Introduction
Oncocytoma is a rare benign neoplasm that may arise
in the glandular epithelium of salivary glands, thyroid,
parathyroid, kidney, adrenal cortex, and in the ocular region
(1–3).
The oncocytic cells (oncocytes, from the Greek onkoustai,
meaning to swell) have a swollen appearance, and are characterised
by a large number of morphologically abnormal and possibly
dysfunctional mitochondria (2,4). Ocular
adnexal oncocytoma is rare and occurs most frequently in the
caruncle, although lesions have also been identified on the eyelid
margin, the conjunctiva, in the lacrimal sac, and in the lacrimal
gland (1). In 1959, the first case of
a lacrimal gland oncocytoma was reported by Beskid and Zarzycka
(5), since then 12 additional cases
have been reported (Table I)
(5–16). The molecular pathogenesis of lacrimal
gland oncocytoma, and the mechanisms leading to the combined
intracellular mitochondrial proliferation and the proliferation of
the oncocyte itself remains unclear (2). It has been suggested that mitochondria
may be involved in oncocytoma pathogenesis, and several studies
have identified multiple sequence variants in mitochondrial DNA
(mtDNA), particularly in genes involved in the cellular respiration
complex I (2,4,16–18). In the present study, a case of
lacrimal gland oncocytoma was described, and a detailed
immunohistochemical profile was presented along with a genetic
profile of the tumor based on high-resolution array comparative
genomic hybridisation (aCGH) of nuclear DNA and next-generation
sequencing of the mitochondrial genome.
 | Table I.Previously published cases of
oncocytoma of the lacrimal gland. |
Table I.
Previously published cases of
oncocytoma of the lacrimal gland.
| Authors | Age (years) | Gender | Symptoms | Duration | Size (mm) | Treatment | Follow-up
(months) | Recurrence | (Refs.) |
|---|
| Beskid and Zarzycka
(1959) | 39 | F | Proptosis | 8 years | N/A | Excision of a
previously, partially excised lacrimal gland tumor | 20 | No | (5) |
| Riedel et al
(1983) | 1.5 | F | Proptosis | 2 months | N/A | Lateral
orbitotomy | 3 | No | (6) |
| Riedel et al
(1983) | 76 | F | Lid swelling | 3 months | 10×10 | Anterior
orbitotomy | 42 | No | (6) |
| Hartman et al
(2003) | 72 | M | Lid swelling,
diplopia | 9 months | 28×30×19 | Lateral
orbitotomy | 18 | No | (7) |
| Calle et al
(2006) | 40 | F | Oedema, pain | 7 months | 24×13 | Lateral
orbitotomy | 21 | No | (8) |
| Archondakis et
al (2009) | 83 | M | Orbital mass | 3 months | 10 Ø | Complete
excision | N/A | N/A | (9) |
| Economou et al
(2007) | 68 | M | Proptosis | 6 months | 10×10×10 | Anterior
orbitotomy | 24 | No | (10) |
| Kim et al
(2010) | 64 | F | Lid swelling,
ptosis | 7 years | 17×24×21 | Lateral
orbitotomy | 13 | No | (11) |
| Aghaji et al
(2011) | 60 | F | Lid swelling | 3 years | 50×50 | Modified
exenteration | N/A | N/A | (12) |
| Limb et al
(2013) | 19 | M | Proptosis | 10 years | Giant, NOS | Subtotal
fronto-orbitozygomatic craniotomy | N/A | N/A | (13) |
| Ferté et al
(2016) | 57 | M | Lid swelling | 6 months | N/A | Anterior
orbitotomy | 4 | No | (14) |
| Jittapiromsak et
al (2017) | 37 | F | Proptosis | 12 months | 33×16 | Lateral
orbitotomy | 2 | No | (15) |
| Present case | 20 | M | Proptosis | 2> years | 25×22×17 | Lateral
orbitotomy | 4 | No | – |
Materials and methods
Clinical history
A 20-year-old male was hospitalised from January 3rd
to January 21st 2015 (Rigshospitalet, Copenhagen University
Hospital, Denmark) due to encephalitic symptoms. Unexpectedly,
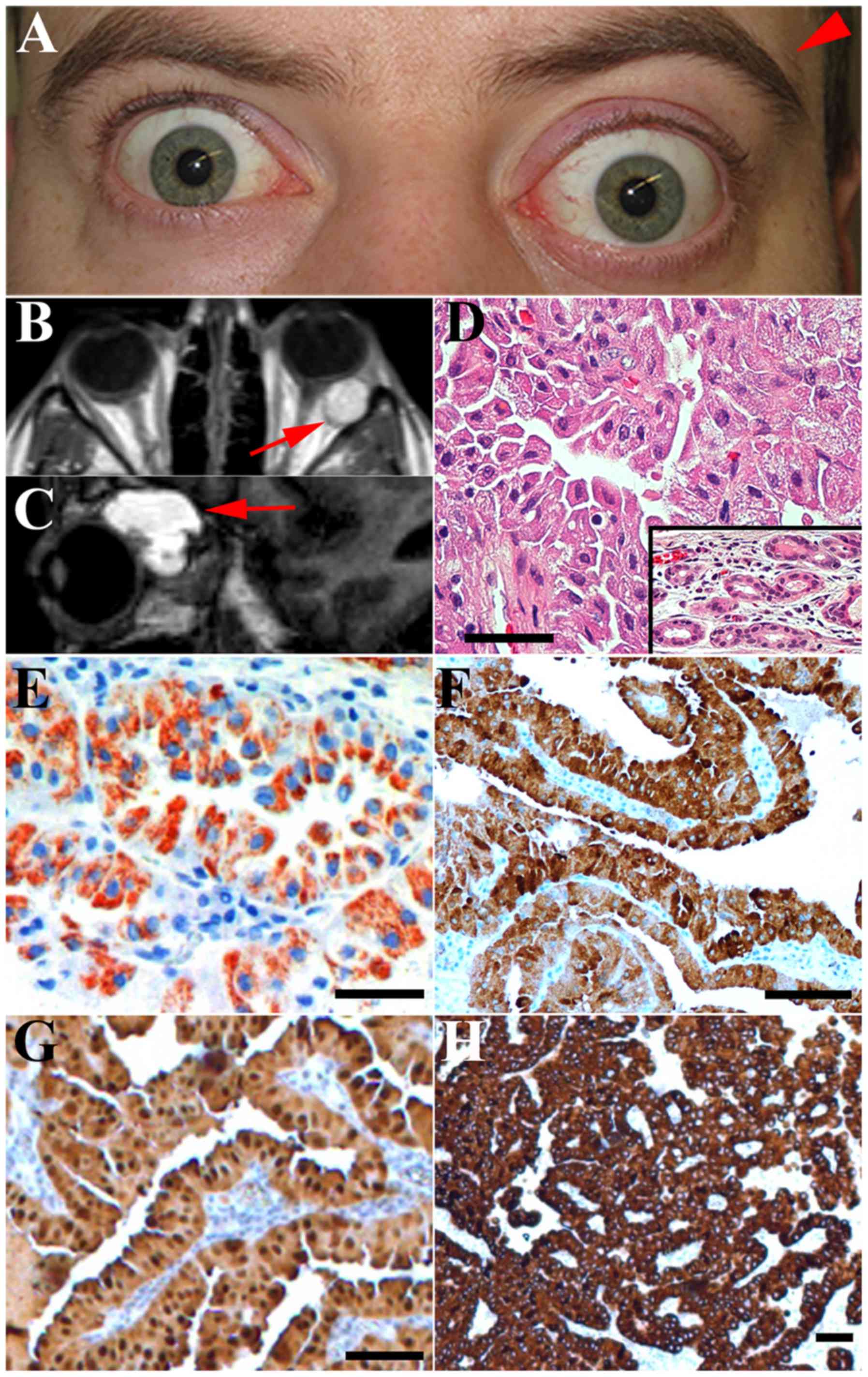
magnetic resonance imaging revealed an ~2×2×2 cm multicystic tumor
in the lacrimal gland of the left orbit along with a 5-mm
protrusion of the left eye causing pronounced asymmetry of the
orbital region (Fig. 1A-C). Computer
tomography confirmed that the orbital roof was intact without tumor
infiltration. The orbital roof was remodelled consistent with a
slow growing benign tumor. The tumor expanded posteriorly in the
orbit due to cystic areas in the tumor. The patient received
treatment for encephalitis, and was subsequently referred for
ophthalmic evaluation of the lacrimal gland mass. No visual
symptoms, pain, or cosmetic changes had been noticed by the
patient. Images revealed that the patient had symmetry of the
orbital region two years previously. On examination prior to
operation, visual acuity measured with the Snellen chart was normal
(6/6 s.c.) in the right eye and reduced (6/30 s.c.) in the left
eye. The patient was not amblyopic and the reduced visual acuity in
the left eye was explained by the refraction error caused by the
tumor mass deforming the eyeball (Fig.
1B). Proptosis (Hertel 18/23-95) of the left eye was present,
and the eye was displaced 2 mm medially and downwards. When
examined the patient reported vertical diplopia. The intraocular
pressure was 12 mmHg in the right eye and 15 mmHg in the left eye
measured with Goldman applanation tonometry. Palpation of the
lacrimal fossa revealed a smooth mass. The patient had decreased
superolateral movement of the left eye due to the space-occupying
lesion. Slit-lamp microscopy, including ophthalmoscopy was normal.
Pupillary reflexes, colour vision and visual fields were normal. A
lateral orbitotomy was performed, and the tumor was completely
excised. Four months following surgery, the visual acuity was 6/6
s.c. in both eyes and the patient was free of any symptoms.
Histopathology and
immunohistochemistry
Formalin-fixed paraffin-embedded (FFPE) tissue from
the resected orbital tumor was sectioned and stained with
haematoxylin and eosin, Alcian blue, periodic acid-Schiff (PAS),
and phosphotungstic acid-haematoxylin (PTAH) according to standard
protocols as previously described (1). Immunohistochemical stainings of 4 µm
sections were performed using the following antibodies:
Mitochondrial antibody MU213-UC (monoclonal, clone no. 113-1; cat
no. MU2130506; mouse anti-human; 1:10; BioGenex Laboratories, Inc.,
San Ramon, CA, USA), Ki-67 (monoclonal, clone MIB-1, cat no.
M724001, mouse anti-human; 1:100), S-100 (polyclonal, cat no.
Z0311, rabbit anti-human; 1:4,000), cytokeratin (CK) 5/6
(monoclonal, clone D5/16 B4, cat no. M723701, mouse anti-human;
1:20), CK 7 (monoclonal, clone OV-TL 12/30, cat no. M701801, mouse
anti-human; 1:1,000), CK 8/18 (monoclonal, clone EP17/EP30, cat no.
M365201-2, rabbit anti-human; 1:50) (all from Dako; Agilent
Technologies, Inc., Santa Clara, CA, USA), CK 14 (monoclonal, clone
LL002, cat no. NCL-LL002; mouse anti-human; 1:40; Novocastra; Leica
Biosystems Newcastle, Newcastle, UK), CK 17 (monoclonal, clone E3,
cat no. M 7046; mouse anti-human; 1:20), CK 19 (monoclonal, clone
RCK 108, cat no. M0888; mouse anti-human; 1:100), CK 20
(monoclonal, clone Ks 20.8, cat no. M 7019; mouse anti-human),
cluster of differentiation (CD)117 (polyclonal, cat no. A450229,
rabbit anti-human; 1:100), smooth muscle actin (SMA; monoclonal,
clone 1A4, cat no. M0851, mouse anti-human, 1:100),
carcinoembryonic antigen (CEA; monoclonal, clone IL-7, cat no.
M7072; mouse anti-human; 1:50), and epithelial membrane antigen
(EMA; monoclonal, clone E29, cat no. M0613; mouse antihuman;
1:1,000) (all from Dako; Agilent Technologies, Inc.). The blocking
reagent was peroxidase (37°C, 10 min incubation). Incubation with
primary antibody occurred at 37°C for 32 min. Secondary antibodies
were used as previously described (1). Immunohistochemistry was performed using
the Ventana BenchMark ULTRA platform (Ventana Medical Systems,
Inc., Tucson, AZ, USA). The MU213-UC staining was performed using a
biotin-free method (EnVision Flex+; Dako; Agilent Technologies,
Inc.) to avoid a false-positive reaction caused by endogenous
biotin in the mitochondrial-rich tissue as previously described
(1). Appropriate controls were
included. All slides were assessed using an Axioplan 2 Imaging
light microscope (Zeiss, Oberkochen, Germany). The investigation
adheres to the tenets of the Declaration of Helsinki (19) and the patient provided informed
consent.
aCGH
Genomic DNA was isolated from FFPE tumor tissue
using the QIAamp® DNA FFPE Tissue kit (Qiagen GmbH,
Hilden, Germany) according to the manufacturer's protocol. aCGH
analysis was performed using the 180K oligonucleotide CGH
microarray (G4449A; Agilent Technologies Inc.) as previously
described (20,21). The slide was scanned on an Agilent
High-Resolution C Microarray scanner, followed by data extraction
and normalisation using Feature Extraction v10.7.1 with linear
normalisation (protocol CGH_107_Sep09) (both from Agilent
Technologies Inc.). Data analysis was performed using Nexus Copy
Number software® Discovery Edition v8.0 (BioDiscovery
Inc., El Segundo, CA, USA) as previously described (21). The FASST2 segmentation algorithm was
used to define non-random regions of CNAs across the genome with a
significance threshold set to P=1.0×10−6. The
log2 ratio thresholds for aberration calls were set to
1.5 for high copy number gain/amplification, 0.2 for gain, −0.2 for
loss, and −1.5 for homozygous deletion. Each aberration was checked
manually to confirm the accuracy of the call. Sex chromosomes and
regions partially or completely covered by a previously reported
copy number variation (Database of Genomic Variants; http://dgvbeta.tcag.ca/dgv/app/news?ref=NCBI37/hg19)
were excluded from the analysis.
Mitochondrial DNA sequencing
Whole mitochondrial genome sequencing was performed
on the Ion PGM™ system with the Precision ID mtDNA Whole Genome
Panel (both Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Library preparation was performed according to the Ion AmpliSeq Kit
for Chef DL8 with minor modifications. After PCR of panel pool 1
and pool 2, the products were pooled for library preparation. Ion
PGM IC 200 kit was used as the template kit for the IonChef (both
from Thermo Fisher Scientific, Inc.). Sequencing was carried out
using Ion PGM IC 200 Sequencing kit (TRS) with the Ion
318™ Chip v2 (Thermo Fisher Scientific, Inc.). Variant
calling was carried out using the Torrent Variant Caller v4.6 of
the Torrent Suite™ software (Thermo Fisher Scientific, Inc.).
PrecisionID_mtDNA_rCRS.fasta was used as the reference genome with
PrecisionID_mtDNA_WG_targets.bed as panel the BED (Thermo Fisher
Scientific, Inc.), and
PrecisionID_mtDNA_TVCv4.6_AnalysisParams.json as the analysis
parameter settings (Thermo Fisher Scientific, Inc.).
Variants were discarded if the coverage was below
×25 and the minimum heteroplasmy threshold level was set to 10% of
the coverage. Data was analysed using the MitoMaster tool (22) and the MitImpact version 2.7 (23). The Polyphen-2 tool was used to predict
harmful single nucleotide polymorphisms (SNP) with a frequency
<0.5% in the GenBank® (24).
Results
Histopathology
Macroscopically, the tumor measured 25×22×17 mm. The
tumor was non-encapsulated. The colour was deep red with blue
cystic areas. Microscopic examination revealed large, cylindrical,
eosinophilic and basal oriented tumor cells (Fig. 1D). It was not possible to identify
normal ducts between the tumor cells, but normal ducts were located
in the periphery of the specimen, possibly consistent with lacrimal
gland oncocytoma, arising from a glandular duct. PAS staining was
positive in a small fraction of the tumor cells and in the cystic
content. Staining with Alcian blue was negative. PTAH was positive
in the tumor cells. An extended immunoprofile included strong
reactivity for CK 5/6, CK 7, CK 8/18, CK 17, CK 19, S-100, and EMA.
CD117 was slightly positive and CK 14 was positive in ~30% of the
basal-type cells. The cytokeratin profile was similar to that of
normal lacrimal gland tissue and other ocular adnexal oncocytomas
(1). The tumor cells stained
positively for the mitochondrial antigen, MU213-UC (Fig. 1E). Staining with anti-Ki67 revealed
positivity in <1% of the tumor cell nuclei, indicating a low
proliferative index. The tumor cells were negative for CK 20, CEA
and SMA.
Genetic profile
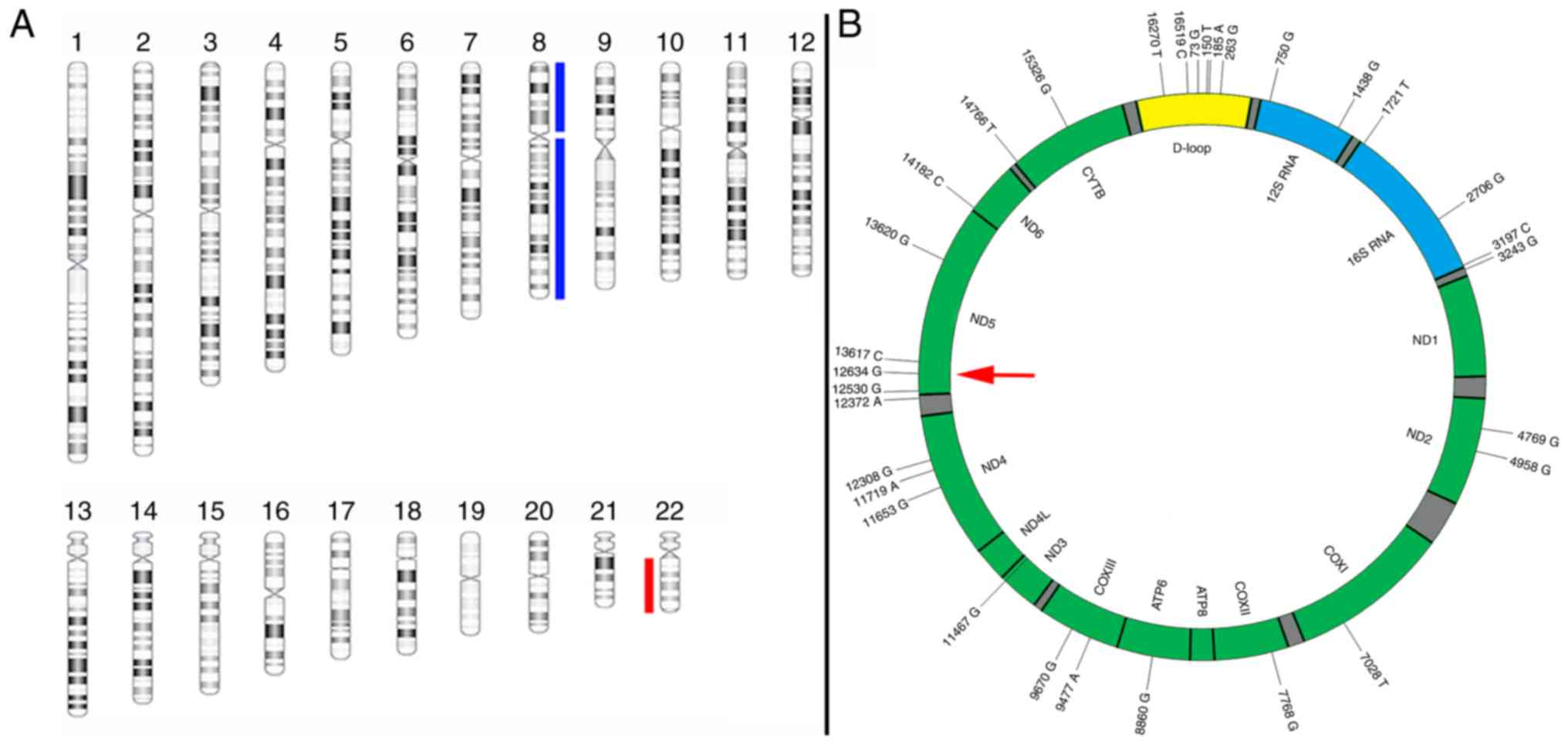
Genome-wide aCGH analysis revealed a genomic profile
characterised by gain of one copy of chromosome 8 and loss of one
copy of chromosome 22 as the sole imbalances (Fig. 2). There was no evidence of gene
amplifications or homozygous deletions.
mtDNA sequencing
Sequencing of the mtDNA revealed 33 sequence
variants (Table II). Five SNPs were
recognised in <0.5% of the samples in the GenBank®.
Four non-synonymous SNPs were identified in the NADH-ubiquinone
oxidoreductase chain 5 (ND5) gene, which is part of the respiratory
complex I. Additionally, an insertion at site 524 was identified in
the displacement-loop (D-loop). One non-synonymous SNP (A12634G)
possibly had a damaging effect. Additionally, SNPs were also
identified in the ND2, ND4 and ND6 genes of the respiratory complex
I. However, these SNPs were synonymous. Two non-synonymous SNPs
(A9670G and G9477A) were identified in the cytochrome oxidase
subunit III gene, which is part of the respiratory complex IV. None
of the synonymous SNPs were involved in splice sites. No
transversions were identified.
 | Table II.Mitochondrial DNA sequencing. |
Table II.
Mitochondrial DNA sequencing.
| SNP | MT-locus | Amino acid
change | Frequency | GenBank frequency
(%) | Polyphen-2.2
(score) |
|---|
| A73G | D-loop | Non-coding | 100.0 | 23631 (73.71) |
|
| C150T | D-loop | Non-coding | 98.3 | 3787 (11.81) |
|
| G185A | D-loop | Non-coding | 100.0 | 1274 (3.97) |
|
| A263G | D-loop | Non-coding | 100.0 | 29979 (93.51) |
|
| 323_324insC | D-loop | Non-coding | 100.0 | 0 (0.00) |
|
| A750G | RNR1 (12s-RNA) | rRNA | 100.0 | 31410 (97.98) |
|
| A1438G | RNR1 (12s-RNA) | rRNA | 100.0 | 30179 (94.14) |
|
| C1721T | RNR2 (16s-RNA) | rRNA | 100.0 | 225 (0.70) |
|
| A2706G | RNR2 (16s-RNA) | rRNA | 100.0 | 24784 (77.31) |
|
| T3197C | RNR2 (16s-RNA) | rRNA | 100.0 | 1350 (4.21) |
|
| A3243Ga | L(UUA/G)/TER | tRNA | 94.5 | 8 (0.02) |
|
| A4769G | ND2 | Synonymous | 100.0 | 31182 (97.26) |
|
| A4958Ga | ND2 | Synonymous | 100.0 | 115 (0.36) |
|
| C7028T | CO1 | Synonymous | 100.0 | 25290 (78.89) |
|
| A7768G | CO2 | Synonymous | 100.0 | 592 (1.85) |
|
| A8860G | ATP6 |
Non-syn:Thr→Ala | 100.0 | 31527 (98.34) | Benign (0) |
| G9477A | CO3 |
Non-syn:Val→Ile | 100.0 | 1344 (4.19) | Benign (0) |
| A9670Ga | CO3 | Non-syn:
Asn→Ser | 100.0 | 28 (0.09) | Benign (0.01) |
| A11467G | ND4 | Synonymous | 100.0 | 4213 (13.14) |
|
| A11653G | ND4 | Synonymous | 100.0 | 194 (0.61) |
|
| G11719A | ND4 | Synonymous | 100.0 | 24160 (75.36) |
|
| A12308G | L(CUN) | tRNA | 97.5 | 4193 (13.08) |
|
| G12372A | ND5 | Synonymous | 100.0 | 4519 (14.10) |
|
|
A12530Ga | ND5 |
Non-syn:Asn→Ser | 100.0 | 20 (0.06) | Benign (0.06) |
|
A12634Ga | ND5 |
Non-syn:Ile→Val | 100.0 | 91 (0.28) | Probably damaging
(1) |
| T13617C | ND5 | Synonymous | 100.0 | 1315 (4.10) |
|
|
A13630Ga | ND5 |
Non-syn:Thr→Ala | 100.0 | 62 (0.19) | Benign (0.05) |
| A13637G | ND5 |
Non-syn:Gln→Arg | 100.0 | 287 (0.90) | Benign (0.1) |
| T14182C | ND6 | Synonymous | 100.0 | 843 (2.63) |
|
| C14766T | CYTB |
Non-syn:Thr→Ile | 100.0 | 24091 (75.15) | Benign (0.01) |
| A15326G | CYTB |
Non-syn:Thr→Ala | 100.0 | 31512 (98.29) | Benign (0.02) |
| C16270T | D-loop | Non-coding | 100.0 | 8147 (25.41) |
|
| T16519C | D-loop | Non-coding | 100.0 | 1694 (5.28) |
|
Discussion
The present study described the immunohistochemical
and genetic profile of a rare case of lacrimal gland oncocytoma in
a 20-year-old male. This is the 13th case of lacrimal gland
oncocytoma in the literature. Men and women are equally affected,
with a median age at the time of diagnosis of 57 years (range, 1–83
years; Table I). The symptoms of
lacrimal gland oncocytoma range from mild lid swelling without pain
to proptosis with severe pain (10,13). The
majority of lacrimal gland oncocytomas were surgically removed
through a lateral orbitotomy or more rarely through a
fronto-orbitozygomatic craniotomy (13). All patients with lacrimal gland
oncocytoma were alive at the time of the last follow-up (3–42
months) (8). Malignant transformation
or recurrence of lacrimal gland oncocytoma following complete
surgical excision has not been reported. Hence, a possible
association between oncocytoma and the exceedingly rare oncocytic
carcinoma is unclear.
Little is known about the genetic changes leading to
oncocytoma formation. In the present case, a gain of one copy of
chromosome 8 and loss of one copy of chromosome 22 were identified
as the sole genomic imbalances. To the best of our knowledge, none
of these alterations have previously been described in oncocytoma
of any sites. The significance of the findings of the current study
is unclear. However, it appears that genomic instabilities are
typical for oncotoma that occurs in males (16). Exome sequencing of renal oncocytoma
has identified two main types. A diploid oncocytoma that has no sex
predilection and another type that has a male predilection, and is
a hypodiploid oncocytoma with complete loss of chromosome 1, 14,
21, X, or Y (16). Other chromosomal
aberrations have been described in oncocytic lesions of the
thyroid, including losses and gains of both arms of chromosomes 1,
2, 5, 7, 12, 17, 19, 20 and 22 (4).
However, only gains involving chromosome 22 have been reported,
contrasting with the loss of chromosome 22 in the present case
(4). The trisomy 8 in the current
case is similarly unprecedented, but trisomy 7 has been
demonstrated in a case of salivary gland oncocytoma (25). There may be a site specific difference
in the alterations identified in oncocytoma, as the frequent
chromosomal aberrations observed in renal, thyroid and salivary
oncocytoma are different, and diffuse (16–18,25). This
supports the idea that the results of the present study may be
specific to lacrimal gland oncocytoma, since these changes have not
been observed elsewhere. Deletions, SNPs and rearrangements have
been described in mtDNA in oncocytoma of the thyroid, kidney,
salivary glands, and adrenal cortex (2). In line with the apparent association
between the oncocytic phenotype and mtDNA gene aberrations,
specifically in genes involved in complex I function, several
sequence variants in the mitochondrial genome of the present case
were identified. The majority of these were transitions, but one
insertion was identified at mtDNA position 524. Insertions in the
D-loop have previously been reported in thyroid adenoma (18). In addition, six SNPs were identified
in <0.5% of the 32,000 mitochondrial genomes registered in the
GenBank®. Notably, two synonymous and four
non-synonymous SNPs were detected in the ND5 gene, which is
a part of the respiratory complex I. Of note, the consequence of
one SNP in ND5 was termed ‘probably damaging’ with the
PolyPhen-2 tool for functional annotation of genetic variants,
thereby supporting a fundamental pathogenic similarity between
lacrimal gland oncocytoma and oncocytic lesions in other anatomical
sites. Overall, these mtDNA variants may potentially impair cell
respiration, and be responsible for the slow-proliferating nature
of the tumor.
It is thought that mutations in the mitochondrial
genes encoding proteins involved in oxidative phosphorylation may
result in compensatory mitochondrial proliferation, while mutations
in nuclear genes encoding oxidative phosphorylation proteins are
less frequently involved (2). The
cause of mitochondrial proliferation and the possible association
with proliferation of the oncocyte itself remains unclear.
In conclusion, a gain of one copy of chromosome 8
and loss of one copy of chromosome 22 were identified in a rare
case of lacrimal gland oncocytoma. In addition to these gross
alterations of nuclear DNA, a peculiar involvement of apparently
damaging mitochondrial point mutations resulting in impairment of
respiratory function was reported, similar to what has previously
been reported in oncocytoma from other anatomical sites, suggesting
a potential involvement of mitochondria in oncocytoma
pathogenesis.
Oncocytoma of the ocular adnexa is more common in
the lacrimal caruncle (1). It would
be of interest in future studies to compare CGH and mitochondrial
sequencing results of a selection of these lesions to the current
case to evaluate the molecular changes in ocular adnexal
oncocytoma.
Acknowledgements
The present study was supported by the Swedish
Cancer Society (grant no. 160509).
References
|
1
|
Østergaard J, Prause JU and Heegaard S:
Oncocytic lesions of the ophthalmic region: A clinicopathological
study with emphasis on cytokeratin expression. Acta Ophthalmol.
89:263–267. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Máximo V, Rios E and Sobrinho-Simoes M:
Oncocytic lesions of the thyroid, kidney, salivary glands, adrenal
cortex and parathyroid glands. Int J Surg Pathol. 22:33–36. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Andreasen S, Esmaeli B, Holstein SL,
Mikkelsen LH, Rasmussen PK and Heegaard S: An update on tumors of
the lacrimal gland. Asia Pac J Ophthalmol (Phila). 6:159–172. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gasparre G, Bonora E, Tallini G and Romeo
G: Molecular features of thyroid oncocytic tumors. Mol Cell
Endocrinol. 321:67–76. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Beskid M and Zarzycka M: A case of
onkocytoma of the lacrimal gland. Klin Oczna. 29:311–315. 1959.(In
Polish). PubMed/NCBI
|
|
6
|
Riedel K, Stefani FH and Kampik A:
Oncocytoma of the ocular adnexa. Klin Monatsbl Augenheilkd.
182:544–548. 1983.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hartman LJ, Mourits MP and Canninga-van
Dijk MR: An unusual tumour of the lacrimal gland. Br J Ophthalmol.
87:3632003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Calle CA, Castillo IG, Eagle RC and Daza
MT: Oncocytoma of the lacrimal gland: Case report and review of the
literature. Orbit. 25:243–247. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Archondakis S, Skagias L, Tsakiris A,
Sambaziotis D and Daskalopoulou D: Oncocytoma of the lacrimal gland
diagnosed initially by fine-needle aspiration cytology. Diagn
Cytopathol. 37:443–445. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Economou M, Seregard S and Sahlin S:
Oncocytoma of the lacrimal gland. Acta Ophthalmologica. 85:576–577.
2007. View Article : Google Scholar
|
|
11
|
Kim JY, Park HY, Paik JS, Kim DC and Yang
SW: Oncocytoma of the lacrimal gland: An Asian case. Jpn J
Ophthalmol. 54:239–241. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Aghaji AE, Olushina DB and Okoye OI:
Oxyphil cell adenoma in a Nigerian: Case report and review of the
literature. Niger J Clin Pract. 14:373–376. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Limb RJ, Rosenfeld JV and McLean C: Giant
orbital oncocytoma. Asian J Neurosurg. 8:192–194. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ferte A, Trechot F, Cloche V, Busby H,
Maalouf T, Angioi K and George JL: Oncocytoma: An uncommon lesion
of the lacrimal gland. J Fr Ophtalmol. 39:e231–e233. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jittapiromsak N, Hou P, Williams MD and
Chi TL: Orbital oncocytoma: Evaluation with dynamic
contrast-enhanced magnetic resonance imaging using a time-signal
intensity curve and positive enhancement integral images. Clin
Imaging. 42:161–164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Joshi S, Tolkunov D, Aviv H, Hakimi AA,
Yao M, Hsieh JJ, Ganesan S, Chan CS and White E: The genomic
landscape of renal oncocytoma identifies a metabolic barrier to
tumorigenesis. Cell Rep. 13:1895–1908. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gasparre G, Porcelli AM, Bonora E, Pennisi
LF, Toller M, Iommarini L, Ghelli A, Moretti M, Betts CM,
Martinelli GN, et al: Disruptive mitochondrial DNA mutations in
complex I subunits are markers of oncocytic phenotype in thyroid
tumors. Proc Natl Acad Sci USA. 104:9001–9006. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Maximo V, Lima J, Soares P, Botelho T,
Gomes L and Sobrinho-Simões M: Mitochondrial D-loop instability in
thyroid tumours is not a marker of malignancy. Mitochondrion.
5:333–340. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
World Medical Association, . World Medical
Association Declaration of Helsinki: Ethical principles for medical
research involving human subjects. JAMA. 310:2191–2194. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Andreasen S, Persson M, Kiss K, Homøe P,
Heegaard S and Stenman G: Genomic profiling of a combined large
cell neuroendocrine carcinoma of the submandibular gland. Oncol
Rep. 35:2177–2182. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Persson F, Winnes M, Andrén Y, Wedell B,
Dahlenfors R, Asp J, Mark J, Enlund F and Stenman G:
High-resolution array CGH analysis of salivary gland tumors reveals
fusion and amplification of the FGFR1 and PLAG1 genes in ring
chromosomes. Oncogene. 27:3072–3080. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lott MT, Leipzig JN, Derbeneva O, Xie HM,
Chalkia D, Sarmady M, Procaccio V and Wallace DC: mtDNA variation
and analysis using mitomap and mitomaster. Curr Protoc
Bioinformatics. 44:1.23–1.16. 2013.
|
|
23
|
Castellana S, Rónai J and Mazza T:
MitImpact: An exhaustive collection of pre-computed pathogenicity
predictions of human mitochondrial non-synonymous variants. Hum
Mutat. 36:E2413–E2422. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Adzhubei I, Jordan DM and Sunyaev SR:
Predicting functional effect of human missense mutations using
PolyPhen-2. Curr Protoc Hum Genet: Chapter 7. Unit 7.20. 2013.
View Article : Google Scholar
|
|
25
|
Mark J, Dahlenfors R, Havel G and Böckmann
P: Benign parotid oncocytoma with the chromosomal abnormality
trisomy 7. Anticancer Res. 11:1735–1737. 1991.PubMed/NCBI
|