recurrence of primary NMIBC at a lower or
equivalent pathological stage, and progression was defined as
muscular invasion, increased tumor grade or metastatic disease.
Good clinical practice protocols
The present study was performed in agreement with
the applicable laws and regulations, good clinical practice and the
ethical principles described in the Declaration of Helsinki. The
study protocol was approved by the Ethics Committee of Chungbuk
National University (IRB approval no. 2010-01-001; Cheongju,
Korea). Written informed consent was obtained from all patients
prior to enrollment in the present study. Sample collection and
analysis procedures were also approved by the Institutional Review
Board of Chungbuk National University.
Analysis of FGFR3 mutations
Genomic DNA was isolated from frozen tumor tissue
specimens using the Wizard Genomic DNA Purification System kit
(Promega Corporation, Madison, WI, USA), according to the
manufacturers protocol. The FGFR3 gene sequence was obtained
from the NCBI database (http://www.ncbi.nlm.nih.gov/gene/2261). Three regions
(exons 7, 9 and 14) harboring 11 frequent oncogenic FGFR3
mutations were simultaneously amplified by polymerase chain
reaction (PCR). Detailed PCR methods were performed as previously
described (25). The PCR products
were purified and sequenced using the BigDye Terminator v3.1 Cycle
Sequencing kit and an ABI 3730xl automatic sequencer (both from
Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA,
USA).
Analysis of FGFR3 mRNA expression
level
Total RNA was extracted from tissue samples using
TRIzol® reagent (Invitrogen, Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. cDNA was prepared
from 1 µg RNA using random primers and a First-Strand cDNA
Synthesis kit (GE Healthcare Life Sciences, Chalfont, UK). To
quantify the expression levels of FGFR3, RT-qPCR
amplification was performed using a Rotor Gene 6000 instrument
(Corbett Life Science; Qiagen, Inc., Valencia, CA, USA). RT-qPCR
assays were performed in micro-reaction tubes (Corbett Life
Science; Qiagen, Inc.) containing SYBR Premix EX Taq (Takara
Biotechnology Co., Ltd., Dalian, China). The following primers were
used to amplify FGFR3 (146 base pairs): Sense,
5′-CGTACTGTGCCACTTCAGTG-3′ and antisense,
5′-CCAGCAGCTTCTTGTCCATC-3′. The PCR reaction was performed in a
final volume of 10 µl, comprising 5 µl of 2X SYBR Premix EX Taq
buffer, 0.5 µl of each 5′ and 3′primer (10 pM/µl) and 1 µl sample
cDNA. The products were purified using a QIAquick Extraction kit
(Corbett Life Science; Qiagen), quantified in a spectrometer (MBA
2000; Perkin Elmer, Inc., Waltham, MA, USA) and sequenced using an
automated laser fluorescence sequencer (ABI PRISM 3100 Genetic
Analyzer; Applied Biosystems, Foster City, CA, USA). A known
concentration of the PCR product was then 10-fold serially diluted
from 100 to 0.1 pg/µl and used to establish a standard curve. The
RT-qPCR conditions were 1 cycle at 96°C for 20 sec, followed by 40
cycles of 2 sec at 96°C for denaturation, 15 sec at 60°C for
annealing and 15 sec at 72°C for extension. The melting program was
performed at 72–95°C with a heating rate of 1°C per 45 sec.
Spectral data were captured and analyzed using Rotor-Gene Real-Time
Analysis Software 6.0 Build 14 (Qiagen, Inc.). All samples were run
in triplicate. GAPDH was used as an endogenous RNA reference gene.
Relative quantification of gene expression was performed using the
2−ΔΔCq calculation formula, based on Cq values for
target and reference genes (26). The
gene expression was normalized to the expression of GAPDH.
Statistical analysis
Continuous variables are expressed as the median and
interquartile range (IQR). Differences between variables
demonstrating a continuous distribution across dichotomous
categories were assessed using the Mann-Whitney U test. The
Fisher's exact and χ2 tests were used to evaluate
associations between categorical variables. The Kaplan-Meier method
was used to estimate time to recurrence and progression, and
differences were assessed using the log-rank test. The prognostic
value of FGFR mutation status and gene expression level was
analyzed using univariate and multivariate Cox's regression test.
FGFR3 mRNA expression level was classified according to the
quartiles of the range, and the lowest quartile
(<107.70×104 copies/µg) was assigned to the reference
group for regression analysis. P<0.05 was considered to indicate
a statistically significant difference. All reported P-values are
two-sided. All statistical analyses were performed using SPSS
version 20.0 software (IBM SPSS, Armonk, NY, USA).
Results
Baseline characteristics
The baseline characteristics of the 120 patients
with primary pT1 BC are presented in Table I. The study cohort included 97 males
and 23 females. The mean age of patients was 65.93 years (range,
24–88 years). The histological grade distribution was as follows:
14.2% grade I; 65.8% grade II and 20.0% grade III. A total of 61
patients (50.8%) exhibited recurrent disease and progression was
observed in 20 patients (16.7%) during a median follow-up period of
69.3 months (IQR, 29.2–103.1 months). The median intervals for
recurrence and progression were 20.7 months (range, 6.4–133.6) and
43.0 months (range, 6.6–115.4), respectively.
 | Table I.Baseline characteristics of the
patients. |
Table I.
Baseline characteristics of the
patients.
| Parameters | Number (%) |
|---|
| Mean age ± SD,
years (range) | 65.93±12.93
(24–88) |
| Median follow-up,
months (IQR) | 69.3
(29.2–103.1) |
| Gender |
|
|
Male | 97 (80.8) |
|
Female | 23 (19.2) |
| Smoking (ex-or
current) | 51 (42.5) |
| Tumor size
(cm) |
|
| ≤3 | 49 (40.8) |
| ≥3 | 71 (59.2) |
| Multiplicity |
|
|
Single | 54 (45.0) |
|
Multiple | 66 (55.0) |
| Grade |
|
| I | 17 (14.2) |
| II | 79 (65.8) |
|
III | 24 (20.0) |
| Recurrence | 61 (50.8) |
| Progression | 20 (16.7) |
Of the 20 progressive cancers, 4 cases demonstrated
an increased tumor grade within the equivalent pathological stage
and 16 cases progressed to MIBC. A total of 15 cases underwent
radical cystectomy and the other cases received palliative
chemotherapy or radiation therapy: Of those, 8 patients succumbed
to BC.
Association between FGFR3 mutation
status and mRNA expression level in pT1 BC tissues
FGFR3 mutations were identified in 48/120
(40.0%) patients with pT1 BC. The most common mutations were Y373C,
R249C and R248C, which were observed in 16, 13 and 11 cases,
respectively. FGFR3 mRNA expression level was significantly
higher in FGFR3 mutant BC compared with in FGFR3
wild-type BC (P<0.001). The median FGFR3 mRNA expression
levels for mutant and wild-type BC were 728.38×104 (IQR,
282.23–1287.61) copies/µg and 154.23×104 (IQR,
61.16–419.49) copies/µg, respectively (Table II).
 | Table II.Association between FGFR3
mutation status and mRNA expression level in pT1 BC. |
Table II.
Association between FGFR3
mutation status and mRNA expression level in pT1 BC.
| FGFR3
mutation | Number (%) | mRNA expression
level, median (IQR; ×104 copies/µg) | P-value |
|---|
| Wild-type | 72 (60.0) | 154.23 |
<0.001a |
|
|
| (61.16–419.49) |
|
| Mutant | 48 (40.0) | 728.38 |
|
|
|
|
(282.23–1287.61) |
|
|
R248C | 11 |
|
|
|
S249C | 13 |
|
|
|
G370C | 2 |
|
|
|
S371C | 2 |
|
|
|
Y373C | 16 |
|
|
|
A391E | 1 |
|
|
|
K650M | 1 |
|
|
|
K650E | 2 |
|
|
|
K650T | 1 |
|
|
Association between FGFR3 mutation
status, mRNA expression level and clinicopathological features in
pT1 BC tissues
BC harboring wild-type FGFR3 and low
FGFR3 expression level was associated with high-grade tumors
(P=0.006). However, there were no significant differences in
FGFR3 mutation status or mRNA expression level according to
other clinicopathological parameters, including age, tumor size and
multiplicity (all P>0.05; Table
III).
 | Table III.Association between FGFR3
mutation status, mRNA expression level and clinicopathological
features in pT1 BC. |
Table III.
Association between FGFR3
mutation status, mRNA expression level and clinicopathological
features in pT1 BC.
|
| FGFR3
mutation |
|
|
|
|---|
|
|
|
|
|
|
|---|
| Parameters | Wild-type
(n=72) | Mutation
(n=48) | P-value | mRNA expression
level, median (IQR; ×104 copies/µg) | P-value |
|---|
| Gender |
|
| 0.350a |
| 0.772b |
|
Male | 56 (77.8) | 41 (85.4) |
| 304.04
(100.58–848.86) |
|
|
Female | 16 (22.2) | 7 (14.6) |
| 263.43
(128.46–514.87) |
|
| Tumor size |
|
| 0.349a |
| 0.056b |
| <3
cm | 32 (44.4) | 17 (35.4) |
| 219.23
(69.98–545.59) |
|
| ≥3
cm | 40 (55.6) | 31 (64.6) |
| 369.27
(146.93–956.35) |
|
| Multiplicity |
|
| 0.708a |
| 0.945b |
|
Single | 31 (43.1) | 23 (47.9) |
| 272.10
(107.73–1116.69) |
|
|
Multiple | 41 (56.9) | 25 (52.1) |
| 336.62
(98.00–727.14) |
|
| Grade |
|
| 0.001a |
| 0.006c |
| I | 6
(8.3) | 11
(22.9) |
| 453.92
(242.84–1076.38) |
|
| II | 44 (61.1) | 35 (72.9) |
| 342.25
(119.95–1038.87) |
|
|
III | 22 (30.6) | 2
(4.2) |
| 130.04
(33.47–306.05) |
|
| Recurrence |
|
| 0.264a |
| 0.856b |
| No | 32 (44.4) | 27 (56.2) |
| 304.04
(127.82–685.67) |
|
|
Yes | 40 (55.6) | 21 (43.8) |
| 286.62
(78.73–869.24) |
|
| Progression |
|
| 0.050a |
| 0.001b |
| No | 56 (77.8) | 44 (91.7) |
| 367.78
(132.51–883.11) |
|
|
Yes | 16 (22.2) | 4 (8.3) |
| 78.73
(22.57–302.03) |
|
Prognostic value of FGFR3 mutation
status and mRNA expression level in pT1 BC tissues
There were no significant differences in
FGFR3 mutation status or mRNA expression level in terms of
tumor recurrence (P=0.264 and P=0.856, respectively). Patients who
experienced cancer progression exhibited significantly lower
expression levels of FGFR3 mRNA compared with patients who
did not (P=0.001; Table III).
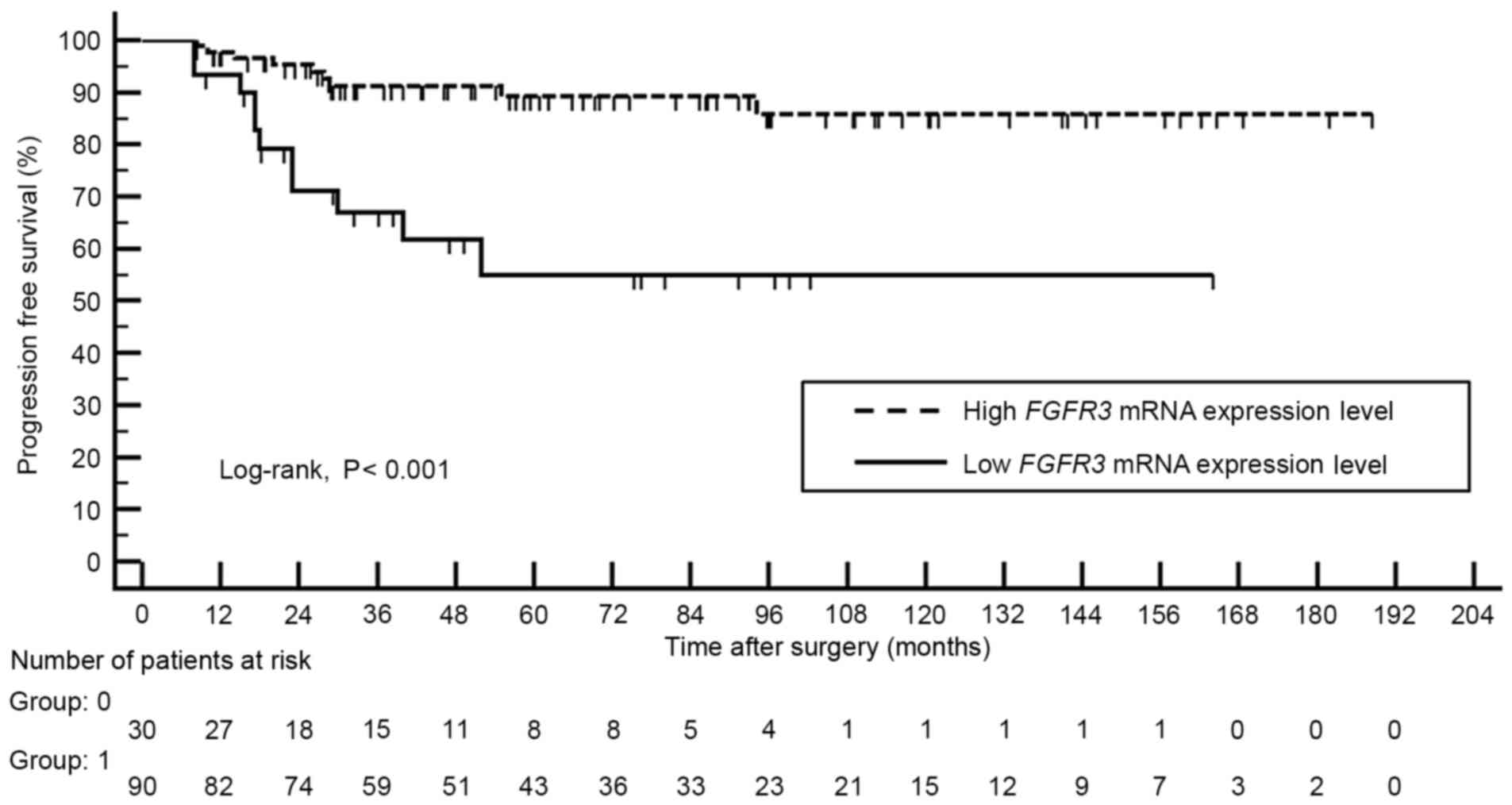
Kaplan-Meier analysis revealed that patients with high FGFR3
mRNA expression level demonstrated better progression-free survival
compared with those with lower expression levels of FGFR3
mRNA (log-rank, P<0.001; Fig.
1).
Multivariate Cox regression analysis identified low
FGFR3 expression level (odds ratio, 3.300; 95% confidence
interval, 1.310–8.313; P=0.011) and tumor grade III (odds ratio,
2.623; 95% confidence interval, 1.161–5.927; P=0.020) as an
independent predictor of cancer progression (Table IV).
 | Table IV.Univariate and multivariate Cox
regression models for the risk of progression in primary T1 BC. |
Table IV.
Univariate and multivariate Cox
regression models for the risk of progression in primary T1 BC.
|
| Univariate
analysis | Multivariate
analysis of FGFR3 mutation | Multivariate
analysis of FGFR3 expression level |
|---|
|
|
|
|
|
|---|
| Parameters | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age | 1.020
(0.983–1.058) | 0.289 |
|
|
|
|
| Gender (male) | 0.996
(0.332–2.984) | 0.994 |
|
|
|
|
| Smoking history
(yes) | 1.263
(0.522–3.057) | 0.605 |
|
|
|
|
| Size (>3
cm) | 1.185
(0.484–2.902) | 0.711 |
|
|
|
|
| Multiplicity
(multiple) | 1.676
(0.668–4.205) | 0.271 |
|
|
|
|
| Grade (I–II vs.
III) | 3.448
(1.401–8.488) | 0.007 | 3.014
(1.290–7.043) | 0.011 | 2.623
(1.161–5.927) | 0.020 |
| FGFR3
mutation (Wt) | 2.643
(0.883–7.917) | 0.082 | 1.549
(0.468–5.125) | 0.473 | Not applicable |
|
| Low FGFR3
expression level (<107.70×104 copies/µg) | 4.586
(1.890–11.127) | 0.001 | Not applicable |
| 3.300
(1.310–8.313) | 0.011 |
Association between FGFR3 mutation
site, mRNA expression level and cancer progression in pT1 BC
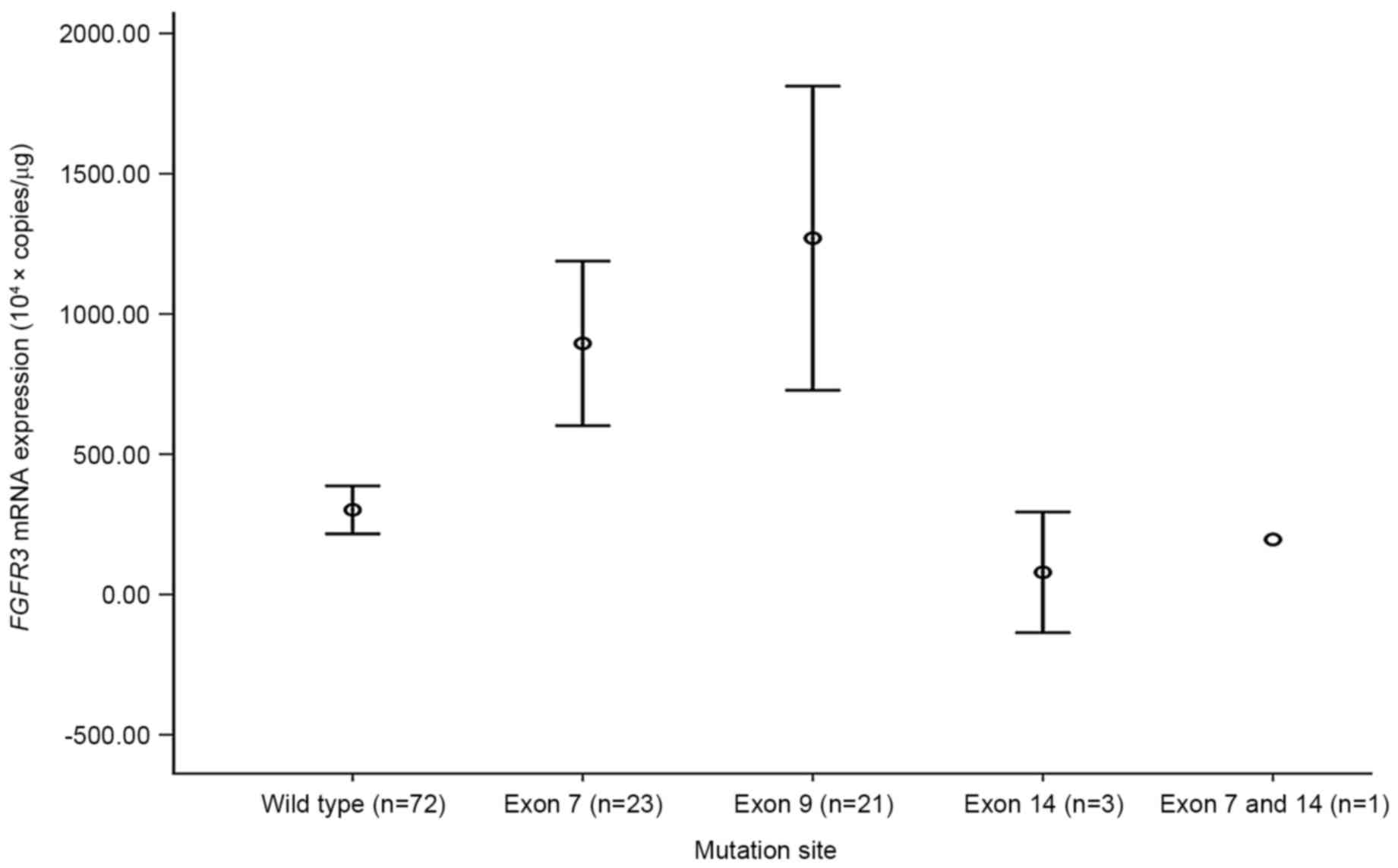
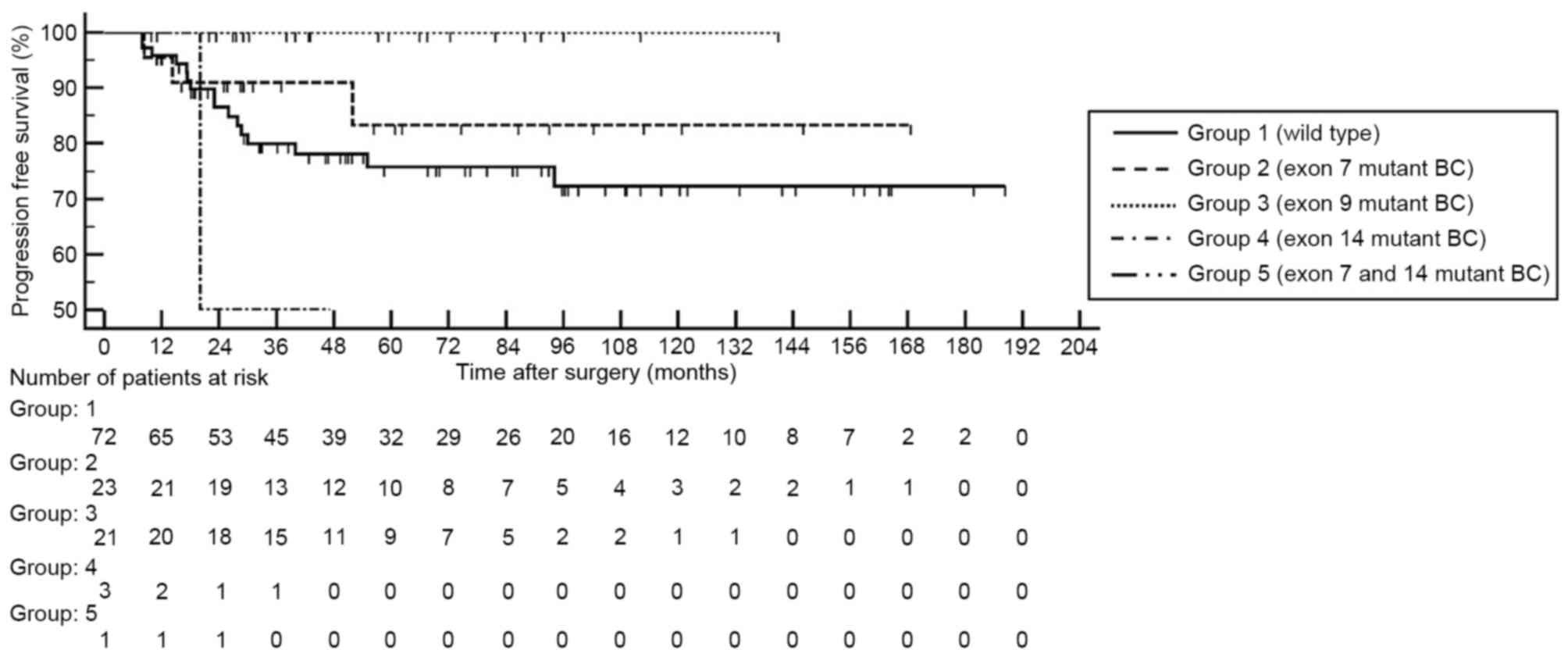
When FGFR3 mutations were categorized by exon
site, mutations in exons 7 and 9 demonstrated significantly high
mRNA expression levels compare with the wild type BC (each
P<0.001; Fig. 2). By contrast,
mutations located in exon 14 did not reveal a significant
difference in FGFR3 mRNA expression level compared with in
the wild type BC. None of the patients with BC harboring
FGFR3 mutation in exon 9 demonstrated disease progression or
metastasis (Fig. 3).
Discussion
The present study examined the utility of
FGFR3 mutations and FGFR3 gene expression as
prognostic markers in primary pT1 BC. FGFR3 mRNA expression
was associated with the presence of FGFR3 mutation.
FGFR3 mRNA expression level was an independent predictor of
progression. FGFR3 mutation was significantly associated
with tumor grade but not with cancer progression.
FGFR3 is a receptor tyrosine kinase
implicated in the tumorigenesis of numerous types of myeloma,
cervical cancer and urothelial carcinoma (13). There are two mechanisms that cause
abnormal activation of FGFR3: Translocation of chromosome 4
to chromosome 14 (leading to overexpression) and activation of
point mutations in the FGFR3 gene (11). Activating mutations of FGFR3
are observed in the majority of NMIBCs (35.5–78.1%), and the
overexpression of a wild-type receptor has been identified in ~40%
of MIBC (21). Constitutive
(ligand-independent) receptor activation occurs most commonly by
substitution of a wild-type residue within the extracellular domain
of FGFR3 with a cysteine residue, resulting in dimerization
and subsequent stimulation of tyrosine kinase activity (11,27). This
in turn induces a number of different oncogenic signaling pathways,
including the RAS/MAPK, PLCc1, PI3K and STAT pathways (7,17,18). FGFR3 point mutations are found
almost exclusively in exons 7, 10 and 15 (19). The most frequent extracellular
domain-activating mutations are R248C and S249C, and transmembrane
domain mutations include G372C and Y375C; other mutations occur at
low frequencies (6,16). The frequency of FGFR3 mutations
at these hot spots in the present study's cohort were similar to
those described in previous studies (23). Cappellen et al (14) conducted the first study examining
FGFR3 involvement in bladder tumors. Since then, numerous
studies have been performed to better understand the potential role
of mutant FGFR3 as an oncogenic driver, particularly in BC
(18–21,23,28).
Previous studies also demonstrated that FGFR3 mutations are
associated with genetically stable Ta and low-grade BC, which
represents the favorable BC pathway (20). Activating mutations in the
FGFR3 gene have been reported in ≤75% of low-grade and
low-stage BC, but are absent or rare in carcinoma in situ
and MIBC (29). The results presented
in the present study confirm previous studies demonstrating that
the presence of FGFR3 mutations is significantly associated
with low tumor grade (23). The
association between FGFR3 mutations and pathological
phenotype has been well established, but the prognostic
significance of FGFR3 mutations in BC remains poorly defined
(20). A previous study by van Rhijn
et al (19) reported that
FGFR3 mutations were an independent predictor of recurrence
in NMIBC. BC recurrence was more common in patients whose initial
tumor was classified as wild-type rather than as harboring a mutant
FGFR3 gene. Conversely, a large prospective study of 772
patients revealed a significantly higher rate of recurrence in
patients harboring an FGFR3 mutation compared with in those
with a FGFR3 wild-type tumor (22). Following stratification according to
tumor stage and grade, the prognostic value of the FGFR3
mutation in terms of tumor recurrence appeared to be restricted to
pTaG1 tumors, and a previous study suggested that additional
molecular alterations within higher grade/stage tumors overrode the
association between FGFR3 mutation and prognosis (22). In addition, there is certain evidence
supporting the prognostic value of FGFR3 mutations for
predicting the risk of progression (23,30). The
exact prognostic role of these mutations with respect to NMIBC
progression has not yet been fully elucidated; however, two
recently published studies suggested the possibility of a
progression-associated prognostic indicator for NMIBC (4,23,30). A study by van Rhijn et al
(23) examined the distribution and
clinical outcome of FGFR3 and P53 alterations in 132
patients with primary pT1 BC. Multivariate analyses revealed that
FGFR3 mutation status was a significant prognostic factor
for progression. Another study by Burger et al (30) revealed that FGFR3 status did
discriminate progressors from non-progressors within a subset of
patients with high-grade BC. Although the design and outcome
evaluations of the present study were similar to previous studies,
the present study demonstrated a different result in which
FGFR3 mutation status did not have prognostic significance
in terms of tumor recurrence or progression. Numerous factors may
account for these discrepant results. Firstly, the resent study
adopted strict exclusion criteria to eliminate possible
interference. To delineate a more homogenous study population,
patients who received intravesical chemotherapy or BCG maintenance
therapy or those diagnosed with a concomitant carcinoma in
situ were excluded from the study. Although van Rhijn et
al (23) specifically analyzed
patients with primary pT1 BC who received BCG, 35% of BC cases were
concomitant carcinoma in situ, which frequently resembles a
muscle invasive disease due to its aggressive biological features.
It is also possible that the participants in the present study had
different tumor characteristics. In the study by Burger et
al (30), the majority of
patients exhibited a relatively favorable tumor characteristic, 81%
of pTa tumor and 89% of G1-2 tumor, whereas van Rhijn et al
(23) enrolled patients with a
primary diagnosis of pT1 and majority of the patients exhibited
high-grade tumors (80%). The results of the present study were also
acquired from a homogenous population with a primary diagnosis of
pT1 and 65% of T1 BC was Grade II. Tumor staging and grading were
reassigned by one genitourinary pathologist; however, only 20% of
T1 BC was assigned to grade III. The progression rate of the BC
cohort was lower compared with in the study by van Rhijn et
al (23) and this may be due to
these tumor characteristics. In the present study, FGFR3
mutant BC was associated with a favorable tumor grade and high
FGFR3 mRNA expression level, but it did not affect
prognostic impact on progression. Further large cohort
collaboration studies should be performed to confirm the prognostic
role of FGFR3 mutation in pT1 BC.
The majority of previous studies focused on
FGFR3 mutation status and protein expression level with
respect to pathological phenotype and oncological outcome (16,19,21,23,28).
At present, little is known about the association between mutation
status and FGFR3 mRNA expression level in BC (28). A study by Bernard-Pierrot et al
(27) investigated the association
between FGFR3 mRNA expression levels and FGFR3
status, and demonstrated that high expression levels of
FGFR3 correlated with the presence of a mutated FGFR3
gene. However, the level of FGFR3 mRNA was determined by
semi-quantitative radioactive RT-qPCR, and they did not identify a
significant association between FGFR3 mRNA expression levels
and tumor characteristics. Furthermore, as far as can be
ascertained, no previous study has addressed the prognostic
implications of FGFR3 mRNA expression level in BC. The
present study revealed that lower FGFR3 mRNA expression
level was an independent predictor of progression. FGFR3
mRNA expression level may be useful for predicting the outcome of
high-risk refractory tumors in pT1 BC prior to their progression.
The present study further analyzed the FGFR3 mRNA expression
categorized by exon site, which encode various functional domains
of FGFR3 protein, including exon 7 (immunoglobulin-like domain:
Codon 248, 249), exon 9 (transmembrane domain: Codon 370, 371, 373,
and 391) and exon 14 (tyrosine kinase domain: Codon 650). Of note,
the present study demonstrated that mutations in exon 7 and 9
revealed significant high FGFR3 mRNA expression levels
compare with in the wild type BC. Mutations located in exon 14 did
not demonstrate significant difference in FGFR3 mRNA
expression level compare with the wild type BC. The present study
could not conduct survival analysis due to the limited number of
progression events. However, none of the FGFR3 mutations in
exon 9 led to disease progression or metastasis. Conversely, among
the 3 patients with harboring mutant BC located in exon 14, 1
patient demonstrated cancer progression within 2 years of short
interval. It was suggested that prognostic influences of
FGFR3 mutations may be modulated by the mutation site of the
FGFR3 gene, but this requires further investigation.
A possible limitation of the present study is that
FGFR3 protein levels were not evaluated. Further studies
should include these experiments to better understand the
association between activating mutations of FGFR3, mRNA
expression level and protein expression level. In addition, the
sample size was relatively small, which may reduce the statistical
power. Thus, further collaborative studies are required in order to
confirm the prognostic role of FGFR3 mutation and gene
expression in pT1 BC.
In conclusion, the results of the present study
suggested that FGFR3 mRNA expression level may be a useful
tool for providing a more accurate prognosis for individual
patients with pT1 BC. Our preliminary analyses suggested that
prognostic influences of FGFR3 mutations may be modulated by
the mutation site of the FGFR3 gene; however, results are
preliminary and thus require validation.
Acknowledgements
This study was supported by Basic Science Research
Program through the National Research Foundation of Korea(NRF)
funded by the Ministry of Science, ICT & Future Planning (grant
no. NRF-2015R1A2A2A03004100) and by the International Science and
Business Belt Program through the Ministry of Science, ICT and
Future Planning (grant no. 2016K000297). The specimens for this
study were provided by Chungbuk National University Hospital, a
member of the National Biobank of Korea, which is supported by the
Ministry of Health, Welfare and Family Affairs. The authors would
like to thank Ms. Eun-Ju Shim from the National Biobank of Korea at
Chungbuk National University Hospital for sample preparation and
technical assistance.
References
|
1
|
Johansson SL and Cohen SM: Epidemiology
and etiology of bladder cancer. Semin Surg Oncol. 13:291–298. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Knowles MA and Hurst CD: Molecular biology
of bladder cancer: New insights into pathogenesis and clinical
diversity. Nat Rev Cancer. 15:25–41. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
van Rhijn BW, Burger M, Lotan Y, Solsona
E, Stief CG, Sylvester RJ, Witjes JA and Zlotta AR: Recurrence and
progression of disease in non-muscle-invasive bladder cancer: From
epidemiology to treatment strategy. Eur Urol. 56:430–442. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim YH, Kim WT, Jeong P, Ha YS, Kang HW,
Yun SJ, Moon SK, Choi YH, Kim IY and Kim WJ: Novel combination
markers for predicting survival in patients with muscle invasive
bladder cancer: USP18 and DGCR2. J Korean Med Sci. 29:351–356.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brausi M, Witjes JA, Lamm D, Persad R,
Palou J, Colombel M, Buckley R, Soloway M, Akaza H and Böhle A: A
review of current guidelines and best practice recommendations for
the management of nonmuscle invasive bladder cancer by the
International Bladder Cancer Group. J Urol. 186:2158–2167. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lotan Y, Shariat SF, Schmitz-Dräger BJ,
Sanchez-Carbayo M, Jankevicius F, Racioppi M, Minner SJ, Stöhr B,
Bassi PF and Grossman HB: Considerations on implementing diagnostic
markers into clinical decision making in bladder cancer. Urol
Oncol. 28:441–448. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mitra AP and Cote RJ: Molecular
pathogenesis and diagnostics of bladder cancer. Annu Rev Pathol.
4:251–285. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
van Rhijn BW, Zuiverloon TC, Vis AN,
Radvanyi F, van Leenders GJ, Ooms BC, Kirkels WJ, Lockwood GA,
Boevé ER, Jöbsis AC, et al: Molecular grade (FGFR3/MIB-1) and EORTC
risk scores are predictive in primary non-muscle-invasive bladder
cancer. Eur Urol. 58:433–441. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dalbagni G, Presti J, Fair W, Reuter VA
and Cordon-Cardo C: Genetic alterations in bladder cancer. Lancet.
342:469–471. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cheng L, Zhang S, MacLennan GT, Williamson
SR, Lopez-Beltran A and Montironi R: Bladder cancer: Translating
molecular genetic insights into clinical practice. Hum Pathol.
42:455–481. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Iyer G and Milowsky MI: Fibroblast growth
factor receptor-3 in urothelial tumorigenesis. Urol Oncol.
31:303–311. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jebar AH, Hurst CD, Tomlinson DC, Johnston
C, Taylor CF and Knowles MA: FGFR3 and Ras gene mutations are
mutually exclusive genetic events in urothelial cell carcinoma.
Oncogene. 24:5218–5225. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pandith AA, Shah ZA and Siddiqi MA:
Oncogenic role of fibroblast growth factor receptor 3 in
tumorigenesis of urinary bladder cancer. Urol Oncol. 31:398–406.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cappellen D, De Oliveira C, Ricol D, de
Medina S, Bourdin J, Sastre-Garau X, Chopin D, Thiery JP and
Radvanyi F: Frequent activating mutations of FGFR3 in human bladder
and cervix carcinomas. Nat Genet. 23:18–20. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shiang R, Thompson LM, Zhu YZ, Church DM,
Fielder TJ, Bocian M, Winokur ST and Wasmuth JJ: Mutations in the
transmembrane domain of FGFR3 cause the most common genetic form of
dwarfism, achondroplasia. Cell. 78:335–342. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kompier LC, Lurkin I, van der Aa MN, van
Rhijn BW, van der Kwast TH and Zwarthoff EC: FGFR3, HRAS, KRAS,
NRAS and PIK3CA mutations in bladder cancer and their potential as
biomarkers for surveillance and therapy. PLoS One. 5:e138212010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Juanpere N, Agell L, Lorenzo M, de Muga S,
López-Vilaró L, Murillo R, Mojal S, Serrano S, Lorente JA, Lloreta
J and Hernández S: Mutations in FGFR3 and PIK3CA, singly or
combined with RAS and AKT1, are associated with AKT but not with
MAPK pathway activation in urothelial bladder cancer. Hum Pathol.
43:1573–1582. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hernández S, López-Knowles E, Lloreta J,
Kogevinas M, Jaramillo R, Amorós A, Tardón A, García-Closas R,
Serra C, Carrato A, et al: FGFR3 and Tp53 mutations in T1G3
transitional bladder carcinomas: Independent distribution and lack
of association with prognosis. Clin Cancer Res. 11:5444–5450. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
van Rhijn BW, Lurkin I, Radvanyi F,
Kirkels WJ, van der Kwast TH and Zwarthoff EC: The fibroblast
growth factor receptor 3 (FGFR3) mutation is a strong indicator of
superficial bladder cancer with low recurrence rate. Cancer Res.
61:1265–1268. 2001.PubMed/NCBI
|
|
20
|
Neuzillet Y, van Rhijn BW, Prigoda NL,
Bapat B, Liu L, Bostrom PJ, Fleshner NE, Gallie BL, Zlotta AR,
Jewett MA and van der Kwast TH: FGFR3 mutations, but not FGFR3
expression and FGFR3 copy-number variations, are associated with
favourable non-muscle invasive bladder cancer. Virchows Arch.
465:207–213. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tomlinson DC, Baldo O, Harnden P and
Knowles MA: FGFR3 protein expression and its relationship to
mutation status and prognostic variables in bladder cancer. J
Pathol. 213:91–98. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hernández S, López-Knowles E, Lloreta J,
Kogevinas M, Amorós A, Tardón A, Carrato A, Serra C, Malats N and
Real FX: Prospective study of FGFR3 mutations as a prognostic
factor in nonmuscle invasive urothelial bladder carcinomas. J Clin
Oncol. 24:3664–3671. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
van Rhijn BW, van der Kwast TH, Liu L,
Fleshner NE, Bostrom PJ, Vis AN, Alkhateeb SS, Bangma CH, Jewett
MA, Zwarthoff EC, et al: The FGFR3 mutation is related to favorable
pT1 bladder cancer. J Urol. 187:310–314. 2012.PubMed/NCBI
|
|
24
|
Torloni H: Histologic typing of urinary
bladder tumors, international histological classification of
tumors. World Health Organization; Geneva: 1973
|
|
25
|
van Oers JM, Lurkin I, van Exsel AJ,
Nijsen Y, van Rhijn BW, van der Aa MN and Zwarthoff EC: A simple
and fast method for the simultaneous detection of nine fibroblast
growth factor receptor 3 mutations in bladder cancer and voided
urine. Clin Cancer Res. 11:7743–7748. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bernard-Pierrot I, Brams A, Dunois-Lardé
C, Caillault A, de Diez Medina SG, Cappellen D, Graff G, Thiery JP,
Chopin D, Ricol D and Radvanyi F: Oncogenic properties of the
mutated forms of fibroblast growth factor receptor 3b.
Carcinogenesis. 27:740–747. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guancial EA, Werner L, Bellmunt J, Bamias
A, Choueiri TK, Ross R, Schutz FA, Park RS, O'Brien RJ, Hirsch MS,
et al: FGFR3 expression in primary and metastatic urothelial
carcinoma of the bladder. Cancer Med. 3:835–844. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Billerey C, Chopin D, Aubriot-Lorton MH,
Ricol D, Gil Diez, de Medina S, Van Rhijn B, Bralet MP,
Lefrere-Belda MA, Lahaye JB, Abbou CC, et al: Frequent FGFR3
mutations in papillary non-invasive bladder (pTa) tumors. Am J
Pathol. 158:1955–1959. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Burger M, van der Aa MN, van Oers JM,
Brinkmann A, van der Kwast TH, Steyerberg EC, Stoehr R, Kirkels WJ,
Denzinger S, Wild PJ, et al: Prediction of progression of
non-muscle-invasive bladder cancer by WHO 1973 and 2004 grading and
by FGFR3 mutation status: A prospective study. Eur Urol.
54:835–844. 2008. View Article : Google Scholar : PubMed/NCBI
|