Introduction
Thapsigargin (TG) is a natural product isolated from
the seeds of Thapsia garganica L, which binds tightly to and
inhibits the function of the transmembrane portion of the
sarcoplasmic/endoplasmic reticulum calcium adenosine triphosphatase
pump, inducing apoptosis (1,2). TG initiates endoplasmic reticulum (ER)
stress via the unfolded protein response (UPR), which is initiated
by 3 ER transmembrane proteins termed protein kinase-like
endoplasmic reticulum kinase (PERK), inositol-requiring enzyme 1
(IRE1) and activating transcription factor (ATF) 6 (3,4). An
abnormality of this network affects the progression of various
types of cancer, such as breast cancer, pancreatic adenocarcinoma
and melanoma (3,5).
A TG prodrug that is activated in the vasculature of
solid tumors by tumor endothelial cells has been developed
(6). It is highly selective to tumor
endothelial cells, and the drug toxicity level is expected to be
low. This TG prodrug is useful in the majority of types of solid
tumors with prostate-specific membrane antigen expression,
including hepatocellular carcinoma (HCC) (7). Several factors contribute to HCC, such
as chronic hepatitis B infection, excessive alcohol consumption and
other chronic hepatic damage (8).
These factors may cause oxidative stress, inflammation and
mutation, which transform hepatic cells into HCC cells by inducing
ER stress (9–11). In China, numerous patients with HCC
are hepatitis B virus (HBV) carriers (12). HBV infection causes massive viral
replication and produces a large number of viral proteins in a
short period of time, which results in the disturbance of ER
homeostasis and therefore protein misfolding. The accumulation of
unfolded or misfolded proteins leads to ER stress, followed by UPR
(13). However, only a limited number
of studies have investigated the effect of HBV on UPR gene
expression and apoptosis (14,15). The
present study used two cell lines, HepG2 and HepG2.2.15, which are
HBV negative and positive, respectively, to study whether HBV
affects apoptosis during ER stress induced by TG.
Materials and methods
Cell culture, reagents and
antibodies
The HepG2 cells were purchased from the Chinese
Academy of Medical Sciences Cell Culture Center (Beijing, China)
and the HepG2.2.15 cells were purchased from China Center for Type
Culture Collection (Wuhan, China). The cells were cultured in
Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific
Inc., Waltham, MA, USA) with 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc.). The cell lines were maintained in
an atmosphere with 5% CO2 and saturated humidity at
37°C. Images of the cells were captured using a CKX41 microscope
(Olympus Corporation, Tokyo, Japan) at a magnification of ×100
using the TopTek ToupView version 3.7 (OPTEC, Chongqing, China). TG
was obtained from Sigma-Aldrich; Merck Millipore (Darmstadt,
Germany; cat. no., 586005-1MG), dissolved in dimethylsulfoxide
(DMSO; Amresco, LLC., Solon, OH, USA) and used at a dilution of 500
nM.
A total of 200 units/well recombinant human
interferon (IFN)α-2A (Fangcheng BioTech Co Ltd; Beijing, China;
cat. no., CYT-204) was added 6 h prior to TG treatment. MTT was
purchased from Sigma-Aldrich; Merck Millipore (cat no. M2128) and
was used at a concentration of 5 mg/ml. PBS was purchased from
Tianjin TBD Haoyang BioTech Co Ltd (Tianjin, China; cat. no.,
PB2004Y). A eukaryotic DNA damage inducible transcript 3 (CHOP)
expression plasmid encoding the full-length CHOP protein was
purchased from Tianyi Huiyuan Biotechnology Co., (Wuhan, China).
Small interfering RNA (siRNA) for the CHOP protein was purchased
from Guangzhou RiboBio Co., Ltd., (Guangzhou, China). Lipofectamine
2000 was purchased from Invitrogen; Thermo Fisher Scientific, Inc.,
(cat. no., 1609073). The Ultrapure RNA (cat. no., CW0581), Protein
Extraction (cat. no., cw0889), SYBR Green polymerase chain reaction
(PCR; cat. no., CW2601), Annexin V-FITC/PI Apoptosis Detection
(cat. no., CW2574) and Cell Cycle and Apoptosis Detection (cat.
no., CW2575) kits were purchased from CWBio (Beijing, China). The
ReverTra Ace reverse transcription quantitative (RT-q)PCR Master
Mix was purchased from Toyobo Co., Ltd. (Osaka, Japan; cat. no.,
FSQ-201). Antibodies against CHOP (polyclonal rabbit, cat. no.,
sc-575) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH;
polyclonal rabbit, cat. no., CW0101) were purchased from Santa Cruz
Biotechnology, Inc., (Santa Cruz; Dallas, TX, USA) and CWBIO
(Beijing, China), respectively. Horseradish peroxidase conjugated
goat anti rabbit IgG was purchased from CWBio (cat. no.,
CW103A).
MTT assay
The HepG2 or HepG2.2.15 cells were seeded at a
density of 5×103 cells/well in 96-well plates. The cells
were treated with TG for 24, 48, 72 and 96 h and cells treated with
PBS were used as controls. Subsequent to the end of each time
point, the cells were incubated with 10 µl MTT for 4 h at 37°C in
the dark. The supernatant was removed and 100 µl DMSO was used for
dissolution. A Synergy H1 microplate reader (BioTek; Winooski, VT,
USA) was used to measure absorbance at 490 nm. All experiments were
performed in triplicate and repeated 3 times.
Flow cytometry assay
A flow cytometry assay was used to investigate the
level of apoptosis and the cell cycle of the HepG2 and HepG2.2.15
cells. The HepG2 cells were divided into 2 groups. The cells in the
first group were treated with TG for 24, 48, 72 and 96 h. The
untreated cells in the second group were used as controls. The
HepG2.2.15 cells were divided into 3 groups. The cells in the first
group were treated with IFNα-2A for 6 h and incubated with TG for
24, 48, 72 and 96 h at 37°C. The cells in the second group were
treated with TG for 24, 48, 72 and 96 h directly. The untreated
cells in the third group were used as controls. For the apoptosis
assays, the cells were incubated with propidium iodide (PI) and
Annexin V-FITC at 24°C for 15 min according to the protocol of the
Annexin V-FITC/PI Apoptosis Detection kit. For the cell cycle
assays, the collected cells were treated according to the protocol
provided with the Cell Cycle and Apoptosis Detection kit. Apoptosis
and cell cycle were quantified and analyzed using the Accuri C6
flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Quantitative analysis of gene
expression
The HepG2 cells were seeded in 24-well plates at a
density of 2×105/well for a RT qPCR analysis. Each well
of HepG2 or HepG2.2.15 cells was incubated with 500 nM TG. The
HepG2 or HepG2.2.15 cells were treated with TG for 24, 48, 72 and
96 h at 37°C. Untreated cells were used as controls. All treatments
were performed in triplicate and repeated 3 times. The cells were
collected at each time point and total RNA extraction was performed
using the Ultrapure RNA kit according to the protocol of the
manufacturer. Complementary DNA was synthesized in a 10 µl reaction
volume using ReverTra Ace qPCR RT Master Mix following the protocol
of the manufacturer. The mRNA expression levels of ATF6, ATF4, CHOP
and protein phosphatase 1 regulatory subunit 15A (GADD34) were
measured by a SYBR Green relative quantitative analysis using the
Bio-Rad iQ5 Real-Time PCR Detection System (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). GAPDH was used as an internal control.
The primers and probes used for the qPCR are listed in Table I. The RT qPCR conditions were as
follows: 95°C for 10 min, followed by 40 cycles of 92°C for 15 sec
and 58°C for 1 min. The relative expression levels of genes were
calculated by the 2−∆∆Ct method (16) and normalized by the level of the
internal control. X-box protein 1 (XBP1) microRNA (mRNA) splicing
was detected by RT qPCR as described previously (17). Theoretically, a 289 bp amplicon is
generated from unspliced XBP1, XBP1u, while a 263 bp
amplicon is generated from spliced XBP1, XBP1s (17).
 | Table I.Primers used in the mRNA
quantification. |
Table I.
Primers used in the mRNA
quantification.
| Primers and
probes | Sequence (5′-3′) |
|---|
| ATF6 |
|
| Forward
primer of PCR |
CCCAAGACTCAAACAAACTC |
| Reverse
primer of PCR |
GTGATTAGGGAGCTGTGTGA |
| ATF4 |
|
| Forward
primer of PCR |
GTGTGCGTTTTCCCTCCTC |
| Reverse
primer of PCR |
TGTCGGTTACAGCAACGCT |
| XBP1 |
|
| Forward
primer of PCR |
TTACGAGAGAAAACTCATGGC |
| Reverse
primer of PCR |
GGGTCCAAGTTGTCCAGAATGC |
| CHOP |
|
| Forward
primer of PCR |
TCTAAGGCACTGAGCGTATC |
| Reverse
primer of PCR |
CAGTCTGGAAAAGCACATCT |
| GADD34 |
|
| Forward
primer of PCR |
CCAGAAACCCCTACTCATG |
| Reverse
primer of PCR |
CAGGAAATGGACAGTGACC |
| GAPDH |
|
| Forward
primer of PCR |
TCTGACTTCAACAGCGACAC |
| Reverse
primer of PCR |
CAAATTCGTTGTCATACCAG |
Overexpression or knockdown of
CHOP
A total of 5×103 HepG2 or HepG2.2.15
cells were seeded in each well of a 96-well plate. The cells were
transfected with 0.1 µg of CHOP expression plasmids or 50 nM siRNA
with 0.2 µl Lipofectamine 2000. The cells transfected with only
Lipofectamine 2000 were set as controls. A total of 6 h subsequent
to transfection, 500 nM TG was added to the culture medium. The
cells were then collected at 24, 48, 72 and 96 h for the MTT
assay.
Western blotting
The proteins were extracted using the Mammalian
Protein Extraction kit, separated by 10% SDS-PAGE, and transferred
to a polyvinylidene fluoride membrane (cat. no., ISEQ00010; EMD
Millipore, Billerica, MA, USA). Subsequent to blocking with 5%
non-fat powdered milk, the membranes were incubated with antibodies
against CHOP (dilution, 1:1,000) and GAPDH (dilution, 1:10,000) at
4°C overnight. The membranes were washed with TBS containing 0.1%
Tween 20 and stained with the 1:10,000 IgG at 37°C for 1 h.
Micrographs were captured using the Tanon 5200 Multi (Tanon Science
and Technology Co., Ltd., Shanghai, China).
Statistical analysis
Spliced XBP1 mRNA as a percentage of the total XBP1
mRNA was estimated using ImageJ software 1.4r (National Institutes
of Health, Bethesda, MD, USA). Statistical analyses were performed
using SPSS v.15.0 (SPSS, Inc., Chicago, IL, USA) and all data were
analyzed by a one-way analysis of variance from three independent
experiments. P-values were determined using unpaired Student's
t-test. P<0.05 was considered to indicate a statistically
significant difference.
Results
HBV-positive HepG2.2.15 cells are less
susceptible to apoptosis
HBV-positive HepG2.2.15 and HBV negative HepG2 cells
were seeded in 24-well plates. Subsequent to 1 day, 500 nM TG or
PBS was added to the experimental or control groups respectively.
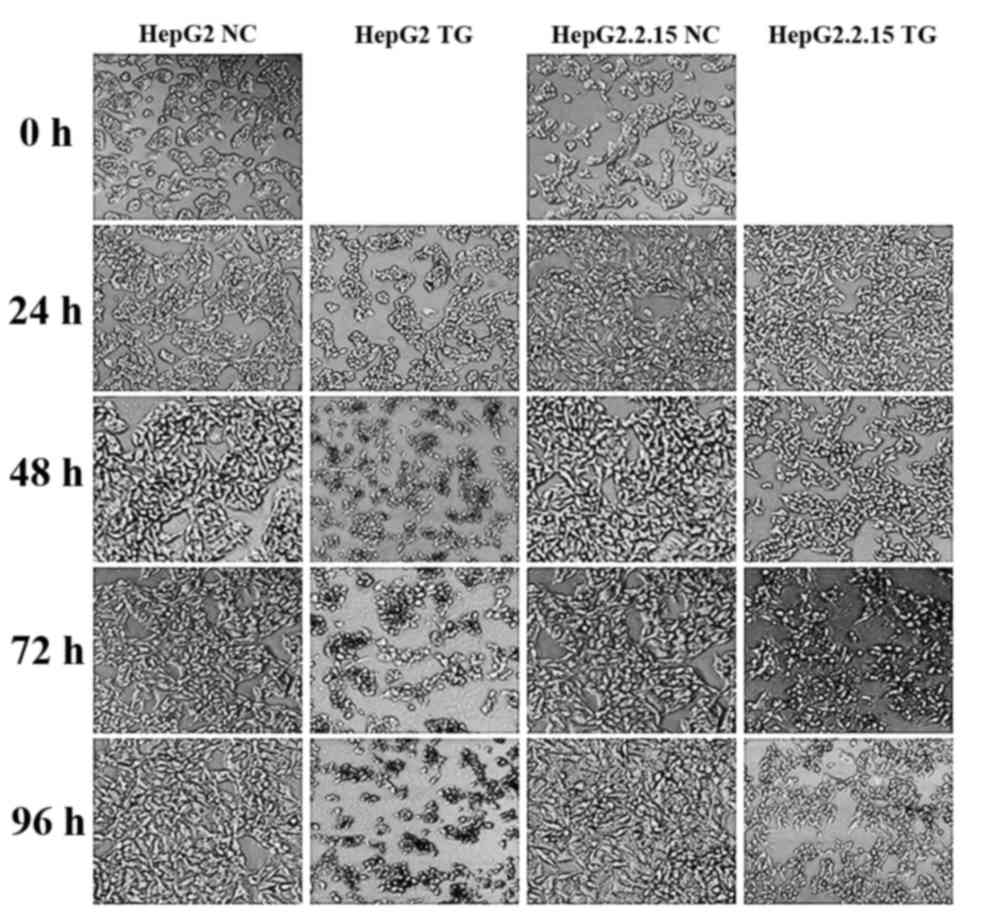
As demonstrated in Fig. 1, the HepG2
and HepG2.2.15 cells treated with PBS exhibited normal
morphologies, with clear cell membrane boundaries and homogeneous
cell cytoplasm densities, whilst the two cell types showed
apoptosis when treated with TG. Notably, the HepG2 cells underwent
more apoptosis than HepG2.2.15 cells, by morphological observation.
Accordingly, it was hypothesized that HBV may alleviate the
apoptosis induced by TG.
HBV may repress apoptosis induced by
ER stress via the CHOP pathway
To determine whether HBV affects cell survival and
apoptosis, the HepG2.2.15 and HepG2 cells were treated with 500 nM
TG or PBS for 24, 48, 72 and 96 h and cell proliferation was
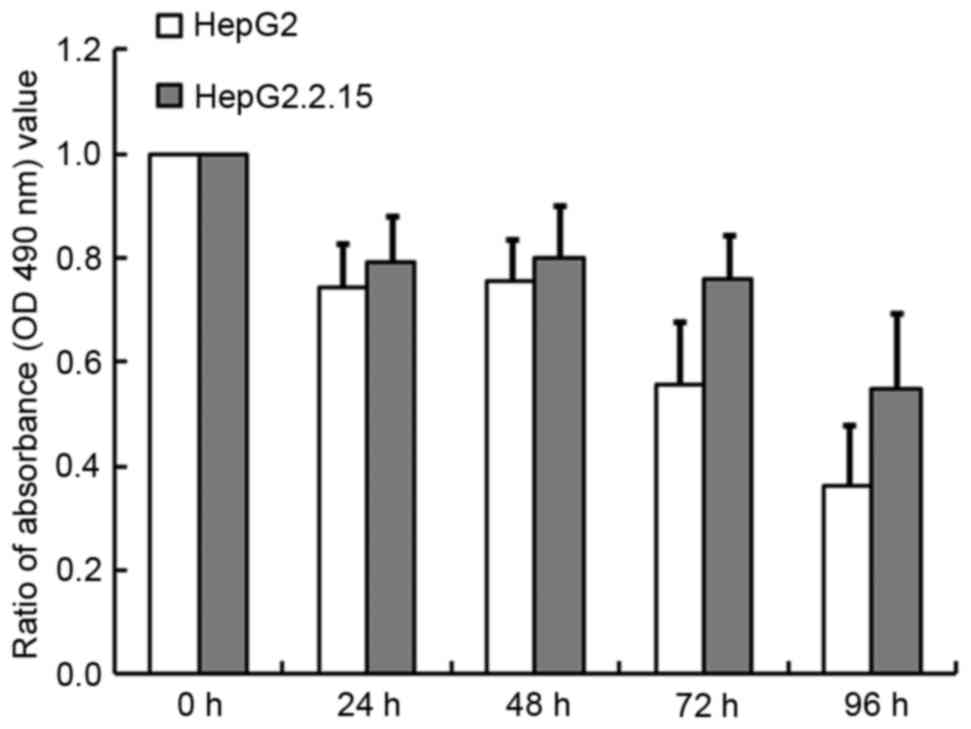
detected using the MTT assay. As illustrated in Fig. 2, TG inhibited cell growth in a
time-dependent manner, especially in the HepG2 cells, for which 64%
cell growth was inhibited at 96 h compared with 45% in the
HepG2.2.15 cells (P<0.001). Similarly, at 24, 48 and 72 h,
greater inhibition was observed for the HepG2 cells than for
HepG2.2.15 cells. These results indicated that HBV may alleviate
the induction of apoptosis by TG.
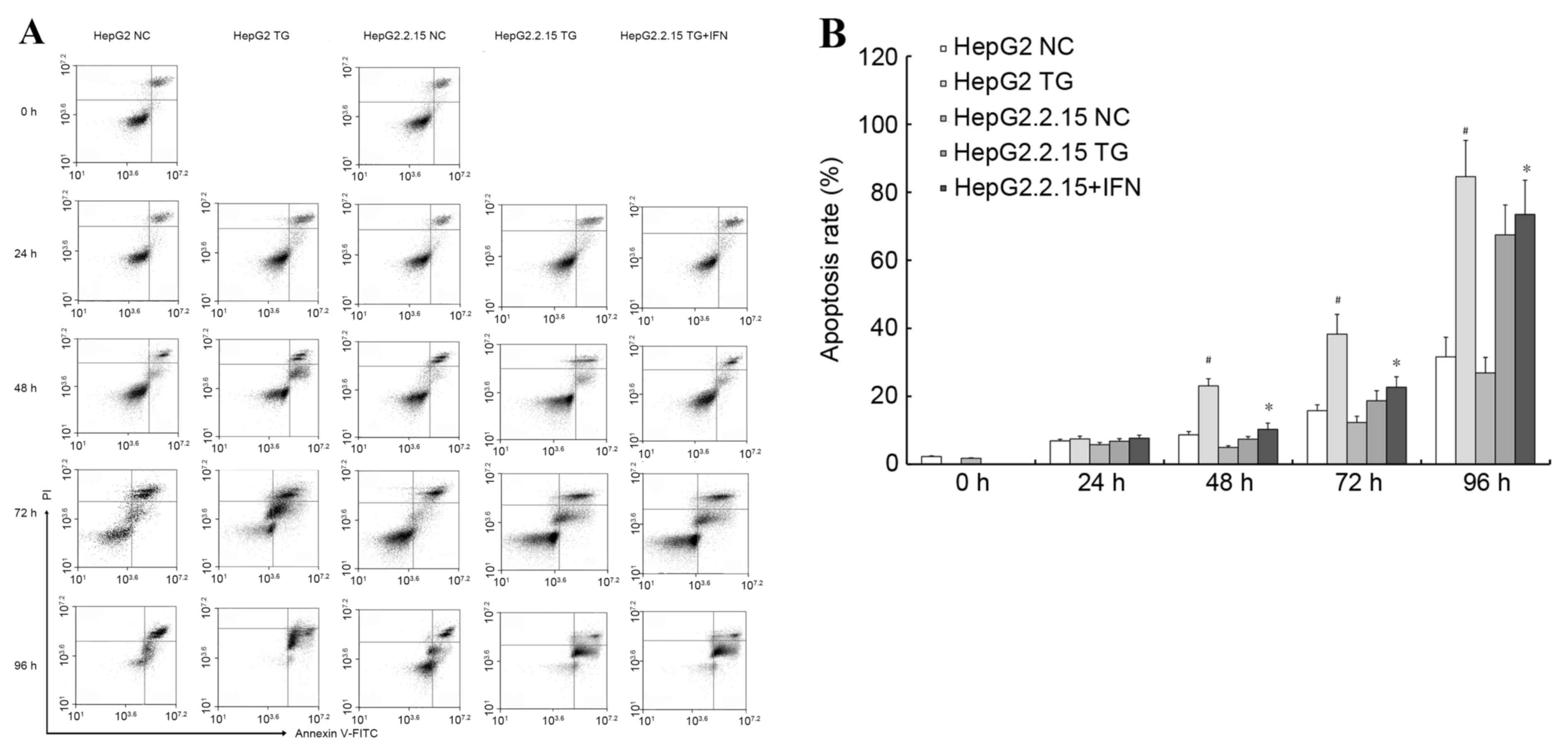
Using annexin V/PI double staining, the effect of TG
on apoptosis in the cells was then examined. Using flow cytometry,
and demonstrated in Fig. 3A, it was
revealed that TG increased the apoptotic populations of HepG2 cells
more significantly (P<0.01) compared with the HepG2.2.15 cells,
as illustrated in Fig. 3B. In
addition, to confirm the role of HBV, the HepG2.2.15 cells were
treated with IFNα-2A, an anti-HBV drug, and the aforementioned
experiments were repeated. When the HepG2.2.15 cells were treated
with IFNα-2A and TG the HBV load decreased and the apoptosis rate
increased compared with the HepG2.2.15 cells treated with TG
(P<0.05), as demonstrated in Fig.
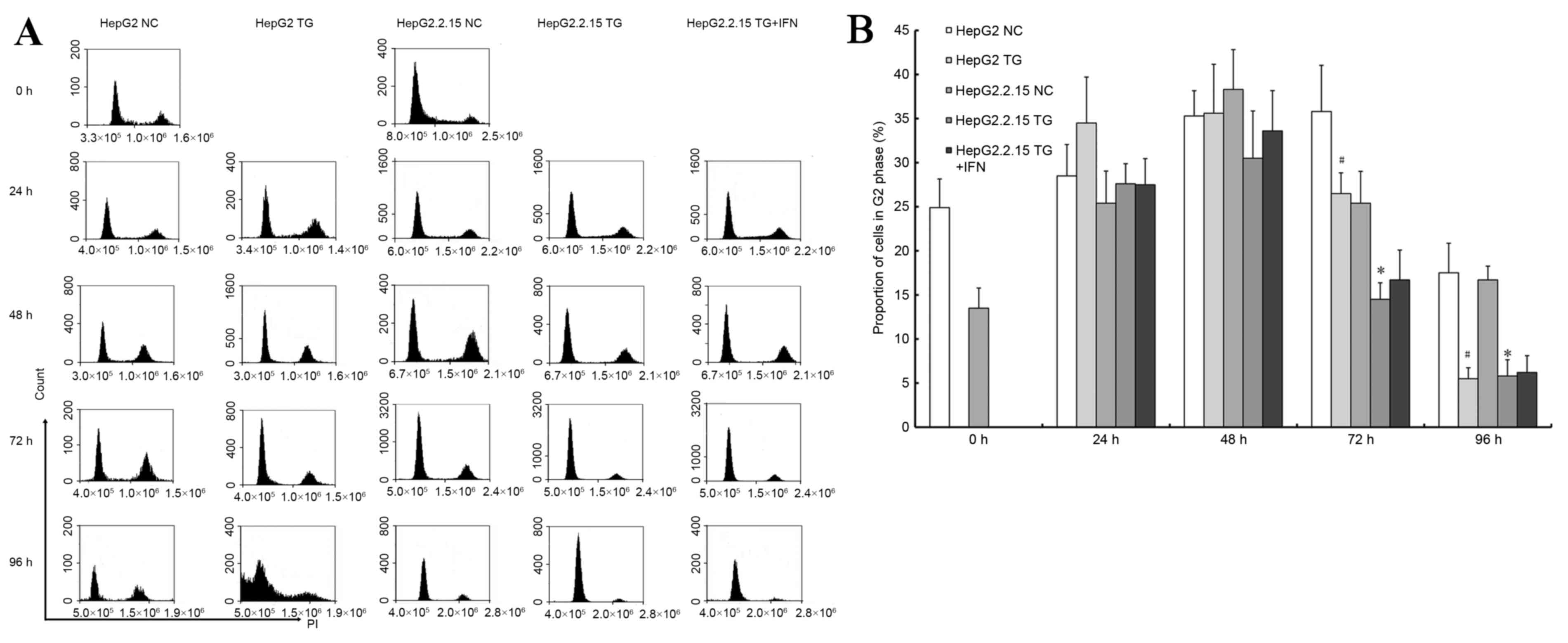
3B. PI staining was used to detect the cell cycle, as
demonstrated in Fig. 4A. TG reduced
the proportion of G2 phase cells (P<0.05), as illustrated in
Fig. 4B. In the HepG2 cells, there
were significant sub-apoptosis peaks at 96 h, which indicated that
HBV may induce apoptosis to a lesser degree. Based on the cell
cycle experiment, the HepG2.2.15 cells treated with IFNα-2A and TG
exhibited sub-apoptosis peaks at 96 h, similar to the HepG2 cells,
as illustrated in Fig. 4B. However,
no difference was observed in the results of the MTT experiment
with respect to IFNα-2A treatment: The ratio of the HepG2.2.15
cells treated with TG and IFNα-2A to the cells treated with TG
alone at each time point was between 0.99 and 1.03. These results
demonstrated that a reduced HBV load in the HepG2.2.15 cells may
increase apoptosis, or HBV may inhibit the apoptosis induced by
TG.
To explore the mechanism underlying the associations
between HBV, TG and apoptosis, the mRNA levels of 5 genes in 3 UPR
pathways: The ATF6 gene in the ATF6 pathway; the XBP1 gene in the
IRE1 pathway and the ATF4, CHOP and GADD34 genes in the PERK
pathway, were examined. The ATF6, ATF4, CHOP and GADD34 genes were
examined by qPCR, and XBP1 mRNA splicing was evaluated by RT-PCR.
The results of these analyses are summarized in Fig. 5. The mRNA levels of CHOP, GADD34 and
XBP1 increased significantly subsequent to TG treatment, as
illustrated in Fig. 5A, and the mRNA
levels of ATF6 and ATF4 exhibited increases, as demonstrated in
Fig. 5B. According to this analysis,
CHOP appeared to be the gene of greatest importance. Subsequent to
TG treatment, the level of CHOP mRNA increased by 42- to 95-fold in
the HepG2 cells, and 10- to 30-fold in the HepG2.2.15 cells. At
each time point, the level of CHOP mRNA expression was
significantly higher in the HepG2 cells compared with the
HepG2.2.15 cells (P<0.001). When the HepG2.2.15 cells were
treated with IFNα-2A and TG, the CHOP mRNA expression levels were
1.16 and 1.11-fold higher than the expression levels without
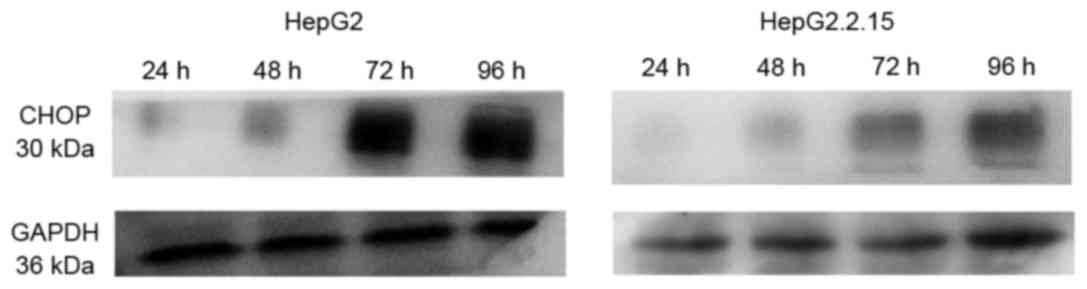
IFNα-2A at 72 and 96 h (P<0.05). Western blotting also confirmed
that the protein levels of CHOP were higher in the HepG2 cells than
in the HepG2.2.15 cells at each time point subsequent to treatment
with TG, as demonstrated in Fig.
6.
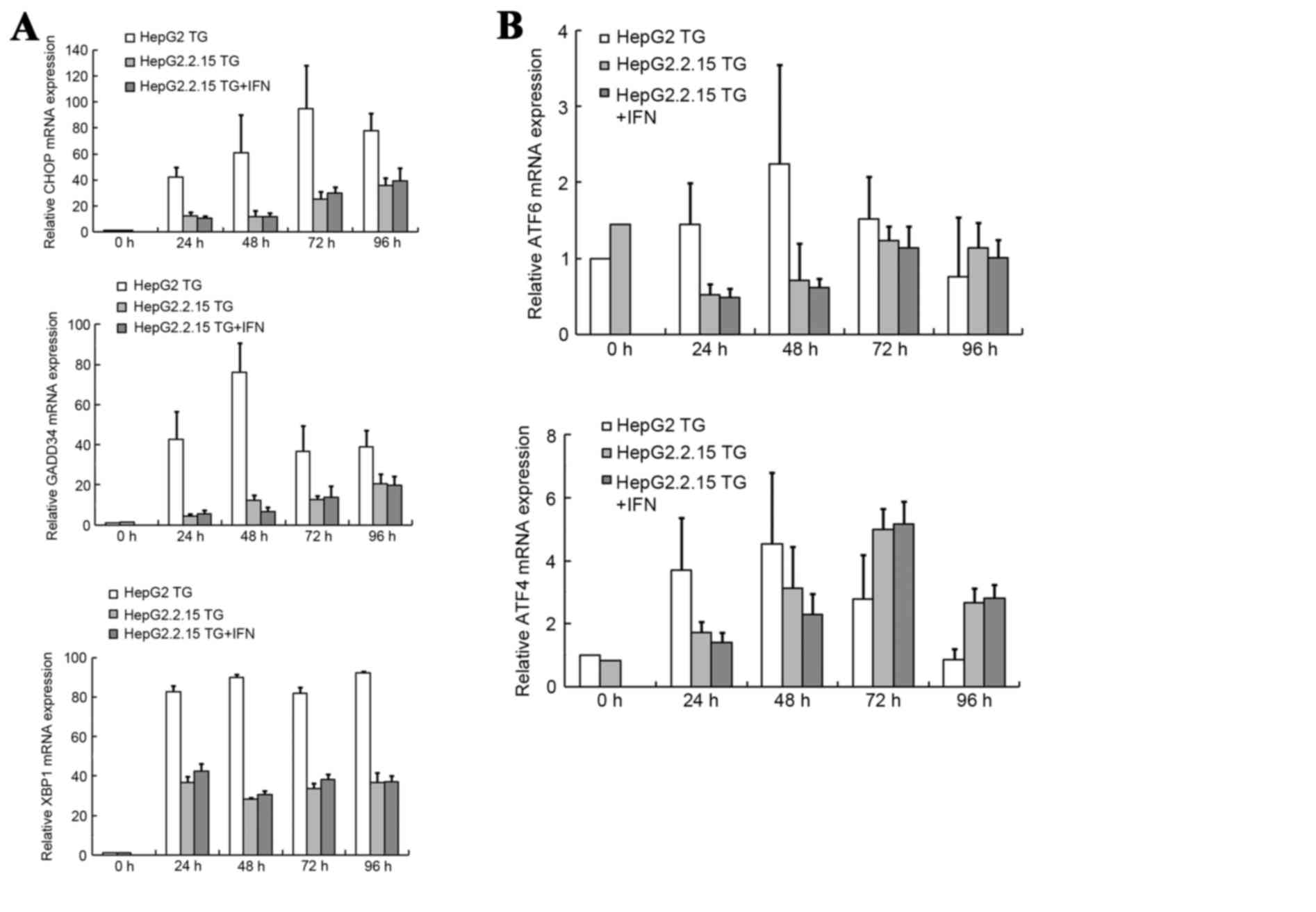 | Figure 5.mRNA expression levels of genes in the
3 UPR pathways. (A) mRNA expression levels of CHOP and GADD34 were
tested by qPCR, and XBP1 mRNA splicing was tested by reverse
transcription PCR. (B) mRNA expression levels of ATF6 and ATF4 were
tested by qPCR. Final abundances were adjusted to yield an
arbitrary value of 1 for each gene in the HepG2 cells. mRNA, micro
RNA; negative control; TG, thapsigargin; IFN, interferon; CHOP, DNA
damage inducible transcript 3; GADD34, protein phosphatase 1
regulatory subunit 15A; XBP1, X-box binding protein; ATF4,
activating transcription factor 4; ATF6, activating transcription
factor 6; qPCR, quantitative polymerase chain reaction. |
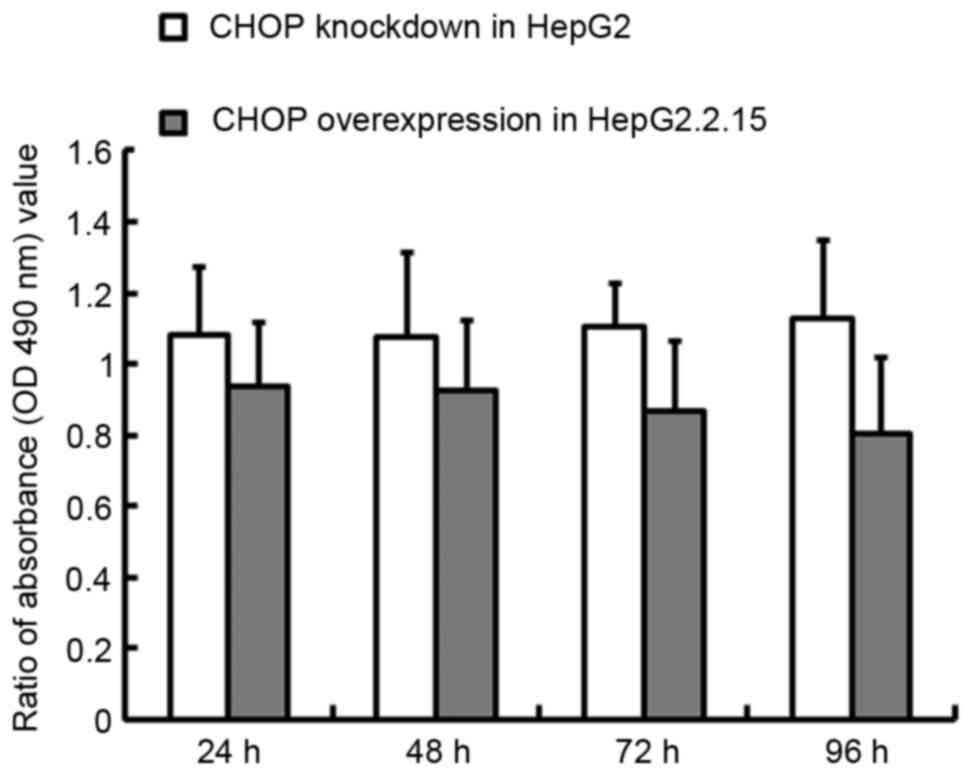
CHOP overexpression in HepG2.2.15
cells or knockdown in HepG2 cells affects proliferation
When CHOP was overexpressed in the HepG2.2.15 cells,
the proliferation rate decreased by 6.5% at 24 h and 19.6% at 96 h,
relative to the wild-type cells (P<0.01), whilst in contrast,
CHOP knockdown in the HepG2 cells increased proliferation 8.1% at
24 h and 12.5% at 96 h (P<0.05), as demonstrated in Fig. 7.
Discussion
The results of the present study indicated that HBV
may inhibit the apoptosis induced by ER stress via the repression
of CHOP. CHOP is a member of the CCAAT/enhancer-binding protein
(C/EBP) family of transcription factors (18), and functions as a dominant-negative
inhibitor by forming heterodimers with other C/EBP members and
preventing their DNA binding activity (19). CHOP is activated by ER stress, and
promotes apoptosis. The activation of PERK increases the level of
the phosphorylation of eukaryotic initiation factor 2 (eIF2),
leading to an increase in the level of ATF4 translation. In turn,
ATF4 induces the expression of CHOP (18). ATF4 and CHOP transactivate GADD34
(20), which selectively
dephosphorylates eIF2α, completing a negative feedback loop and
promoting the translation of other UPR genes. Whether CHOP promotes
or inhibits oncogenesis is controversial. A previous study has
revealed that pharmacological ER stresses that induce CHOP may kill
cancer cells, including hepatomas, in vitro (21). However, in additional studies, CHOP
appears to promote oncogenesis (22,23). In
the present study, it was demonstrated that increased levels of
CHOP expression may have promoted HCC cell apoptosis, as summarized
in Figs. 2–7, suggesting an antitumor role of CHOP.
According to a previous study (13), HBV induces ER stress independently,
but the regulatory mechanisms of HBV-infected cells may be
activated to reduce ER stress. Previous studies have investigated
the pathological effect of HBV surface protein expression on the
liver. In the livers of BALB/c transgenic mice, the expression of
the HBV surface protein activates the PERK pathway and results in
the expression of CHOP, leading to more extensive liver injury and
fibrosis compared with transgenic mice with the C57BL/6 background
(24). In another study using
hepatoma cells, HBV small surface proteins triggered UPR, activated
the PERK pathway and induced the phosphorylation of eIF2α, which
promotes the expression of CHOP (25). In TG treated HepG2.2.15 cells, the
present study demonstrated that HBV reduces the expression of CHOP.
This affects liver cancer cell apoptosis.
The present study contained a number of limitations.
HepG2.2.15 cells were derived from HepG2 cells, and were stably
transformed with 2 copies of the HBV genome (26). The culture medium of the HepG2.2.15
cells stably expressed HBV particles, hepatitis B surface antigen
and Hepatitis B envelope antigen, but at low concentrations.
Therefore, when the cells were treated with IFNα-2A, the antiviral
effect was not measured due to the baseline HBV concentration being
low. Additionally, these HBV markers are encoded by 2 copies of the
HBV genome, stably transformed into the genomes, which is
dissimilar to the natural progression of HBV infection in the human
liver. Previously, the Na+-taurocholate cotransporting
polypeptide (NTCP) was identified as a functional receptor for
human HBV, a topic that requires attention (27). In future studies, HepG2 cells may be
transfected with NTCP to increase the expression of HBV in the
culture medium, simulating the natural history of HBV infection.
In vivo studies should also be conducted to verify the role
of HBV during ER stress.
In conclusion, the present study demonstrated that
HBV may inhibit the cell apoptosis induced by ER stress, which is
important for the development of ER stress based antitumor
therapies for patients with HBV.
Acknowledgements
The present study was supported by grants from the
National Basic Research Program of China (973 Program; grant nos.,
2012CB519005 and 2013CB944903).
Glossary
Abbreviations
Abbreviations:
|
HBV
|
hepatitis B virus
|
|
HCC
|
hepatocellular carcinoma
|
|
ER
|
endoplasmic reticulum
|
|
UPR
|
unfolded protein response
|
|
TG
|
thapsigargin
|
|
PERK
|
protein kinase-like endoplasmic
reticulum kinase
|
|
IRE1
|
inositol-requiring enzyme 1
|
|
ATF4
|
activating transcription factor 4
|
|
ATF6
|
activating transcription factor 6
|
|
CHOP
|
DNA damage inducible transcript 3
|
|
GADD34
|
protein phosphatase 1 regulatory
subunit 15A
|
References
|
1
|
Toyoshima C, Nomura H and Sugita Y:
Crystal structures of Ca2+-ATPase in various physiological states.
Ann N Y Acad Sci. 986:1–8. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Winther AM, Liu H, Sonntag Y, Olesen C, le
Maire M, Soehoel H, Olsen CE, Christensen SB, Nissen P and Møller
JV: Critical roles of hydrophobicity and orientation of side chains
for inactivation of sarcoplasmic reticulum Ca2+-ATPase with
thapsigargin and thapsigargin analogs. J Biol Chem.
285:28883–28892. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rajapaksa G, Nikolos F, Bado I, Clarke R,
Gustafsson JÅ and Thomas C: ERβ decreases breast cancer cell
survival by regulating the IRE1/XBP-1 pathway. Oncogene.
34:4130–4141. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Senft D and Ronai ZA: UPR, autophagy, and
mitochondria crosstalk underlies the ER stress response. Trends
Biochem Sci. 40:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Romero-Ramirez L, Cao H, Regalado MP,
Kambham N, Siemann D, Kim JJ, Le QT and Koong AC: X box-binding
protein 1 regulates angiogenesis in human pancreatic
adenocarcinomas. Transl Oncol. 2:31–38. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Denmeade SR, Mhaka AM, Rosen DM, Brennen
WN, Dalrymple S, Dach I, Olesen C, Gurel B, Demarzo AM, Wilding G,
et al: Engineering a prostate-specific membrane antigen-activated
tumor endothelial cell prodrug for cancer therapy. Sci Transl Med.
4:140ra1862012. View Article : Google Scholar
|
|
7
|
Park SW and Ozcan U: Potential for
therapeutic manipulation of the UPR in disease. Semin Immunopathol.
35:351–373. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen CJ and Chen DS: Interaction of
hepatitis B virus, chemical carcinogen, and genetic susceptibility:
Multistage hepatocarcinogenesis with multifactorial etiology.
Hepatology. 36:1046–1049. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang WA, Groenendyk J and Michalak M:
Endoplasmic reticulum stress associated responses in cancer.
Biochim Biophys Acta. 1843:2143–2149. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu YH, Weng YP, Lin HY, Tang SW, Chen CJ,
Liang CJ, Ku CY and Lin JY: Aqueous extract of Polygonum bistorta
modulates proteostasis by ROS-induced ER stress in human hepatoma
cells. Sci Rep. 7:414372017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tameire F, Verginadis II and Koumenis C:
Cell intrinsic and extrinsic activators of the unfolded protein
response in cancer: Mechanisms and targets for therapy. Semin
Cancer Biol. 33:3–15. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lazar C, Uta M and Branza-Nichita N:
Modulation of the unfolded protein response by the human hepatitis
B virus. Front Microbiol. 5:4332014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sunami Y, Ringelhan M, Kokai E, Lu M,
O'Connor T, Lorentzen A, Weber A, Rodewald AK, Mullhaupt B,
Terracciano L, et al: Canonical NF-κB signaling in hepatocytes acts
as a tumor-suppressor in hepatitis B virus surface antigen-driven
hepatocellular carcinoma by controlling the unfolded protein
response. Hepatology. 63:1592–1607. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yeganeh B, Rezaei Moghadam A, Alizadeh J,
Wiechec E, Alavian SM, Hashemi M, Geramizadeh B, Samali A, Bagheri
Lankarani K, Post M, et al: Hepatitis B and C virus-induced
hepatitis: Apoptosis, autophagy, and unfolded protein response.
World J Gastroenterol. 21:13225–13239. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin JH, Li H, Yasumura D, Cohen HR, Zhang
C, Panning B, Shokat KM, Lavail MM and Walter P: IRE1 signaling
affects cell fate during the unfolded protein response. Science.
318:944–949. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Y, Guo Y, Tang J, Jiang J and Chen Z:
New insights into the roles of CHOP-induced apoptosis in ER stress.
Acta Biochim Biophys Sin (Shanghai). 46:629–640. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sheedy C, Mooney C, Jimenez-Mateos E,
Sanz-Rodriguez A, Langa E, Mooney C and Engel T: De-repression of
myelin-regulating gene expression after status epilepticus in mice
lacking the C/EBP homologous protein CHOP. Int J Physiol
Pathophysiol Pharmacol. 6:185–198. 2014.PubMed/NCBI
|
|
20
|
Marciniak SJ, Yun CY, Oyadomari S, Novoa
I, Zhang Y, Jungreis R, Nagata K, Harding HP and Ron D: CHOP
induces death by promoting protein synthesis and oxidation in the
stressed endoplasmic reticulum. Genes Dev. 18:3066–3077. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Moon DO, Park SY, Choi YH, Ahn JS and Kim
GY: Guggulsterone sensitizes hepatoma cells to TRAIL-induced
apoptosis through the induction of CHOP-dependent DR5: Involvement
of ROS-dependent ER-stress. Biochem Pharmacol. 82:1641–1650. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Crozat A, Aman P, Mandahl N and Ron D:
Fusion of CHOP to a novel RNA-binding protein in human myxoid
liposarcoma. Nature. 363:640–644. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Panagopoulos I, Höglund M, Mertens F,
Mandahl N, Mitelman F and Aman P: Fusion of the EWS and CHOP genes
in myxoid liposarcoma. Oncogene. 12:489–494. 1996.PubMed/NCBI
|
|
24
|
Churin Y, Roderfeld M, Stiefel J, Würger
T, Schröder D, Matono T, Mollenkopf HJ, Montalbano R, Pompaiah M,
Reifenberg K, et al: Pathological impact of hepatitis B virus
surface proteins on the liver is associated with the host genetic
background. PLoS One. 9:e906082014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li J, Liu Y, Wang Z, Liu K, Wang Y, Liu J,
Ding H and Yuan Z: Subversion of cellular autophagy machinery by
hepatitis B virus for viral envelopment. J Virol. 85:6319–6333.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sells MA, Chen ML and Acs G: Production of
hepatitis B virus particles in Hep G2 cells transfected with cloned
hepatitis B virus DNA. Proc Natl Acad Sci USA. 84:1005–1009. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z,
Huang Y, Qi Y, Peng B, Wang H, et al: Sodium taurocholate
cotransporting polypeptide is a functional receptor for human
hepatitis B and D virus. Elife. 1:e000492012. View Article : Google Scholar : PubMed/NCBI
|