Introduction
Triptolide is a bioactive ingredient isolated from
Tripterygium wilfordii (1)
known to exhibit immune-suppressive and anti-inflammatory activity
(2–7).
A number of studies have identified that triptolide also exhibits
antitumor activity, inhibiting proliferation and migration of
cancer cells (8–12). A previous study has demonstrated that
triptolide inhibited ATPase activity of the basal transcription
factor transcription factor II H (TFIIH) and RNA polymerase
II-mediated transcription in nucleotide excision repair (NER)
(13). Triptolide also interfered
with other transcription factors including p53, nuclear factor-κB
and heat-shock factor protein 1 (6,14).
Considering the important role it serves by interfering with the
transcription of tumor-associated factors, triptolide was
demonstrated to be a potent antitumor drug. However, the underlying
molecular mechanisms of triptolide action in DNA damage repair are
not well defined.
The DNA repair protein X-ray repair
cross-complementing protein 1 (XRCC1) serves a key role in base
excision repair (BER) and single strand break (SSB) repair as a
scaffolding protein which recruits a number of DNA
repair-associated factors in the repair pathway, including ligase
III, poly(ADP-ribose) polymerase 1 (PARP1), apurinic/apyrimidinic
endonuclease 1, proliferating cell nuclear antigen (PCNA) and DNA
polymerase-β (15–17). Ligase IV serves to join together the
double-strand breaks (DSBs) in non-homologous end-joining (NHEJ)
repair (18). Rad51 serves vital
roles in homologous recombination (HR), in which the Rad51 filament
seeks a homologous sequence for DNA replication (19). PCNA is a DNA sliding clamp,
functioning in DNA replication (20).
The present study used XRCC1−/− and ligase
IV−/− CH12F3 cells to analyze the DNA breaks induced by
triptolide while also detecting cellular Rad51 and nuclear PCNA
levels following triptolide treatment to investigate the effect of
triptolide in cellular DNA repair.
PARPs are a family of proteins that transfer
mono(ADP-ribose) or poly(ADP-ribose) (PAR) groups onto their target
proteins (21). As the PARylation of
DNA repair factors is required for recruitment to DNA breaks, PARPs
are essential for DNA repair (22–25). The
function of PARP1 in DNA repair contributes to cancer cell survival
in response to genotoxic agents, thus PARP1 is a promising
therapeutic target in cancer therapy, particularly for breast
cancer 1-deficient cancer cells. The phosphoinositide 3-kinase
(PI3K) pathway is a critical signaling pathway frequently activated
in numerous types of cancer, and a number of PI3K-targeted
compounds are used therapeutically in the clinic (26–28). The
present study investigated the combination of triptolide with PARP1
inhibitors and PI3K inhibitors to analyze the potential use of
triptolide in treatment of lymphoma. The present study contributes
to the current understanding of the role served by triptolide in
DNA damage and apoptosis, as well as in clinical therapeutics in
combination with chemotherapeutic agents to treat patients with
lymphoma.
Materials and methods
Antibodies and reagents
Triptolide, PARP inhibitor, PI3K inhibitor, protease
inhibitor and phosphatase inhibitor were purchased from Selleck
Chemicals (Shanghai, China). Anti-Rad51 (catalog no. 14961-1-AP)
and anti-H3 (catalog no. 17168-1-AP) antibodies were purchased from
ProteinTech Group, Inc. (Chicago, IL, USA). RPMI-1640 medium, fetal
bovine serum (FBS) and penicillin and streptomycin were purchased
from Hyclone (GE Healthcare Life Sciences, Logan, UT, USA).
Dimethyl sulfoxide (DMSO) and β-mercaptoethanol (BME) were
purchased from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany). MTT
was purchased from Tiangen (Beijing, China). The Cell Cytoplasmic
and Nuclear Protein Extraction kit was purchased from Sangon
Biotech Co. Ltd. (Shanghai, China). Antibodies against
phospho-histone H2AX (γH2AX; Ser139) were purchased from
CST Biological Reagents Company Limited (Shanghai, China; catalog
no. 2577). Antibodies against caspase-3 (catalog no. 19677-1-AP),
caspase-9 (catalog no. 10380-1-AP), PCNA (catalog no. 10205-1-AP),
PARP1 (catalog no. 13371-1-AP) and β-actin, used as the reference
gene (catalog no. 60008-1), were purchased from Wuhan Sanying
Biotechnology (Wuhan, China) and all used at a dilution of 1:300 in
western blot analysis. Goat anti-Rabbit immunoglobulin G (IgG)
secondary antibody conjugated to Alexa Fluor-488 (catalog no.
A-11034) was purchased from Thermo Fisher Scientific Inc. (Waltham,
MA, USA). HRP-conjugated goat anti-rabbit and anti-mouse IgG
secondary antibody (catalog nos. SA00001-2 and SA00001-1,
respectively) were from purchased from Wuhan Sanying Biotechnology,
Inc. (Wuhan, China).
Cell culture
The murine B-cell lymphoma cell line CH12F3 was
obtained from Professor T. Honjo (Kyoto University, Kyoto, Japan).
Ligase IV−/− and XRCC1−/− CH12F3 cells were
obtained from Dr Kefei Yu (Michigan State University, East Lansing,
MI, USA). All cells were cultured in RPMI-1640 medium supplemented
with 10% FBS, 100 U/ml penicillin, 100 ng/ml streptomycin and 50 nM
BME, and incubated at 37°C with 5% CO2.
Western blot analysis
The 6-well plates were plated with 1×106
cells and treated with distinct concentrations of triptolide for 24
h. Cellular whole protein or nuclear protein was extracted using
the cytoplasmic and nuclear protein extraction kit according to the
manufacturer's protocol. Protein concentrations were determined
using a Qubit 2.0 fluorimeter (Thermo Fisher Scientific, Inc.). A
total of 50 µg protein from each sample was separated using
SDS-PAGE in a 10 or 15% gel for 2 h at 120 V and transferred onto
polyvinylidene difluoride (PVDF) membranes. PVDF membranes were
blocked with 5% skimmed milk powder (diluted in Tris-Buffered
Saline-Tween 20) for 1 h at room temperature prior to incubation
with the mentioned primary antibodies (1:300 dilution) overnight at
4°C. HRP-conjugated goat anti-rabbit and anti-mouse IgG secondary
antibody with appropriate dilution (1:2,000) were used against
primary antibodies (dilution, 1:300) for 1 h at room temperature.
Finally, the proteins were detected using enhanced chemiluminescent
substrate (Thermo Fisher Scientific, Inc.). Histone 3 (H3) was used
as the control.
Flow cytometric analysis
CH12F3 cells were treated with triptolide (0, 10,
20, 30, 40 and 50 nM) for 4 h before collection, and 0.5 ml 4%
formaldehyde was added for 10 min at 37°C. Cells were collected by
centrifugation (1,000 × g, 3 min, room temperature) and ice-cold
100% methanol was slowly added until a final concentration of 90%
methanol was achieved. Cells were incubated for 30 min on ice. A
total of 0.5×106 cells of each sample were washed with 2
ml wash buffer including 1X PBS with 0.5% bovine serum albumin
(Sangon Biotech Co. Ltd.). Cells were suspended in 100 µl of
anti-rabbit antibody against γH2AX (1:200 dilution in 1X PBS) and
incubated for 1 h at room temperature. Cells were washed in 2 ml
wash buffer. Cells were suspended in 100 µl goat anti-rabbit IgG
secondary antibody Alexa Fluor 488 for 30 min at room temperature
prior to a repeat wash with wash buffer as aforementioned. Finally,
cells were suspended in 0.5 ml PBS for flow cytometry (FCM) and the
results were analyzed using BD CellQuest 5.1 software (BD
Biosciences, San Jose, CA, USA).
Cell viability assay
A total of 5,000 cells were placed in a 96-well
plate and incubated in triptolide (10, 20, 30, 40 and 50 nM) at
37°C for 24 h. Following incubation, MTT was added to each well at
a final concentration of 0.5 mg/ml. Cells were incubated at 37°C
for 4 h before collection by centrifugation (1,000 × g for 3 min)
at room temperature. A total of 200 µl DMSO was added to each well
and the plate was incubated for 15 min at 37°C. Finally, the
absorbance values were detected at 589 nm using a microplate
photometer (Thermo Fisher Scientific, Inc.).
Apoptosis assay
Cellular apoptosis was detected using FCM. Annexin V
and propidium iodide (PI) were used to stain cells that were
treated with 5 nM triptolide for 12 h and incubated for 15 min at
room temperature. The apoptotic cells were quantified using FCM and
the results were analyzed using BD CellQuest 5.1 software (BD
Biosciences).
Statistical analysis
SPSS software (version 6.0; SPSS, Inc., Chicago, IL,
USA) was used for statistical analysis. A Student's t-test was
performed to compare differences between groups. P<0.05 was
considered to indicate a statistically significant difference, with
P<0.01 considered to be highly significant.
Results
Triptolide suppresses CH12F3 cell
viability
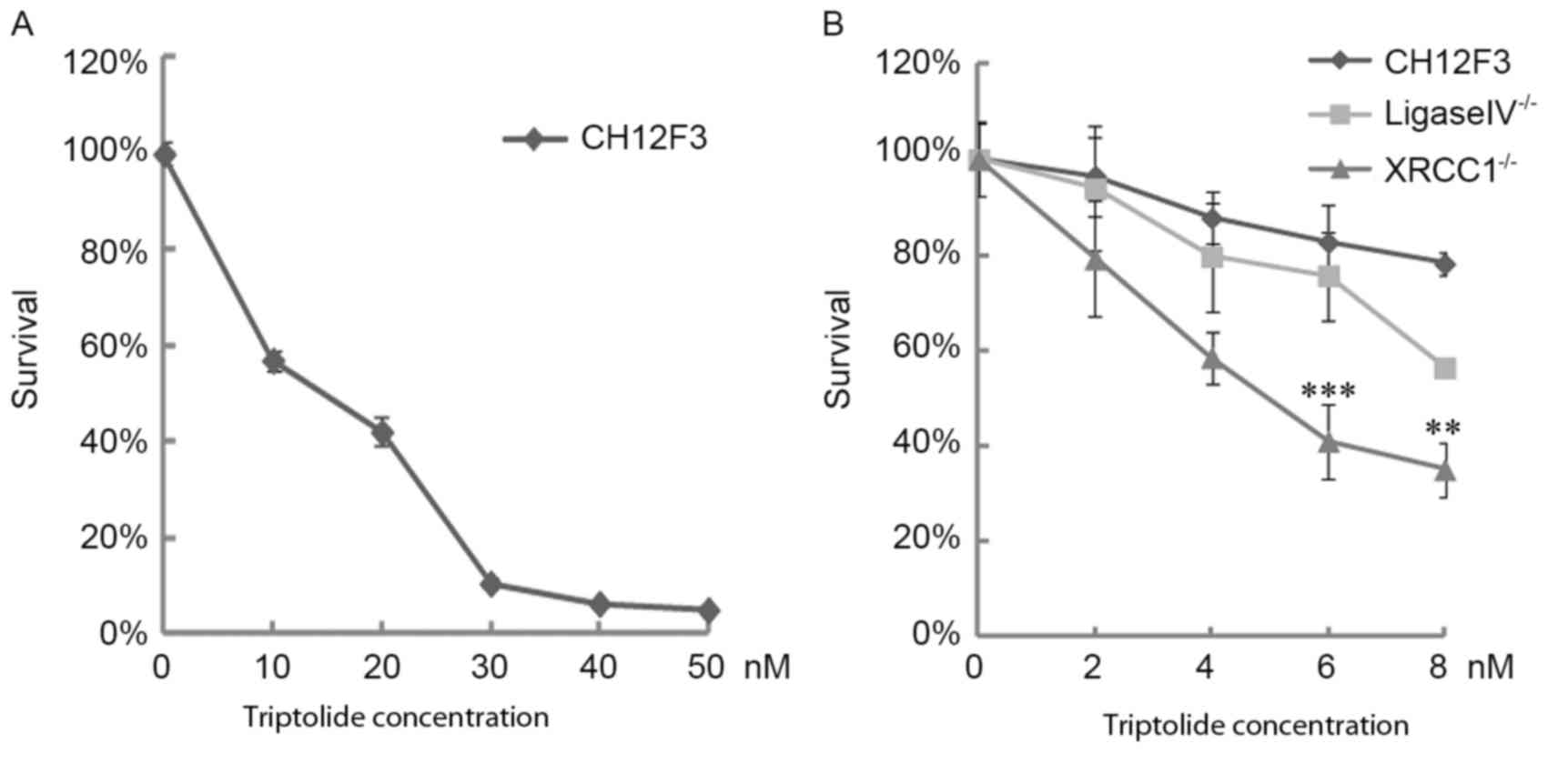
The effects of triptolide on CH12F3 cell viability
were analyzed using an MTT assay which revealed that triptolide
suppressed CH12F3 cell proliferation. Following treatment with
triptolide doses ranging between 0 and 50 nM for 24 h, cell
viability was almost completely inhibited at 30 nM (Fig. 1A). To assess the effects of triptolide
on DNA damage, the viability of ligase IV−/− and
XRCC1−/− CH12F3 cells was analyzed. Results of the MTT
assay demonstrated that 6 nM triptolide suppressed
XRCC1−/− viability to 40%; however, the viability of
ligase IV−/− and control CH12F3 cells were increased,
compared with XRCC1−/− cells at the same dose of
triptolide (Fig. 1B). As XRCC1 is
vital for the BER SSB pathway and ligase IV is essential for NHEJ
DSB repair, these results suggested that triptolide induces DNA
damage, primarily dependent on XRCC1-mediated repair.
Triptolide induces DNA breaks and
regulates Rad51 and PCNA levels
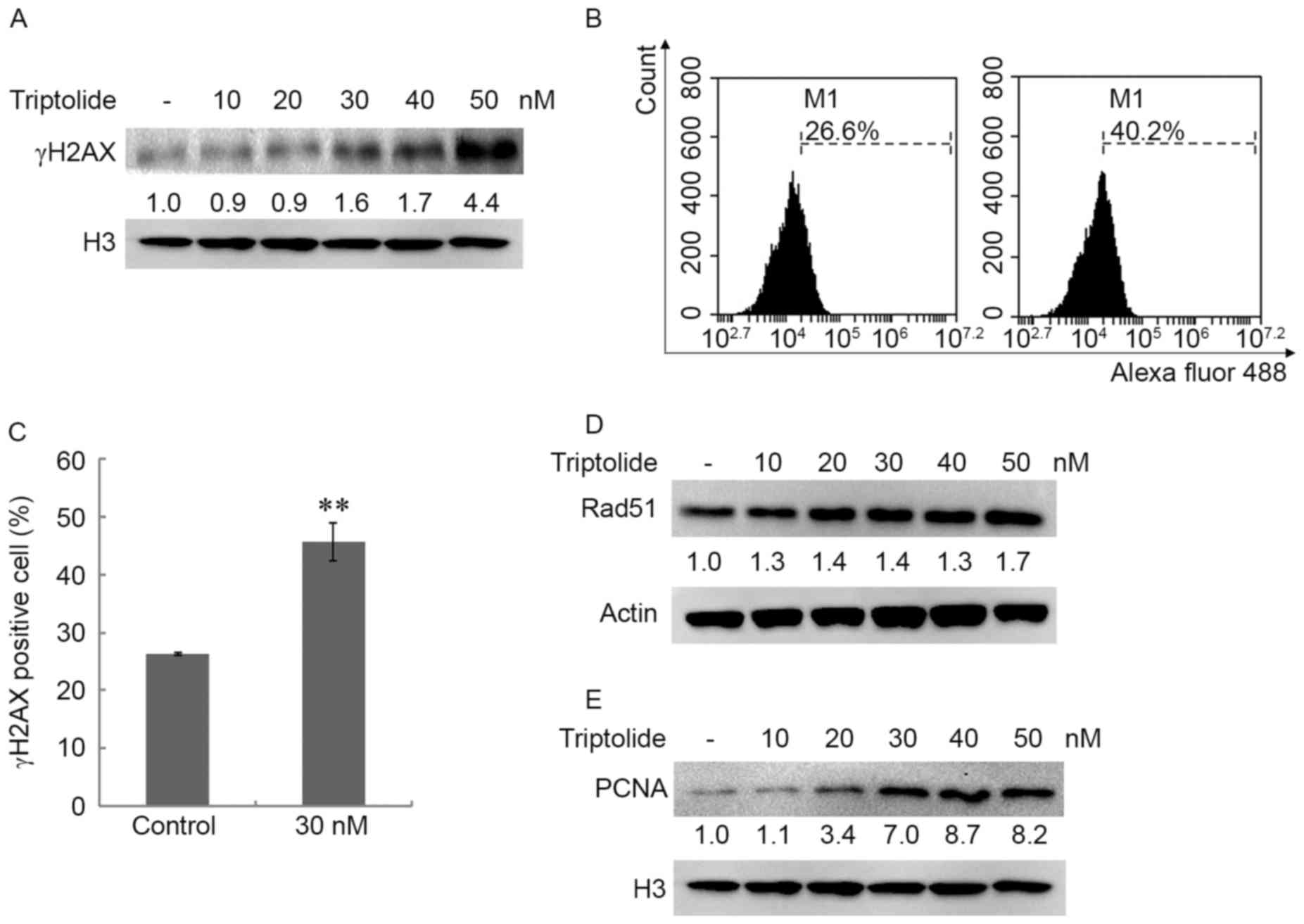
The present study investigated the DNA damage of
CH12F3 cells following triptolide exposure. The CH12F3 cells were
treated with triptolide (0, 10, 20, 30, 40 and 50 nM) for 4 h and
the nuclear proteins were extracted. γH2AX was detected using
western blotting (Fig. 2A). The
expression of nuclear γH2AX was upregulated in a dose-dependent
manner following triptolide treatment, which suggested that a high
dose of triptolide induced DSBs (29,30). To
confirm this result, the γH2AX level was detected using FCM, which
revealed that triptolide increased the γH2AX level (Fig. 2B and C). These results further
suggested that a high dose of triptolide resulted in cellular
DSBs.
To illustrate the cellular response to triptolide,
the expression of Rad51 and nuclear PCNA was detected in cells
treated with triptolide. Following treatment with triptolide (0,
10, 20, 30, 40 and 50 nM) for 4 h, Rad51 levels were increased
(Fig. 2D) and nuclear PCNA was
markedly upregulated by triptolide (Fig.
2E). PCNA is a DNA sliding clamp that functions in DNA
replication. These results suggest the effect of PCNA in DNA
replication is important for repairing DNA damage caused by
triptolide.
Triptolide induces caspase-3-dependent
apoptosis
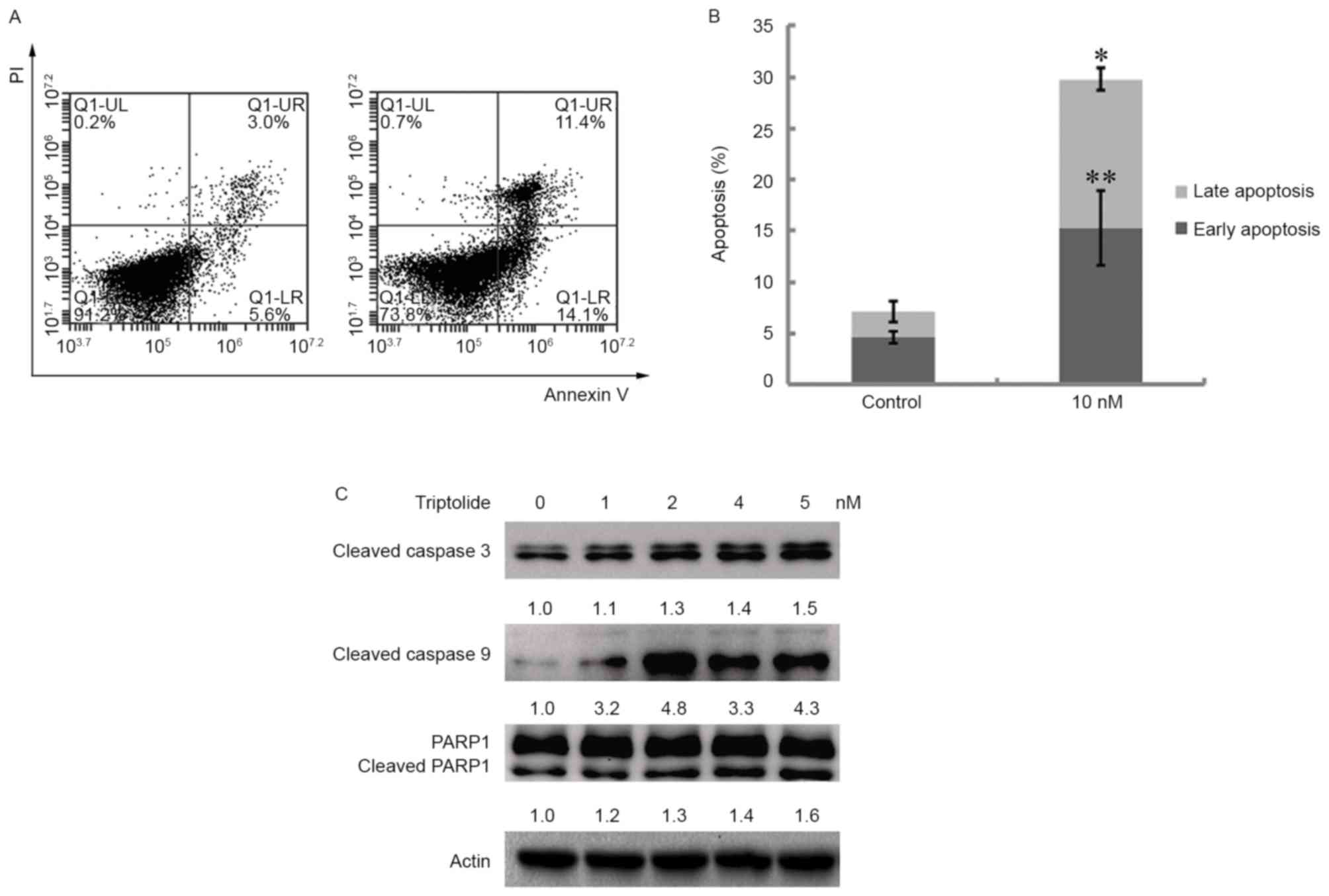
Apoptosis of cells treated with triptolide was
analyzed. CH12F3 cells were treated with 10 nM triptolide for 12 h.
The apoptotic cells were analyzed through annexin V/PI staining.
Triptolide caused apoptosis at early (annexin V-positive and
PI-negative) and late (annexin V-positive and PI-positive) stages
(Fig. 3A and B). The underlying
molecular mechanism of apoptosis was revealed through analyzing the
expression of apoptotic proteins. Cells were treated with
triptolide at 1, 2, 4 and 5 nM for 12 h. The whole cellular lysate
was extracted for western blot assay. Cleaved caspase-3, cleaved
caspase-9 and cleaved PARP1 were up-regulated in a dose-dependent
manner (Fig. 3C). These results
demonstrated that triptolide induces caspase-3-dependent
apoptosis.
Triptolide sensitizes CH12F3 cells to
PARP1 and PI3K inhibitors
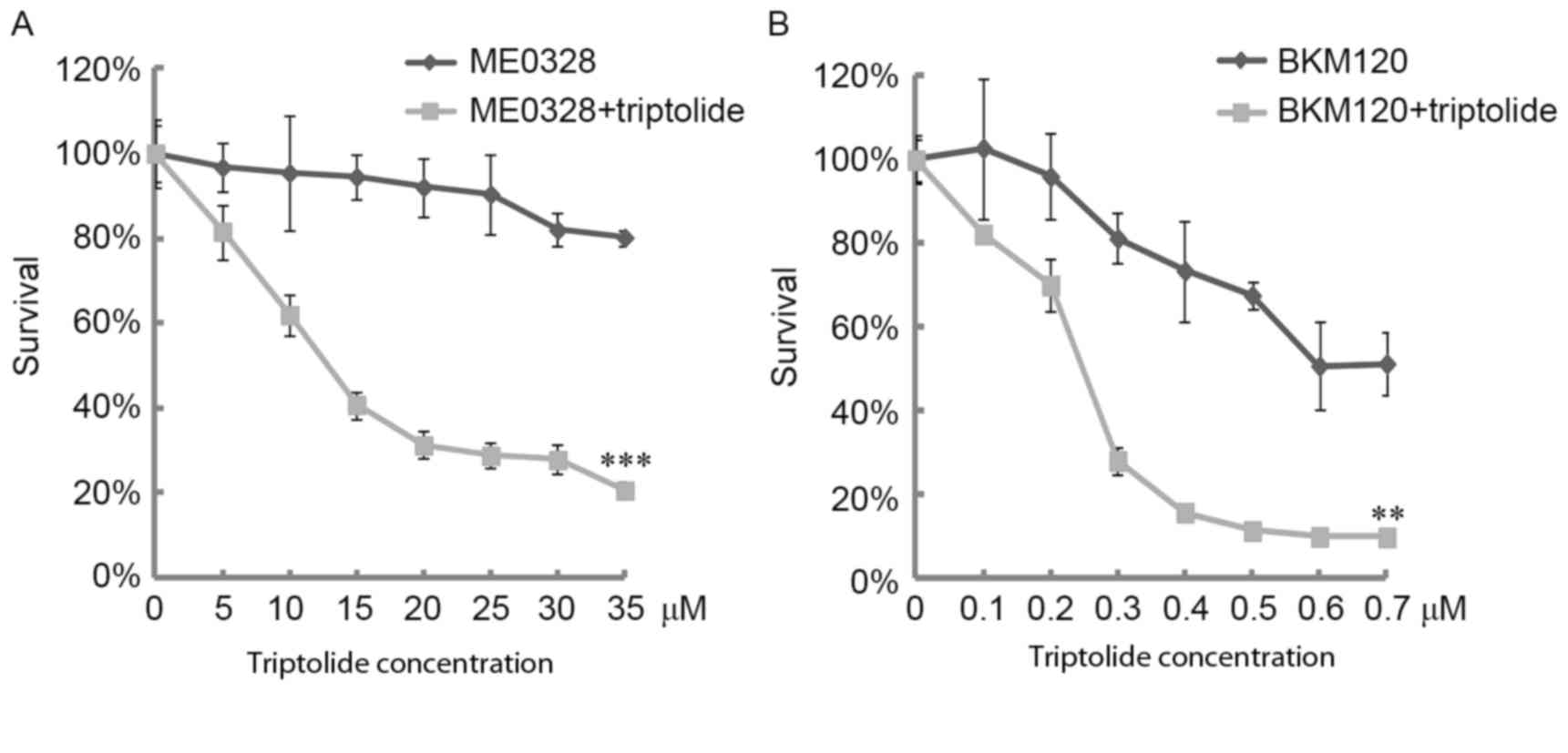
Following the aforementioned results demonstrating
that triptolide induced DNA damage and apoptosis in CH12F3 cells,
the sensitivity of CH12F3 cells to triptolide in combination with
PARP1 and PI3K inhibitors was analyzed. At a dose of 5 nM
triptolide, the cell viability was >80%; however, this dose of
triptolide significantly sensitized CH12F3 cells to PARP1 (ME0328)
and PI3K inhibitors (Fig. 4A and B).
These results demonstrate that triptolide sensitizes lymphoma to
PARP1 and PI3K inhibitors, which supports the use of triptolide as
an antitumor drug to sensitize lymphoma cells to genotoxic
agents.
Discussion
Triptolide was previously reported to inhibit cancer
cell growth and induce apoptosis in different types of tumor
(1–3).
However, the majority of its antitumor activity was observed at
high concentrations, at which adverse effects were noted (31). The present study investigated the
effects of low doses of triptolide on DNA breaks and apoptosis as
well as in combination with chemotherapeutic agents to treat B
lymphoma cells. The results of the present study demonstrated that
triptolide inhibits CH12F3 cell viability and low doses of
triptolide (6 nM) suppressed XRCC1−/− CH12F3 cells,
indicating that triptolide induces DNA breaks dependent on the
XRCC1-mediated repair pathway. Additionally, γH2AX expression in
CH12F3 cells treated with various triptolide doses demonstrated
that triptolide induced DSBs in a dose-dependent manner. The
present study also identified that triptolide induced
caspase-3-dependent apoptosis.
A previous study demonstrated that triptolide
specifically binds to TFIIH basal transcription factor and, through
which, inhibits NER (13). This
result suggested that triptolide exhibits DNA damage effects;
however, the underlying molecular mechanism of triptolide in DNA
damage repair are not well defined. The present study demonstrated
that low doses of triptolide inhibited XRCC1−/− CH12F3
cells, but not ligase IV−/− CH12F3 cells or wild-type
CH12F3 cells. XRCC1 is a scaffolding protein which recruits DNA
repair-associated factors involved in BER and SSB repair. Thus, the
results of the present study suggest that triptolide induces base
damage or SSBs. Analyzing the expression of Rad51 and PCNA in cells
treated with triptolide, the results of the present study revealed
an 8-fold increase in PCNA expression following triptolide
treatment. Rad51 serves a role in HR and PCNA is a sliding clamp
that functions in DNA replication. This result suggests that
triptolide may highly regulate the transcription of PCNA in
response to DNA breaks. The present study analyzed the apoptosis
induced by triptolide in CH12F3 cells and identified that low doses
of triptolide induced caspase-3-dependent apoptosis. Considering
that low doses of triptolide induced DNA breaks, it may be
concluded that the DNA damage induced by triptolide triggers
caspase-3-dependent apoptosis.
PARP1 inhibitors are promising clinical therapeutic
agents in the treatment cancer cells, particularly for HR-deficient
cancer cells. Activation of PI3K signaling contributes to cancer
cell proliferation, thus PI3K inhibitors are used clinically to
treat tumors with aberrant PI3K signaling. Following the results of
the present study revealing that low doses of triptolide induced
DNA breaks and apoptosis, the application of low doses of
triptolide in combination with PARP1 and PI3K inhibitors to treat
lymphoma cells was analyzed. The present study demonstrated that 5
nM triptolide may efficiently sensitize CH12F3 cells to PARP1 and
PI3K inhibitors. Therefore, triptolide may be a potent antitumor
drug to sensitize lymphoma cells to chemotherapeutic agents.
Further studies are required to explore the underlying molecular
mechanism of triptolide function in regulating cellular DNA repair
and sensitizing tumor cells to antitumor genotoxic agents.
Acknowledgements
The present study was supported by the Chinese
National Scientific Foundation (grant nos. 81201563 and
31371254).
References
|
1
|
Canter PH, Lee HS and Ernst E: A
systematic review of randomised clinical trials of Tripterygium
wilfordii for rheumatoid arthritis. Phytomedicine. 13:371–377.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cheng X, Shi W, Zhao C, Zhang D, Liang P,
Wang G and Lu L: Triptolide sensitizes human breast cancer cells to
tumor necrosis factor-α-induced apoptosis by inhibiting activation
of the nuclear factor-κB pathway. Mol Med Rep. 13:3257–3264. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ding X, Zhou X, Jiang B, Zhao Q and Zhou
G: Triptolide suppresses proliferation, hypoxia-inducible factor-1α
and c-Myc expression in pancreatic cancer cells. Mol Med Rep.
12:4508–4513. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li J, Zhu W, Leng T, Shu M, Huang Y, Xu D,
Qiu P, Su X and Yan G: Triptolide-induced cell cycle arrest and
apoptosis in human renal cell carcinoma cells. Oncol Rep.
25:979–987. 2011.PubMed/NCBI
|
|
5
|
Yan X, Ke XX, Zhao H, Huang M, Hu R and
Cui H: Triptolide inhibits cell proliferation and tumorigenicity of
human neuroblastoma cells. Mol Med Rep. 11:791–796. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhu W, Hu H, Qiu P and Yan G: Triptolide
induces apoptosis in human anaplastic thyroid carcinoma cells by a
p53-independent but NF-kappaB-related mechanism. Oncol Rep.
22:1397–1401. 2009.PubMed/NCBI
|
|
7
|
Yang SX, Gao HL, Xie SS, Zhang WR and Long
ZZ: Immunosuppression of triptolide and its effect on skin
allograft survival. Int J Immunopharmacol. 14:963–969. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jiang QW, Cheng KJ, Mei XL, Qiu JG, Zhang
WJ, Xue YQ, Qin WM, Yang Y, Zheng DW, Chen Y, et al: Synergistic
anticancer effects of triptolide and celastrol, two main compounds
from thunder god vine. Oncotarget. 6:32790–32804. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reno TA, Kim JY and Raz DJ: Triptolide
inhibits lung cancer cell migration, invasion, and metastasis. Ann
Thorac Surg. 100:1817–1825. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu J, Shen M, Yue Z, Yang Z, Wang M, Li
C, Xin C, Wang Y, Mei Q and Wang Z: Triptolide inhibits
colon-rectal cancer cells proliferation by induction of G1 phase
arrest through upregulation of p21. Phytomedicine. 19:756–762.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brincks EL, Kucaba TA, James BR, Murphy
KA, Schwertfeger KL, Sangwan V, Banerjee S, Saluja AK and Griffith
TS: Triptolide enhances the tumoricidal activity of TRAIL against
renal cell carcinoma. FEBS J. 282:4747–4765. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jao HY, Yu FS, Yu CS, Chang SJ, Liu KC,
Liao CL, Ji BC, Bau DT and Chung JG: Suppression of the migration
and invasion is mediated by triptolide in B16F10 mouse melanoma
cells through the NF-kappaB-dependent pathway. Environ Toxicol.
31:1974–1984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Titov DV, Gilman B, He QL, Bhat S, Low WK,
Dang Y, Smeaton M, Demain AL, Miller PS, Kugel JF, et al: XPB, a
subunit of TFIIH, is a target of the natural product triptolide.
Nat Chem Biol. 7:182–188. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Westerheide SD, Kawahara TL, Orton K and
Morimoto RI: Triptolide, an inhibitor of the human heat shock
response that enhances stress-induced cell death. J Biol Chem.
281:9616–9622. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Breslin C, Hornyak P, Ridley A, Rulten SL,
Hanzlikova H, Oliver AW and Caldecott KW: The XRCC1
phosphate-binding pocket binds poly (ADP-ribose) and is required
for XRCC1 function. Nucleic Acids Res. 43:6934–6944. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Caldecott KW, McKeown CK, Tucker JD,
Ljungquist S and Thompson LH: An interaction between the mammalian
DNA repair protein XRCC1 and DNA ligase III. Mol Cell Biol.
14:68–76. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sossou M, Flohr-Beckhaus C, Schulz I,
Daboussi F, Epe B and Radicella JP: APE1 overexpression in
XRCC1-deficient cells complements the defective repair of oxidative
single strand breaks but increases genomic instability. Nucleic
Acids Res. 33:298–306. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Critchlow SE, Bowater RP and Jackson SP:
Mammalian DNA double-strand break repair protein XRCC4 interacts
with DNA ligase IV. Curr Biol. 7:588–598. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Baumann P and West SC: Role of the human
RAD51 protein in homologous recombination and double-stranded-break
repair. Trends Biochem Sci. 23:247–251. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bravo R: Synthesis of the nuclear protein
cyclin (PCNA) and its relationship with DNA replication. Exp Cell
Res. 163:287–293. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Altmeyer M, Messner S, Hassa PO, Fey M and
Hottiger MO: Molecular mechanism of poly(ADP-ribosyl)ation by PARP1
and identification of lysine residues as ADP-ribose acceptor sites.
Nucleic Acids Res. 37:3723–3738. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
El-Khamisy SF, Masutani M, Suzuki H and
Caldecott KW: A requirement for PARP-1 for the assembly or
stability of XRCC1 nuclear foci at sites of oxidative DNA damage.
Nucleic Acids Res. 31:5526–5533. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Herceg Z and Wang ZQ: Functions of
poly(ADP-ribose) polymerase (PARP) in DNA repair, genomic integrity
and cell death. Mutat Res. 477:97–110. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Javle M and Curtin NJ: The role of PARP in
DNA repair and its therapeutic exploitation. Br J Cancer.
105:1114–1122. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schreiber V, Amé JC, Dollé P, Schultz I,
Rinaldi B, Fraulob V, Ménissier-de Murcia J and de Murcia G:
Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient
base excision DNA repair in association with PARP-1 and XRCC1. J
Biol Chem. 277:23028–23036. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Follo MY, Manzoli L, Poli A, McCubrey JA
and Cocco L: PLC and PI3K/Akt/mTOR signalling in disease and
cancer. Adv Biol Regul. 57:10–16. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
D'Amato V, Rosa R, D'Amato C, Formisano L,
Marciano R, Nappi L, Raimondo L, Di Mauro C, Servetto A, Fusciello
C, et al: The dual PI3K/mTOR inhibitor PKI-587 enhances sensitivity
to cetuximab in EGFR-resistant human head and neck cancer models.
Br J Cancer. 110:2887–2895. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hu Y, Guo R, Wei J, Zhou Y, Ji W, Liu J,
Zhi X and Zhang J: Effects of PI3K inhibitor NVP-BKM120 on
overcoming drug resistance and eliminating cancer stem cells in
human breast cancer cells. Cell Death Dis. 6:e20202015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Valdiglesias V, Giunta S, Fenech M, Neri M
and Bonassi S: γH2AX as a marker of DNA double strand breaks and
genomic instability in human population studies. Mutat Res.
753:24–40. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Garcia-Canton C, Anadón A and Meredith C:
γH2AX as a novel endpoint to detect DNA damage: Applications for
the assessment of the in vitro genotoxicity of cigarette smoke.
Toxicol In Vitro. 26:1075–1086. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu J, Jiang Z, Liu L, Zhang Y, Zhang S,
Xiao J, Ma M and Zhang L: Triptolide induces adverse effect on
reproductive parameters of female Sprague-Dawley rats. Drug Chem
Toxicol. 34:1–7. 2011. View Article : Google Scholar : PubMed/NCBI
|