Introduction
Tongue squamous cell carcinoma (TSCC) is the most
commonly occurring type of oral cancer (1). Due to the high risk of occult metastasis
and neck nodal metastasis, patients with TSCC exhibit a
significantly poorer prognosis compared with those with other
cancers of the oral cavity (2). The
prognosis of TSCC remains reliant on the Tumor Node Metastasis
(TNM) staging (3) of the tumor;
however, the outcome of patients at the same stage may vary
considerably. Thus, novel prognostic indicators are required.
Proline rich 11 (PRR11) was first identified during
a screen for novel cancer-associated genes (4). PRR11 is a 360-amino acid protein that is
encoded by a gene located on human chromosome 17q22 (5). Human chromosome 17 hosts a number of
other cancer-associated genes, including the essential tumor
suppressor genes tumor protein (p)53 and breast
cancer 1 (6,7). PRR11 comprises 10 exons, and the encoded
protein typically serves as a ligand for SRC Homology 3 (SH3), WW
and enabled/VASP homology 1 domains (8). PRR11 expression is elevated in lung and
breast cancer, and numerous types of tumors of the digestive system
(5,8,9). PRR11 is
also suggested to be associated with tumor development and
progression (5,8–10).
However, whether PRR11 is involved in tongue squamous cell
carcinoma has not been determined. The present study aimed to
investigate the expression of PRR11 in tongue squamous cell
carcinoma, and to examine its association with clinical parameters
and prognosis in patients with TSCC.
Materials and methods
The cancer genome atlas (TCGA) TSCC
data mining
PRR11 mRNA expression data from 126 TSCC and 12
non-cancerous tongue tissue samples were downloaded from the TCGA
database (http://cancergenome.nih.gov/) in December 2014, along
with overall patient survival data. The association between PRR11
expression and overall survival was evaluated by comparing the top,
and bottom 50% of the specimens, using the log-rank test.
Tissue specimens and patient
information
Fresh tumor specimens were collected from patients
with TSCC who had undergone surgery at The First Affiliated
Hospital of Sun Yat-sen University (Guangzhou, China) from March
2014 to October 2014 and used for quantitative reverse
transcription polymerase chain reaction (RT-qPCR) and western
blotting. For immunohistochemistry, 72 TSCC paraffin-embedded
specimens were prepared at The First Affiliated Hospital of Sun
Yat-sen University between January 2007 and September 2010 from
patients who were histopathologically, and clinically diagnosed
with TSCC. The male: female ratio of the patients included in the
present study was 38:34, and the median age was 54 (age range,
28–80 years). No patients received any additional therapy prior to
surgery. Patients with apparent distant metastasis were excluded.
Tumor grade and stage were defined according to the 6th edition of
the TNM classification of the Union for International Cancer
Control (UICC, 2002) (3). Written
informed consent and approval from the First Affiliated Hospital of
Sun Yat-sen University Institutional Review Board were obtained
from all participants prior to any experiments. Sample clinical
information is summarized in Table
I.
 | Table I.Clinicopathological characteristics
and proline rich 11 expression in patients tongue squamous cell
carcinoma. |
Table I.
Clinicopathological characteristics
and proline rich 11 expression in patients tongue squamous cell
carcinoma.
| Variable | Number of cases
(%) |
|---|
| Sex |
|
| Male | 38 (52.8) |
|
Female | 34 (47.2) |
| Age, years |
|
| ≥54 | 36 (50.0) |
|
<54 | 36 (50.0) |
| Clinical stage |
|
| I | 11 (15.3) |
| II | 27 (37.5) |
| III | 25 (34.7) |
| IV | 9 (12.5) |
| T classification |
|
| T1 | 16 (22.2) |
| T2 | 46 (63.9) |
| T3 | 7 (9.7) |
| T4 | 3 (4.2) |
| N classification |
| N0 | 46 (63.9) |
| N1 | 19 (26.4) |
| N2 | 7 (9.7) |
| M
classification |
|
| No | 72 (100.0) |
|
Yes | 0 (0.0) |
| Differentiation
grade |
|
|
Well | 40 (55.6) |
|
Moderate | 26 (36.1) |
|
Poor | 6 (8.3) |
| Vital status (at
follow-up) |
|
|
Alive | 42 (58.3) |
|
Succumbed | 30 (41.7) |
| Expression of
PRR11 |
|
|
Low | 27 (37.5) |
|
High | 45 (62.5) |
|
Detectable | 71 (98.6) |
|
Undetectable | 1 (1.4) |
RT-qPCR
Total RNA was isolated using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the manufacturer's protocol. The concentration and
quality of RNA were measured spectrophotometrically at 260, and 280
nm. RNA was reverse-transcribed by heating at 25°C for 10 min, then
at 55°C for 30 min, and at 85°C for 5 min to produce cDNA using the
Oligo (dT) 15 primer and M-MLV Reverse Transcriptase kit (Promega
Corporation, Madison, WI, USA). Primers were as follows: PRR11
forward, 5′-GACTTCCAAAGCTGTGCTTCC-3′ and reverse,
5′-CTGCATGGGTCCATCCTTTTT-3′; 18S rRNA forward,
5′-CCTGGATACCGCAGCTAGGA-3′ and reverse,
5′-GCGGCGCAATACGAATGCCCC-3′. qPCR was performed with FastStart
Universal SYBR Green Master (Rox; Roche Diagnostics GmbH, Mannheim,
Germany) as follows: 2 min at 95°C; followed by 40 cycles of 10 sec
at 95°C, 30 sec at 60°C, and 30 sec at 72°C. qPCR was performed
using the ABI Prism 7900 HT real-time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The 2−ΔΔCq
method was used to calculate gene expression relative to the
18SrRNA housekeeping control (11).
All experiments were repeated in triplicate.
Western blotting
Fresh tissue samples were ground to powder in liquid
nitrogen and lysed with 10 times the tissue volume of the
pre-cooled radioimmunoprecipitation assay buffer (Wuhan Boster
Biological Technology, Ltd., Wuhan, China) containing phosphatase
inhibitors (Phosphatase Inhibitor Cocktails Set II, Calbiochem;
Merck KGaA, Darmstadt, Germany), proteinase inhibitors (Protease
Inhibitor Cocktails Set I, Calbiochem; Merck KGaA) and 1 mmol/l
phenylmethanesulfonyl fluoride (Sigma-Aldrich; Merck KGaA). Lysate
protein concentration was measured by bicinchoninic protein assay
(Sigma-Aldrich; Merck KGaA). Proteins (40 µg/lane) were separated
by 10% SDS-PAGE and transferred to a polyvinylidene fluoride
membrane (EMD Millipore, Billerica, MA, USA). To avoid unspecific
binding, the membrane was blocked with 5% non-fat milk (Merck KGaA)
in phosphate buffered saline (PBS)/Tween (0.05%) at room
temperature for 1 h. Subsequently, the membrane was incubated with
a polyclonal rabbit anti-human PRR11 antibody (dilution, 1:250;
cat. no. NBP1-83784; Novus Biologicals, LLC, Littleton, CO, USA)
overnight at 4°C, and anti-β-actin monoclonal antibody (dilution,
1:1,000; cat. no. ab8226, Abcam Inc., Cambridge, MA, UK) was used
as the loading control, and then incubated with a horseradish
peroxidase-conjugated affiniPure goat anti-rabbit secondary
antibody (dilution, 1:10,000; cat. no. 111–035-003, Jackson
ImmunoResearch Inc., West Grove, PA, USA) at room temperature for 1
h. Immunoreactive bands were visualized with an enhanced
chemiluminescence detection system (EMD Millipore, Billerica, MA,
USA).
Immunohistochemistry (IHC)
IHC was performed on 72 human TSCC tissues. Antigen
retrieval was performed by heating these sections in 10 mmol/l
citric acid buffer (pH 6.0). The sections were blocked with 5%
normal goat serum (Wuhan Boster Biological Technology, Ltd.) for 30
min at 25°C, and incubated in 3% hydrogen peroxide at 25°C.
Sections were then incubated with a polyclonal rabbit anti-human
PRR11 antibody (1:100) at 4°C overnight, followed by incubation
with a horseradish peroxidase-conjugated goat anti-rabbit secondary
antibody (dilution, 1:1,000; cat. no. 111-035-003, Jackson
ImmunoResearch Laboratories Inc., West Grove, PA, USA) for 1 h at
25°C. Finally, slides were treated with chromogen
3,3′-diaminobenzidine (Dako; Agilent Technologies, Inc., Santa
Clara, CA, USA) for 1 min and counterstained with 5% hematoxylin
for 20 sec at 25°C. The degree of immunostaining of the sections
was viewed and scored separately by two independent investigators
who were blind to the histopathological features, and patient data.
Scores were determined by combining the proportion of positively
stained tumor cells: 0, no positive tumor cells; 1, <10%
positive tumor cells; 2, 10–50% positive tumor cells; and 3,
>50% positive tumor cells. The intensity of staining was
determined as follows: 0, no staining; 1, weak staining/light
yellow; 2, moderate staining/yellowish brown; and 3, strong
staining/brown. The staining index was calculated as the product of
the proportion of positive cells and the staining intensity score.
Using this method of assessment, the protein expression was
evaluated by determining the staining index (0, 1, 2, 3, 4, 6, 9).
Cut-off values were chosen based on heterogeneity of the log-rank
test score with respect to overall survival. The optimal cut-off
value was determined: A staining index score ≥6 was used to define
tumors with high PRR11 expression; and a score ≤4 indicated low
PRR11 expression (12).
IHC was also performed on tumor lesions and normal
tissues to measure protein expression in using an AxioVision
Rel.4.6 computerized image analysis system and an automatic
measurement program (Carl Zeiss AG, Oberkochen, Germany).
Specifically, stained sections were evaluated using a light
microscope at magnification, ×200. A total of 10 representative
staining fields of each section were analyzed to verify the mean
optical density (MOD), which represents the strength of staining
signal (number of positive pixels) (13).
Statistical analysis
Data collection and statistical analysis were
performed using SPSS17.0 software (SPSS Inc., Chicago, IL, USA).
Differences in PRR11 expression were compared using a student's
t-test for comparisons between two groups or one-way analysis of
variance with Newman Keul's multiple comparison test for
comparisons between ≥2 groups. The χ2 test and Fisher's
exact test were used to analyze the association between PRR11
expression, and clinicopathological characteristics. MOD data were
statistically analyzed using an unpaired Student's t-test to
compare the average MOD difference between different groups of
tissues (13). Survival curves were
plotted using the Kaplan-Meier method and compared with the
log-rank test. The significance of survival variables was analyzed
using univariate and multivariate Cox's regression analysis.
P<0.05 (two-tailed) was considered to indicate a statistically
significant difference.
Results
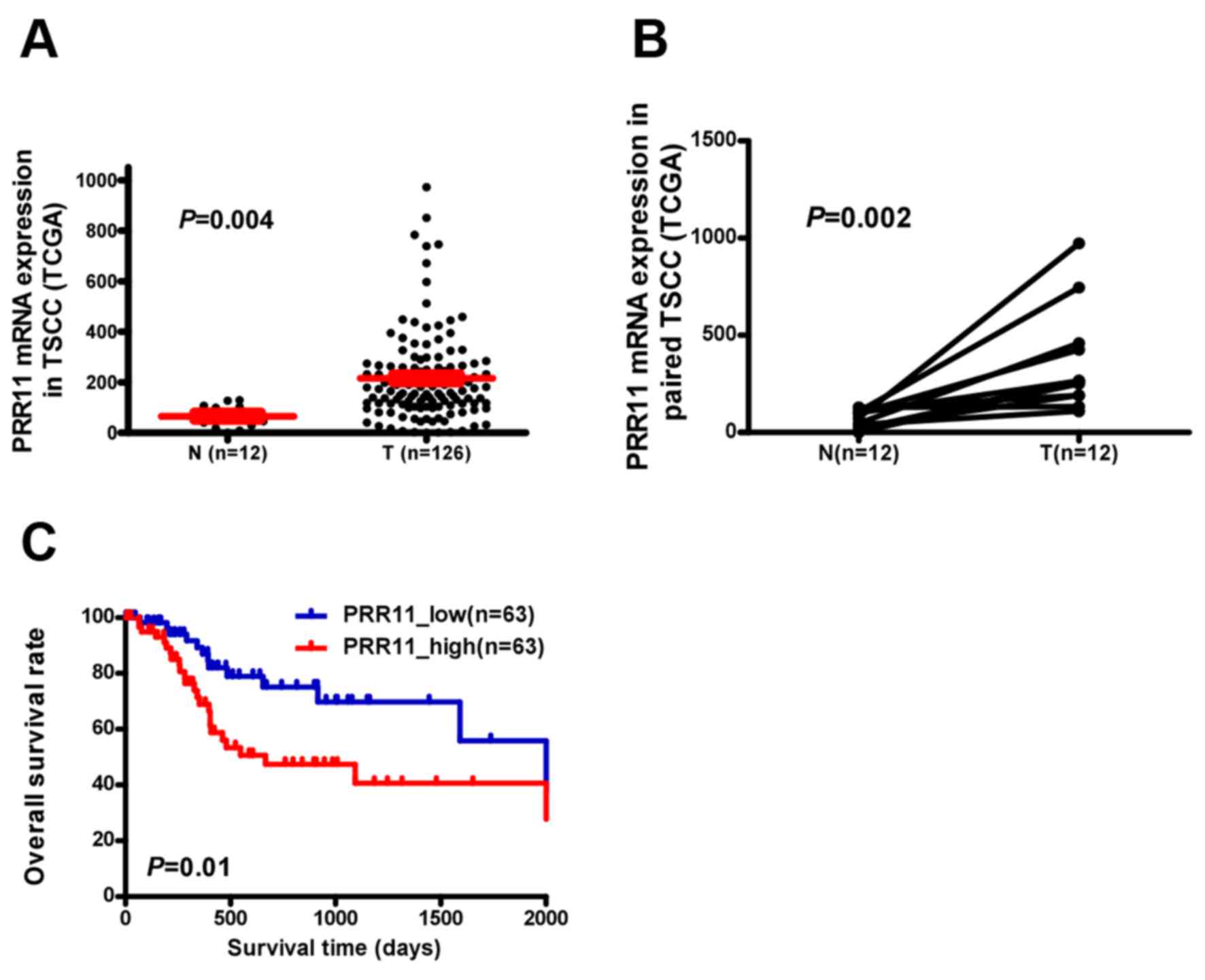
PRR11 is overexpressed in TSCC tissues
and is associated with patient survival
PRR11 transcription was examined in an independent
TCGA cohort and a significantly higher expression was observed in
TSCC tissues compared with non-cancerous tongue tissue (P=0.004;
Fig. 1A). PRR11 was identified to be
significantly upregulated at the mRNA level in 12 human TSCC
tissues compared with the equivalent non-cancerous tissues
(P=0.002; Fig. 1B). Additionally,
assessment of patient survival using Kaplan-Meier analysis and
log-rank test indicated an inverse correlation between PRR11
expression and overall survival time of patients with TSCC (P=0.01;
Fig. 1C).
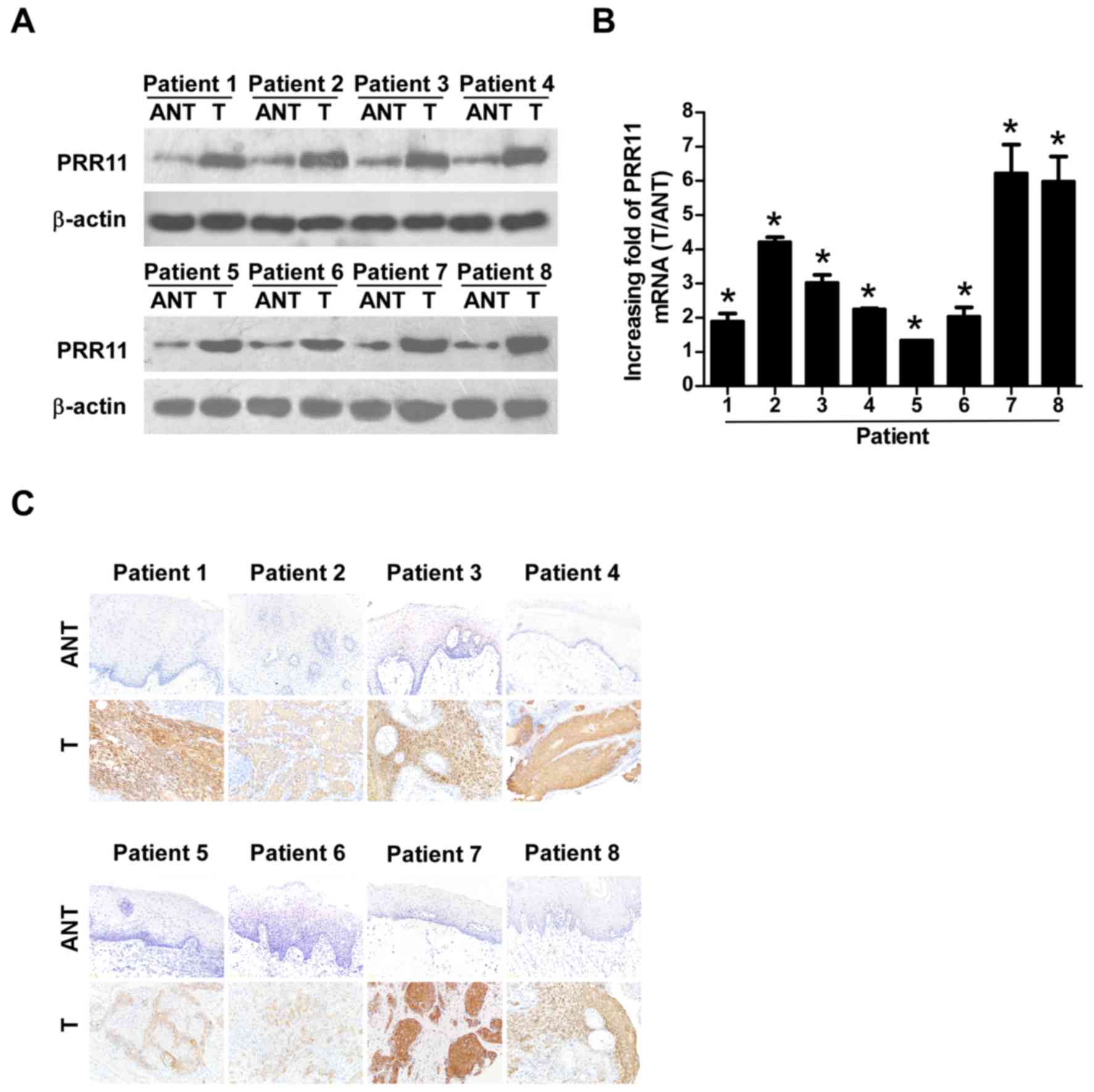
PRR11 is upregulated in TSCC tissues
and is associated with TSCC progression
To verify the results of the TCGA analysis, 8 TSCC
tissues and their equivalent noncancerous counterparts were
subjected to IHC, western blotting, and RT-qPCR analysis (Fig. 2). PRR11 was markedly upregulated at
the protein level in all 8 human TSCC tissues according to western
blotting (Fig. 2A) and IHC (Fig. 2C) analyses. In addition, PRR11 mRNA
levels, as measured by the tumor/normal tissue ratio, were between
1.9–6.2-fold higher in TSCC tissues compared with their equivalent
noncancerous tissues (Fig. 2B).
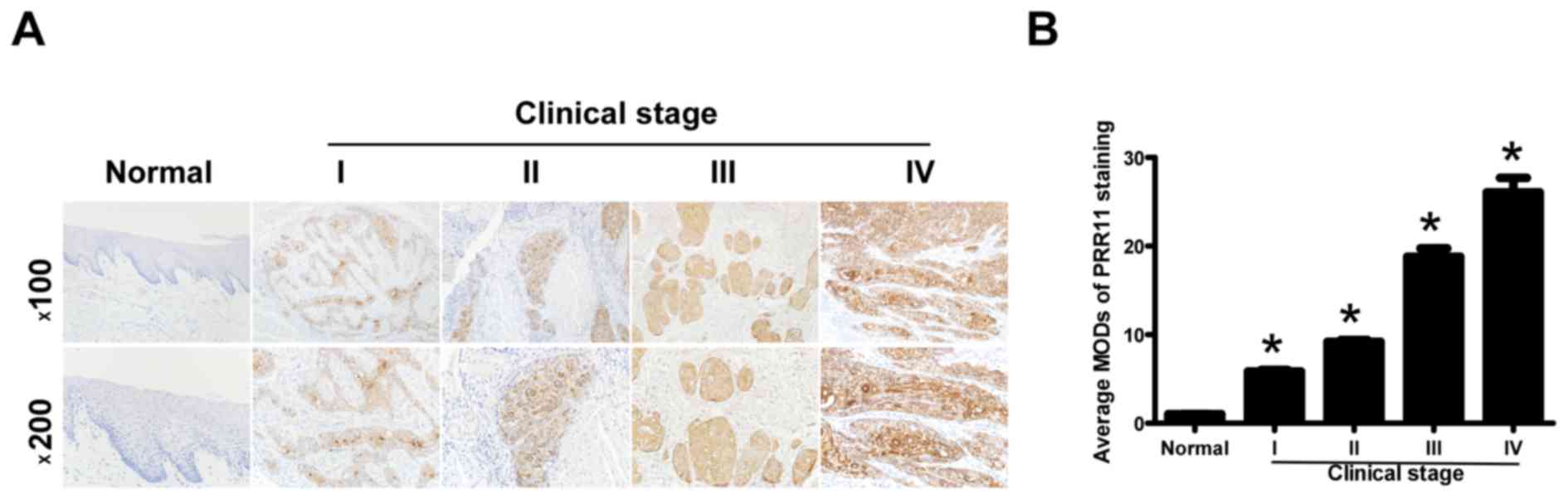
To additionally explore the prevalence of PRR11
upregulation in TSCC, 72 paraffin-embedded archived TSCC tissues
and 5 normal human oral mucosal tissues were subjected to IHC. High
levels of PRR11 expression were observed in areas containing
primary TSCC cells, while PRR11 was undetectable or only marginally
detectable in normal human oral mucosal tissues and equivalent
noncancerous tissues (Fig. 3A).
Quantitative analysis indicated that the average MOD of PRR11
staining in clinical stage I-IV primary tumors was significantly
higher compared with in normal human oral mucosal tissues
(P<0.05), and significantly increased with a progression of
tumor stage from I to IV (P<0.05; Fig.
3B). Taken together, these results clearly demonstrated that
PRR11 expression was elevated in TSCC and was associated with TSCC
progression.
PRR11 expression was identified to be significantly
associated with clinical stage (P<0.001), T classification
(P=0.009), N classification (P=0.017) and vital status (P=0.010),
but not with any other clinicopathological features, including age,
sex, and differentiation grade (Table
II). In conclusion, these data suggest that the upregulation of
PRR11 is associated with clinical stage, T, N classification and
vital status, which additionally support the hypothesis that the
overexpression of PRR11 is associated with TSCC progression.
 | Table II.Association between
clinicopathological characteristics and PRR11 expression in
patients with tongue squamous cell carcinoma. |
Table II.
Association between
clinicopathological characteristics and PRR11 expression in
patients with tongue squamous cell carcinoma.
|
|
| PRR11 |
|
|
|---|
|
|
|
|
|
|
|---|
| Variable | Total | Low (%) | High (%) | χ2 test
P-value | Fisher's exact test
P-value |
|---|
| Age, years |
|
|
|
|
|
|
≥54 | 36 | 13 (36.1) | 23 (63.9) | 0.808 | 0.809 |
|
<54 | 36 | 14 (38.9) | 22 (61.1) |
|
|
| Sex |
|
|
|
|
|
|
Male | 38 | 13 (34.2) | 25 (65.8) | 0.542 | 0.545 |
|
Female | 34 | 14 (41.2) | 20 (58.8) |
|
|
| Clinical stage |
|
|
|
|
|
|
I–II | 38 | 22 (57.9) | 16 (42.1) | <0.001 | <0.001 |
|
III–IV | 34 | 5 (14.7) | 29 (85.3) |
|
|
| T
classification |
|
|
|
|
|
|
T1-T2 | 62 | 27 (43.5) | 35 (56.5) | 0.008 | 0.009 |
|
T3-T4 | 10 | 0 (0) | 10 (100) |
|
|
| N
classification |
|
|
|
|
|
| N0 | 46 | 22 (47.8) | 24 (52.2) | 0.016 | 0.017 |
|
N1-N2 | 26 | 5 (19.2) | 21 (80.8) |
|
|
| Grade
(differentiation) |
|
|
|
|
|
|
Well | 40 | 17 (42.5) | 23 (57.5) | 0.327 | 0.331 |
|
Moderate and poor | 32 | 10 (31.3) | 22 (68.7) |
|
|
| Vital status |
|
|
|
|
|
|
Alive | 42 | 21 (50.0) | 21 (50.0) | 0.010 | 0.010 |
|
Succumbed | 30 | 6 (20.0) | 24 (80.0) |
|
|
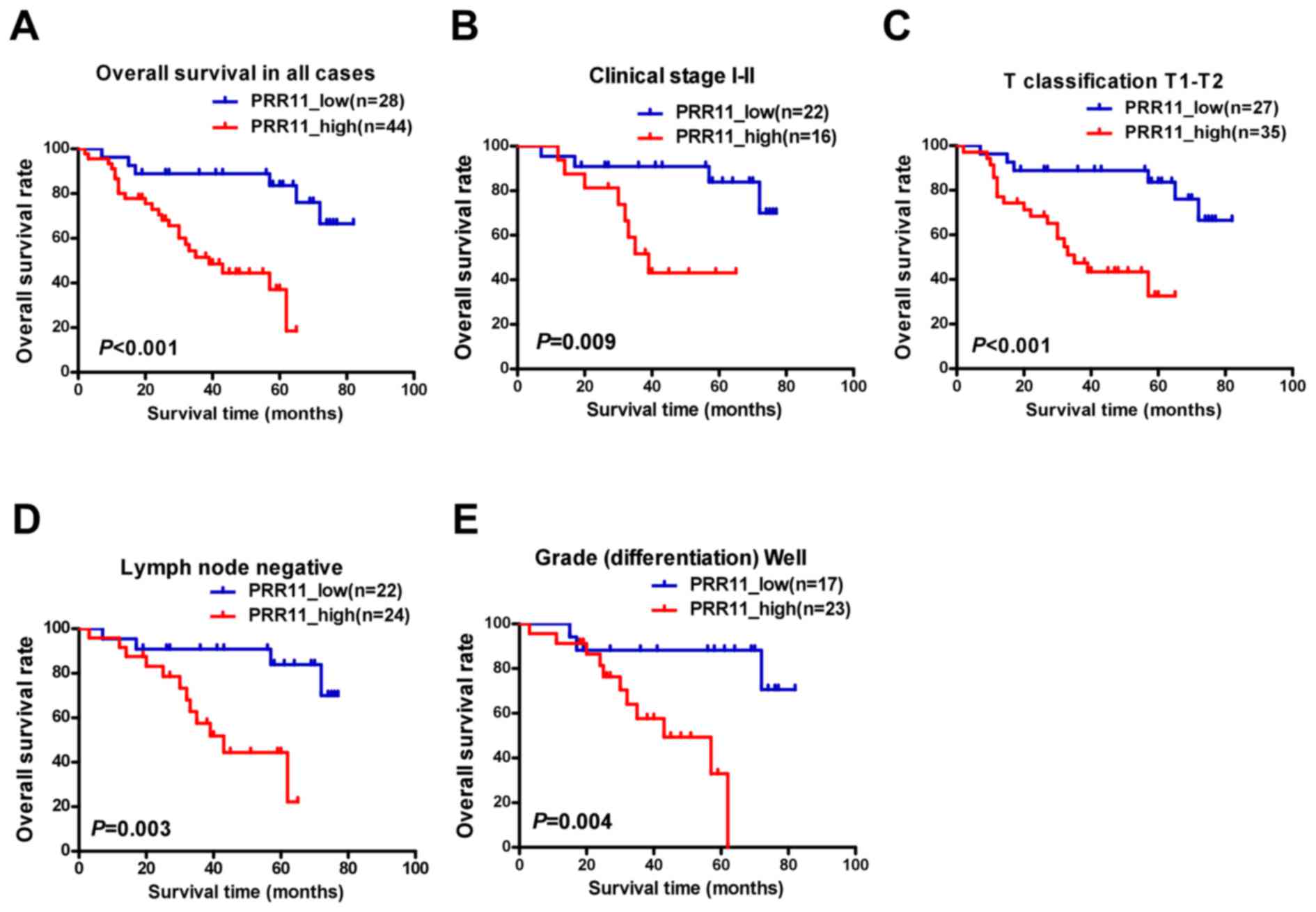
Elevated PRR11 expression is
associated with poor prognosis in patients with TSCC
To assess the clinical significance of elevated
PRR11 expression in patients with TSCC, survival rates were
analyzed using 5-year follow-up data. The 5-year cumulative
survival rates of patients with higher and lower PRR11 expression
were 36.8, and 83.8%, respectively. Kaplan-Meier analysis indicated
that high PRR11 expression was associated with shorter overall
survival time (P<0.001; Fig. 4A).
In addition, the prognostic value of PRR11 expression was assessed
by separating patients according to pathologic primary tumor
(pT)/pathologic regional lymph nodes (pN) status, clinical stage
and differentiation. Upregulation of PRR11 was a strong inverse
prognostic factor for patients with TSCC in clinical stages I–II
(early stage; P=0.009; Fig. 4B).
Similarly, patients with higher PRR11 expression demonstrated a
significantly shorter survival time (pT1-2, P<0.001, Fig. 4C; lymph node metastasis negative,
P=0.003, Fig. 4D; well
differentiated, P=0.004, Fig. 4E).
However, no statistically significant differences were identified
between PRR11 expression and survival time in subsets of clinical
stage III–IV, pT3-4, pN1-2, and moderate to poor differentiation,
which may reflect the limited number of patients recruited in each
subset.
Univariate survival analysis demonstrated that PRR11
expression was significantly associated with poorer overall
survival [hazard ratio (HR), 5.523; 95% confidence interval (CI),
1.977–15.427; P=0.001; Table III].
Multivariate Cox regression analysis revealed that PRR11 expression
was an independent prognostic factor for the overall survival of
patients with TSCC (HR, 5.454; 95% CI, 1.821–16.337; P=0.002;
Table III). Taken together, these
results indicate that PRR11 may be a useful prognostic factor in
patients with TSCC.
 | Table III.Univariate and multivariate analyses
of prognostic parameters in patients with tongue squamous cell
carcinoma assessed using Cox regression analysis. |
Table III.
Univariate and multivariate analyses
of prognostic parameters in patients with tongue squamous cell
carcinoma assessed using Cox regression analysis.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age | 1.460
(0.702–3.035) | 0.311 | – | – |
| Sex | 1.057
(0.516–2.169) | 0.879 | – | – |
| Clinical stage | 2.324
(1.113–4.852) | 0.025 | – | – |
| T
classification | 1.532
(0.581–4.039) | 0.389 | – | – |
| N
classification | 2.033
(0.989–4.176) | 0.054 | – | – |
| Differentiation
grade | 1.590
(0.776–3.260) | 0.206 | – | – |
| Proline rich 11
expression | 5.523
(1.977–15.427) | 0.001 | 5.454
(1.821–16.337) | 0.002 |
Discussion
Previous studies demonstrated that PRR11
overexpression is associated with cancer development and
progression in several tumor types (5,8–10). However, the role of PRR11 in TSCC has
not been addressed. In the present study, it was identified that
PRR11 expression was significantly increased in TSCC tissues
compared with non-tumorous oral mucosal tissues, and PRR11
overexpression was also associated with tumor stage. Univariate and
multivariate Cox regression analyses suggested that PRR11 was an
independent predictor for the prognosis of patients with TSCC.
Although the present study clarifies the pattern of PRR11
expression and potential clinical significance in TSCC, the
potential functions, and exact mechanisms of PRR11 overexpression
remain unclear.
It has been demonstrated that PRR11 contains two
proline-rich motifs and one zinc-finger domain (8). Proline-rich motifs bind SH3 domains and
mediate protein-protein interactions involved in cellular signaling
events (14), while zinc-finger
domains are known to bind double stranded DNA, and modulate gene
transcription (15). Additionally, in
lung, breast and numerous types of digestive system cancer,
silencing of PRR11 expression induced S-phase arrest, and inhibited
cell proliferation, migration, invasion and particularly tumor
growth (5,8,10,16,17).
However, forced expression of PRR11 inhibited cellular
proliferation, and was accompanied by premature chromatin
condensation in lung cancer cells (18). This discrepancy suggests that PRR11
may cooperate with other tumor-associated proteins in order to
exert its tumor-promoting activity. Additional studies are required
to examine this hypothesis.
Lung cancer-associated genes dehydrogenase/reductase
2 (DHRS2), erythrocyte membrane protein band 4.1 like 3
(EPB41L3), cyclin A1 (CCNA1), mitogen-activated
protein kinase kinase kinase kinase 4 (MAP4K4), nuclear
factor I B (NFIB) and ribonucleotide reductase catalytic
subunit M1 (RRM1) are deregulated following PRR11 knockdown
(8,16). Of these, DHRS2, CCNA1, MAP4K4
and RRM1 are important regulators of cell cycle progression,
while CCNA1, MAP4K4, NFIB, and EPB41L3
are involved in tumorigenesis (19–22). In
particular, several studies identified that MAP4K4 and
EPB41L3 are involved in invasiveness and/or metastasis
(23,24). These data indicate that PRR11 may
serve a potential role in proliferation, tumorigenesis,
invasiveness and/or metastasis. Additionally, in breast cancer,
PRR11 depletion reduces the expression of epithelial-mesenchymal
transition (EMT)-associated transcription factors snail family
transcriptional repressor (SNAI) 1, SNAI2, zinc finger-box-binding
homeobox (ZEB) 1 and ZEB2 (9). These
are members of the zinc-finger transcription factor family, and are
direct repressors of epithelial-cadherin transcription and
essential mediators of EMT (25,26). PRR11
may therefore be involved in proliferation, migration, invasion and
tumorigenesis by regulating the expression of these, and other
genes. However, the molecular mechanisms involved in the
association with TSCC patient survival warrant additional
investigation.
Due to its unusual histological makeup (rich
lymphatic network and highly muscularized structure), the tongue is
poorly equipped to protect itself from invasion and metastasis, and
TSCC is more frequently associated with metastasis to draining
lymph nodes compared with any other cancer of the oral cavity
(25,26). As nodal metastasis in the neck is an
important prognostic factor, patients with TSCC exhibit a
significantly poorer prognosis compared with those patients with
cancer in other sites of the oral cavity (27). The clinical course of TSCC is also
unpredictable, due to the relatively high rate of occult metastasis
in patients presenting with a very small primary tumor without
clinical evidence of metastatic disease (28). The use of neck dissection in the
surgical management of clinical stage I–II TSCC has been a source
of debate for this reason (29).
The results of the present study indicated that
patients with high levels of PRR11 expression exhibited a shorter
survival time, and PRR11 expression was also associated with
regional draining lymph nodes metastasis. PRR11 may therefore be a
useful prognostic marker in patients with TSCC, and may indicate
whether the use of neck dissection in clinical stages I–II is a
sensible option in the absence of TNM staging information.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos., 81272948 and
81201548).
Glossary
Abbreviations
Abbreviations:
|
PRR11
|
proline rich 11
|
|
TSCC
|
tongue squamous cell carcinoma
|
References
|
1
|
Chen YK, Huang HC, Lin LM and Lin CC:
Primary oral squamous cell carcinoma: An analysis of 703 cases in
southern Taiwan. Oral Oncol. 35:173–179. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sano D and Myers JN: Metastasis of
squamous cell carcinoma of the oral tongue. Cancer Metastasis Rev.
26:645–662. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Greene FL, Page DL, Fleming ID, et al:
AJCC Cancer Staging Hhandbook. 6th. New York: Springer-Verlag; pp.
35–46. 2002
|
|
4
|
Ota T, Suzuki Y, Nishikawa T, Otsuki T,
Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al:
Complete sequencing and characterization of 21,243 full-length
human cDNAs. Nat Genet. 36:40–45. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen Y, Cha Z, Fang W, Qian B, Yu W, Li W,
Yu G and Gao Y: The prognostic potential and oncogenic effects of
PRR11 expression in hilar cholangiocarcinoma. Oncotarget.
6:20419–20433. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tai YC, Domchek S, Parmigiani G and Chen
S: Breast cancer risk among male BRCA1 and BRCA2 mutation carriers.
J Natl Cancer Inst. 99:1811–1814. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yan J, Jiang J, Lim CA, Wu Q, Ng HH and
Chin KC: BLIMP1 regulates cell growth through repression of p53
transcription. Proc Natl Acad Sci USA. 104:1841–1846. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ji Y, Xie M, Lan H, Zhang Y, Long Y, Weng
H, Li D, Cai W, Zhu H, Niu Y, et al: PRR11 is a novel gene
implicated in cell cycle progression and lung cancer. Int J Biochem
Cell Biol. 45:645–656. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou F, Liu H, Zhang X, Shen Y, Zheng D,
Zhang A, Lai Y and Li H: Proline-rich protein 11 regulates
epithelial-to-mesenchymal transition to promote breast cancer cell
invasion. Int J Clin Exp Pathol. 7:8692–8699. 2014.PubMed/NCBI
|
|
10
|
Song Z, Liu W, Xiao Y, Zhang M, Luo Y,
Yuan W, Xu Y, Yu G and Hu Y: PRR11 is a prognostic marker and
potential oncogene in patients with gastric cancer. PLoS One.
10:e1289432015.
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Song L, Gong H, Lin C, Wang C, Liu L, Wu
J, Li M and Li J: Flotillin-1 promotes tumor necrosis factor-α
receptor signaling and activation of NF-κB in esophageal squamous
cell carcinoma cells. Gastroenterology. 143:995–1005.e12. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li J, Yu L, Zhang H, Wu J, Yuan J, Li X
and Li M: Down-regulation of pescadillo inhibits proliferation and
tumorigenicity of breast cancer cells. Cancer Sci. 100:2255–2260.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ball LJ, Kühne R, Schneider-Mergener J and
Oschkinat H: Recognition of proline-rich motifs by
protein-protein-interaction domains. Angew Chem Int Ed Engl.
44:2852–2869. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sera T: Zinc-finger-based artificial
transcription factors and their applications. Adv Drug Deliv Rev.
61:513–526. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao Q: RNAi-mediated silencing of
praline-rich gene causes growth reduction in human lung cancer
cells. Int J Clin Exp Pathol. 8:1760–1767. 2015.PubMed/NCBI
|
|
17
|
Wang Y, Zhang Y, Zhang C, Weng H, Li Y,
Cai W, Xie M, Long Y, Ai Q, Liu Z, et al: The gene pair PRR11 and
SKA2 shares a NF-Y-regulated bidirectional promoter and contributes
to lung cancer development. Biochim Biophys Acta. 1849:1133–1144.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang C, Zhang Y, Li Y, Zhu H, Wang Y, Cai
W, Zhu J, Ozaki T and Bu Y: PRR11 regulates late-S to G2/M phase
progression and induces premature chromatin condensation (PCC).
Biochem Biophys Res Commun. 458:501–508. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dooley AL, Winslow MM, Chiang DY, Banerji
S, Stransky N, Dayton TL, Snyder EL, Senna S, Whittaker CA, Bronson
RT, et al: Nuclear factor I/B is an oncogene in small cell lung
cancer. Genes Dev. 25:1470–1475. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu AW, Cai J, Zhao XL, Jiang TH, He TF,
Fu HQ, Zhu MH and Zhang SH: ShRNA-targeted MAP4K4 inhibits
hepatocellular carcinoma growth. Clin Cancer Res. 17:710–720. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Malumbres M and Carnero A: Cell cycle
deregulation: A common motif in cancer. Prog Cell Cycle Res.
5:5–18. 2003.PubMed/NCBI
|
|
22
|
Yageta M, Kuramochi M, Masuda M, Fukami T,
Fukuhara H, Maruyama T, Shibuya M and Murakami Y: Direct
association of TSLC1 and DAL-1, two distinct tumor suppressor
proteins in lung cancer. Cancer Res. 62:5129–5133. 2002.PubMed/NCBI
|
|
23
|
Bernkopf DB and Williams ED: Potential
role of EPB41L3 (protein 4.1B/Dal-1) as a target for treatment of
advanced prostate cancer. Expert Opin Ther Targets. 12:845–853.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Collins CS, Hong J, Sapinoso L, Zhou Y,
Liu Z, Micklash K, Schultz PG and Hampton GM: A small interfering
RNA screen for modulators of tumor cell motility identifies MAP4K4
as a promigratory kinase. Proc Natl Acad Sci USA. 103:3775–3780.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cano A, Pérez-Moreno MA, Rodrigo I,
Locascio A, Blanco MJ, del Barrio MG, Portillo F and Nieto MA: The
transcription factor snail controls epithelial-mesenchymal
transitions by repressing E-cadherin expression. Nat Cell Biol.
2:76–83. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lo HW, Hsu SC, Xia W, Cao X, Shih JY, Wei
Y, Abbruzzese JL, Hortobagyi GN and Hung MC: Epidermal growth
factor receptor cooperates with signal transducer and activator of
transcription 3 to induce epithelial-mesenchymal transition in
cancer cells via up-regulation of TWIST gene expression. Cancer
Res. 67:9066–9076. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rusthoven K, Ballonoff A, Raben D and Chen
C: Poor prognosis in patients with stage I and II oral tongue
squamous cell carcinoma. Cancer. 112:345–351. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Byers RM, El-Naggar AK, Lee YY, Rao B,
Fornage B, Terry NH, Sample D, Hankins P, Smith TL and Wolf PJ: Can
we detect or predict the presence of occult nodal metastases in
patients with squamous carcinoma of the oral tongue? Head Neck.
20:138–144. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
D'Cruz AK, Siddachari RC, Walvekar RR,
Pantvaidya GH, Chaukar DA, Deshpande MS, Pai PS and Chaturvedi P:
Elective neck dissection for the management of the N0 neck in early
cancer of the oral tongue: Need for a randomized controlled trial.
Head Neck. 31:618–624. 2009. View Article : Google Scholar : PubMed/NCBI
|