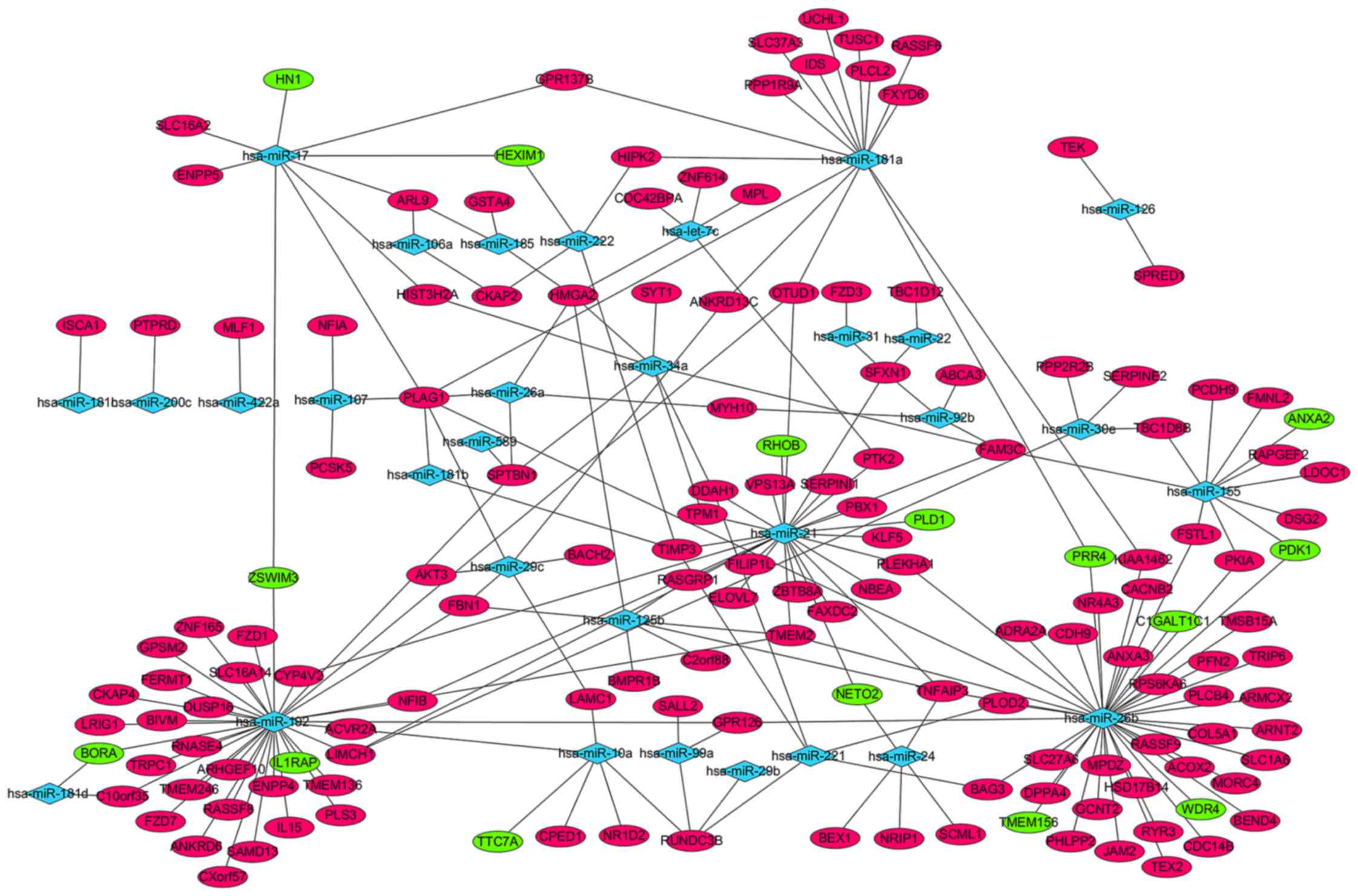
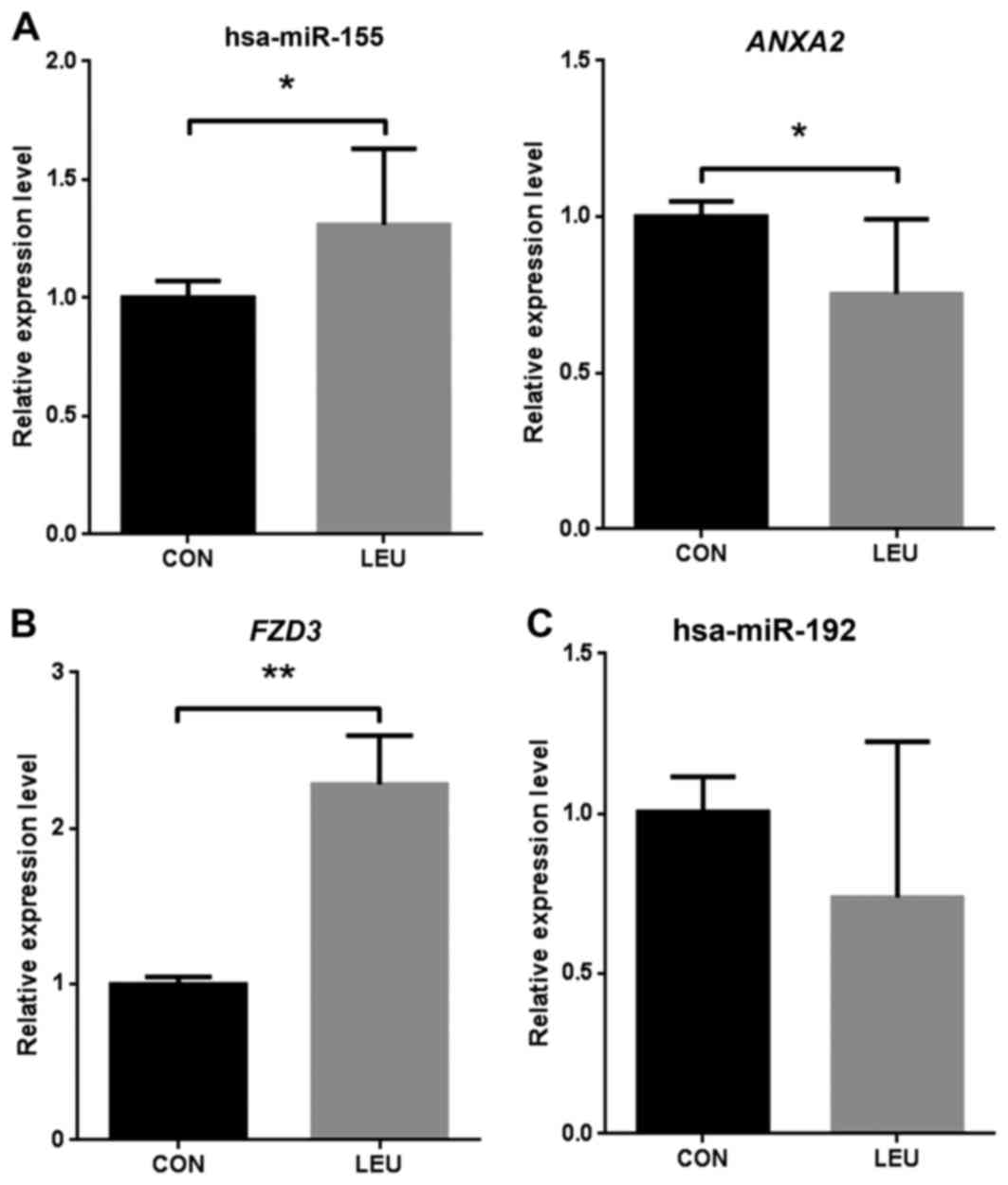
|
1
|
Chen W, Zheng R, Zeng H and Zhang S: The
updated incidences and mortalities of major cancers in China, 2011.
Chin J Cancer. 34:507. 2015. View Article : Google Scholar
|
|
2
|
Cripe LD: Adult acute leukemia. Curr Probl
Cancer. 21:1–64. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marcucci G, Haferlach T and Döhner H:
Molecular genetics of adult acute myeloid leukemia: Prognostic and
therapeutic implications. J Clin Oncol. 29:475–486. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Deguchi K and Gilliland DG: Cooperativity
between mutations in tyrosine kinases and in hematopoietic
transcription factors in AML. Leukemia. 16:740–744. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Finn L, Sproat L, Heckman MG, Jiang L,
Diehl NN, Ketterling R, Tibes R, Valdez R and Foran J: Epidemiology
of adult acute myeloid leukemia: Impact of exposures on clinical
phenotypes and outcomes after therapy. Cancer Epidemiol.
39:1084–1092. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bennett JM, Catovsky D, Daniel MT,
Flandrin G, Galton DA, Gralnick HR and Sultan C: Proposals for the
classification of the acute leukaemias. French-American-British
(FAB) co-operative group. Br J Haematol. 33:451–458. 1976.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vardiman JW, Thiele J, Arber DA, Brunning
RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM,
Hellström-Lindberg E, Tefferi A and Bloomfield CD: The 2008
revision of the World Health Organization (WHO) classification of
myeloid neoplasms and acute leukemia: Rationale and important
changes. Blood. 114:937–951. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Marcucci G, Mrózek K, Ruppert AS, Maharry
K, Kolitz JE, Moore JO, Mayer RJ, Pettenati MJ, Powell BL, Edwards
CG, et al: Prognostic factors and outcome of core binding factor
acute myeloid leukemia patients with t(8;21) differ from those of
patients with inv(16): A Cancer and Leukemia Group B study. J Clin
Oncol. 23:5705–5717. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stone RM: Prognostic factors in AML in
relation to (ab)normal karyotype. Best Pract Res Clin Haematol.
22:523–528. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schanz J and Haase D: Cytogenetic features
in myelodysplastic syndromes. 2014.
|
|
11
|
Care RS, Valk PJM, Goodeve AC, Abu-Duhier
FM, Geertsma-Kleinekoort WM, Wilson GA, Gari MA, Peake IR,
Löwenberg B and Reilly JT: Incidence and prognosis of c-KIT and
FLT3 mutations in core binding factor (CBF) acute myeloid
leukaemias. Br J Haematol. 121:775–777. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Langer C, Radmacher MD, Ruppert AS,
Whitman SP, Paschka P, Mrózek K, Baldus CD, Vukosavljevic T, Liu
CG, Ross ME, et al: High BAALC expression associates with other
molecular prognostic markers, poor outcome, and a distinct
gene-expression signature in cytogenetically normal patients
younger than 60 years with acute myeloid leukemia: A Cancer and
Leukemia Group B (CALGB) study. Blood. 111:5371–5379. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Preudhomme C, Sagot C, Boissel N, Cayuela
JM, Tigaud I, de Botton S, Thomas X, Raffoux E, Lamandin C,
Castaigne S, et al: Favorable prognostic significance of CEBPA
mutations in patients with de novo acute myeloid leukemia: A study
from the acute leukemia French association (ALFA). Blood.
100:2717–2723. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Döhner K, Tobis K, Ulrich R, Fröhling S,
Benner A, Schlenk RF and Döhner H: Prognostic significance of
partial tandem duplications of the MLL gene in adult patients 16 to
60 years old with acute myeloid leukemia and normal cytogenetics: A
study of the acute myeloid leukemia study group Ulm. J Clin Oncol.
20:3254–3261. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ufkin ML, Peterson S, Yang X, Driscoll H,
Duarte C and Sathyanarayana P: miR-125a regulates cell cycle,
proliferation, and apoptosis by targeting the ErbB pathway in acute
myeloid leukemia. Leuk Res. 38:402–410. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Morris VA, Zhang A, Yang T, Stirewalt DL,
Ramamurthy R, Meshinchi S and Oehler VG: MicroRNA-150 expression
induces myeloid differentiation of human acute leukemia cells and
normal hematopoietic progenitors. PLoS One. 8:e758152013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hsu SD, Lin FM, Wu WY, Liang C, Huang WC,
Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al: miRTarBase: A
database curates experimentally validated microRNA-target
interactions. Nucleic Acids Res. 39:(Database Issue). D163–D169.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hsu SD, Tseng YT, Shrestha S, Lin YL,
Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al:
miRTarBase update 2014: An information resource for experimentally
validated miRNA-target interactions. Nucleic Acids Res.
42:(Database Issue). D78–D85. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng L, Lin H, Hu Y, Wang J and Yang Z:
Gene function prediction based on the Gene Ontology hierarchical
structure. PLoS One. 9:e1071872014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kanehisa M, Sato Y, Kawashima M, Furumichi
M and Tanabe M: KEGG as a reference resource for gene and protein
annotation. Nucleic Acids Res. 44:D457–D462. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zheng Q and Wang XJ: GOEAST: A web-based
software toolkit for Gene Ontology enrichment analysis. Nucleic
Acids Res. 36:W358–W363. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Carmona-Saez P, Chagoyen M, Tirado F,
Carazo JM and Pascual-Montano A: GENECODIS: A web-based tool for
finding significant concurrent annotations in gene lists. Genome
Biol. 8:R32007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cramer-Morales K, Nieborowska-Skorska M,
Scheibner K, Padget M, Irvine DA, Sliwinski T, Haas K, Lee J, Geng
H, Roy D, et al: Personalized synthetic lethality induced by
targeting RAD52 in leukemias identified by gene mutation and
expression profile. Blood. 122:1293–1304. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Barreyro L, Will B, Bartholdy B, Zhou L,
Todorova TI, Stanley RF, Ben-Neriah S, Montagna C, Parekh S,
Pellagatti A, et al: Overexpression of IL-1 receptor accessory
protein in stem and progenitor cells and outcome correlation in AML
and MDS. Blood. 120:1290–1298. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kikushige Y, Shima T, Takayanagi S, Urata
S, Miyamoto T, Iwasaki H, Takenaka K, Teshima T, Tanaka T, Inagaki
Y and Akashi K: TIM-3 is a promising target to selectively kill
acute myeloid leukemia stem cells. Cell Stem Cell. 7:708–717. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Majeti R, Becker MW, Tian Q, Lee TL, Yan
X, Liu R, Chiang JH, Hood L, Clarke MF and Weissman IL:
Dysregulated gene expression networks in human acute myelogenous
leukemia stem cells. Proc Natl Acad Sci USA. 106:3396–3401. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tan YS, Kim M, Kingsbury TJ, Civin CI and
Cheng WC: Regulation of RAB5C is important for the growth
inhibitory effects of MiR-509 in human precursor-B acute
lymphoblastic leukemia. PLoS One. 9:e1117772014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Havelange V, Stauffer N, Heaphy CC,
Volinia S, Andreeff M, Marcucci G, Croce CM and Garzon R:
Functional implications of microRNAs in acute myeloid leukemia by
integrating microRNA and messenger RNA expression profiling.
Cancer. 117:4696–4706. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Forrest AR, Kanamori-Katayama M, Tomaru Y,
Lassmann T, Ninomiya N, Takahashi Y, de Hoon MJ, Kubosaki A, Kaiho
A, Suzuki M, et al: Induction of microRNAs, mir-155, mir-222,
mir-424 and mir-503, promotes monocytic differentiation through
combinatorial regulation. Leukemia. 24:460–466. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
O'Connell RM, Rao DS, Chaudhuri AA, Boldin
MP, Taganov KD, Nicoll J, Paquette RL and Baltimore D: Sustained
expression of microRNA-155 in hematopoietic stem cells causes a
myeloproliferative disorder. J Exp Med. 205:585–594. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Marcucci G, Maharry KS, Metzeler KH,
Volinia S, Wu YZ, Mrózek K, Nicolet D, Kohlschmidt J, Whitman SP,
Mendler JH, et al: Clinical role of microRNAs in cytogenetically
normal acute myeloid leukemia: miR-155 upregulation independently
identifies high-risk patients. J Clin Oncol. 31:2086–2093. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chuang MK, Chiu YC, Chou WC, Hou HA,
Chuang EY and Tien HF: A 3-microRNA scoring system for
prognostication in de novo acute myeloid leukemia patients.
Leukemia. 29:1051–1059. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhi F, Cao X, Xie X, Wang B, Dong W, Gu W,
Ling Y, Wang R, Yang Y and Liu Y: Identification of circulating
microRNAs as potential biomarkers for detecting acute myeloid
leukemia. PLoS One. 8:e567182013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Qu YL, Wang HF, Sun ZQ, Tang Y, Han XN, Yu
XB and Liu K: Up-regulated miR-155-5p promotes cell proliferation,
invasion and metastasis in colorectal carcinoma. Int J Clin Exp
Pathol. 8:6988–6994. 2015.PubMed/NCBI
|
|
39
|
Baba O, Hasegawa S, Nagai H, Uchida F,
Yamatoji M, Kanno NI, Yamagata K, Sakai S, Yanagawa T and Bukawa H:
MicroRNA-155-5p is associated with oral squamous cell carcinoma
metastasis and poor prognosis. J Oral Pathol Med. 45:248–255. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Merhautova J, Hezova R, Poprach A,
Kovarikova A, Radova L, Svoboda M, Vyzula R, Demlova R and Slaby O:
miR-155 and miR-484 are associated with time to progression in
metastatic renal cell carcinoma treated with sunitinib. Biomed Res
Int. 2015:9419802015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Park MH, Cho SA, Yoo KH, Yang MH, Ahn JY,
Lee HS, Lee KE, Mun YC, Cho DH, Seong CM and Park JH: Gene
expression profile related to prognosis of acute myeloid leukemia.
Oncol Rep. 18:1395–1402. 2007.PubMed/NCBI
|
|
42
|
Conacci-Sorrell M, Zhurinsky J and
Ben-Ze'ev A: The cadherin-catenin adhesion system in signaling and
cancer. J Clin Invest. 109:987–991. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Giles RH, van Es JH and Clevers H: Caught
up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta.
1653:1–24. 2003.PubMed/NCBI
|
|
44
|
Kühnl A, Valk PJ, Sanders MA, Ivey A,
Hills RK, Mills KI, Gale RE, Kaiser MF, Dillon R, Joannides M, et
al: Downregulation of the Wnt inhibitor CXXC5 predicts a better
prognosis in acute myeloid leukemia. Blood. 125:2985–2994. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang Y, Krivtsov AV, Sinha AU, North TE,
Goessling W, Feng Z, Zon LI and Armstrong SA: The Wnt/beta-catenin
pathway is required for the development of leukemia stem cells in
AML. Science. 327:1650–1653. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Simon M, Grandage VL, Linch DC and Khwaja
A: Constitutive activation of the Wnt/beta-catenin signalling
pathway in acute myeloid leukaemia. Oncogene. 24:2410–2420. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ye M and Zhang J and Zhang J, Miao Q, Yao
L and Zhang J: Curcumin promotes apoptosis by activating the
p53-miR-192-5p/215-XIAP pathway in non-small cell lung cancer.
Cancer Lett. 357:196–205. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jin H, Qiao F, Wang Y, Xu Y and Shang Y:
Curcumin inhibits cell proliferation and induces apoptosis of human
non-small cell lung cancer cells through the upregulation of
miR-192-5p and suppression of PI3K/Akt signaling pathway. Oncol
Rep. 34:2782–2789. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kas K, Voz ML, Hensen K, Meyen E and Van
de Ven WJ: Transcriptional activation capacity of the novel PLAG
family of zinc finger proteins. J Biol Chem. 273:23026–23032. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Landrette SF, Kuo YH, Hensen K, van
Waalwijk van Doorn-Khosrovani Barjesteh S, Perrat PN, Van de Ven
WJ, Delwel R and Castilla LH: Plag1 and Plagl2 are oncogenes that
induce acute myeloid leukemia in cooperation with Cbfb-MYH11.
Blood. 105:2900–2907. 2005. View Article : Google Scholar : PubMed/NCBI
|