Introduction
Lung cancer is a kind of malignancy that arises from
epithelial cells. It is the leading cause of cancer-related death
worldwide (1). There were nearly 1.8
million new patients and caused 159 million deaths in 2012, of
which China accounted for more than one third (2). According to the size and appearance of
the malignant cells, lung cancer is categorized as non small cell
lung cancer (NSCLC) and small cell lung cancer (3). NSCLC represents approximately 85% of
lung cancer (4), and it can be
further subdivided into large cell carcinoma, lung adenocarcinoma
(LUAD) and lung squamous cell carcinoma (LUSC). There are a lot of
differences in molecular profiling, characteristics and therapeutic
methods between LUAD and LUSC (5).
Promoter hypermethylation of the tumor suppressor
genes has been recognized as an important factor in inducing
oncogenesis (6). For example, SRY-box
17 (SOX17) methylation was found in 60.2% of primary lung
cancer samples, and promoter methylation of SOX17 silenced
gene expression, leading to the elimination of cell proliferation
suppression in lung cancer (7).
Identification of specific gene hypermethylation may explain the
genomic instability and complexity of NSCLC and provide a basis for
targeted therapy or risk prediction.
AGTR1 encodes the angiotensin II (Ang II)
type I receptor that belongs to the family of G-protein coupled
receptors (8). Ang II is a major
effector controlling blood pressure in cardiovascular system and
induces diverse signal transduction pathways such as the biphasic
activation of Raf-1, MEK, and ERK via both Gq and Gi proteins
(9). It regulates the aldosterone
secretion and is involved in vascular remodeling, inflammation and
endothelial dysfunction (10).
Hypomethylation in the AGTR1 promoter had been validated to
be inversely correlated with uric acid levels, which can be a
significant risk predictor of essential hypertension (EH) (11). Besides, AGTR1 methylation had
been extensively studied in human cancers, such as oral squamous
cell carcinoma (12), colorectal
cancer (13), breast cancer (14), ovarian cancer (15) and oral cancer (12). For example, several studies support a
possible role for AGTR1 in regulating cell growth and
proliferation during cancer development in breast cancer (16), which was shown to be amplified and
overexpressed in 10–20% of breast cancer cases, and was even
markedly overexpressed more than 100 fold (14). Besides, the AGTR1 promoter
methylation is associated with oral squamous cell carcinoma (OSCC)
development (12). And AGTR1 was also
validated in stool DNA with a high detection sensitivity for
noninvasive diagnosis of colorectal cancer (13). In addition, DNA methylation microarray
datasets showed that methylation of AGTR1 might be an
effective biomarker for NSCLC diagnosis (17).
Since the detection of AGTR1 methylation in
different pathologic subtypes and stages have not been conducted,
we used LUSC and LUAD samples to study the effects of AGTR1
methylation on the risk of the disease in this study.
Materials and methods
Patients
Tumor tissues and paired adjacent non-tumor tissues
from 111 patients were collected from Affiliated Wujiang Hospital
of Nantong University (Jiangsu, China) between August 2010 and
October 2013. There were 73 male and 38 female patients with a mean
age of 63.59±10.19 years (range, 33–82 years), including 42
patients with LUSC and 69 patients with LUAD. Clinical pathological
data and isoforms were obtained from the patients' medical records
and pathology files. Clinical stage was classified by the third
edition of the American College of Chest Physicians (ACCP) Lung
Cancer Guidelines (LC III) (18). The
study protocol was approved by the Ethics Committee of Affiliated
Wujiang Hospital of Nantong University. All the patients had signed
the written informed consent forms.
DNA extraction and bisulphite
conversion
DNA was isolated from the formalin-fixed and
paraffin-embedded (FFPE) cancer sample using QIAamp DNA FFPE Tissue
Kit (Qiagen Inc., Hilden, Germany). DNA concentrations were
measured using the NanoDrop 1000 spectrophotometer (Thermo Fisher
Scientific Inc., Waltham, MA, USA). Then DNA were converted using
the EZ DNA Methylation-Gold Kit™ (Zymo Research Corporation,
Irvine, CA, USA).
Quantitative methylation-specific
PCR
SYBR green-based quantitative methylation-specific
PCR (qMSP) was conducted to detect the methylation level. PCR was
carried out in a final volume of 20 µl containing 5 µl SYBR mix, 4
µl H2O, 0.5 µl primer and 0.5 µl modified DNA. PCR
amplification were run in triplicate for every sample, and the
reactions was performed on Light Cycler 480 system (Roche Applied
Science, Mannheim, Germany) under the following conditions: 10 min
of denaturation at 95°C followed by 45 cycles of 20 sec at 95°C, 20
sec at 58°C and 30 sec at 72°C. Melting curves system was as
follows: 15 sec at 95°C, 1 min at 58°C, 95°C continuous prior to
drop the temperature to 40°C for 4 min. The Actin Beta gene
(ACTB) was used as an internal reference by amplifying
non-CpG sequences. Results with cycle threshold values (Ct values)
of ACTB> 40 were defined as detection failures.
M.SssI catalyzes the deamination of target cytosine to
uracil to generate DNAs that differ only in their CpG methylation
status (19). The methylated DNA was
prepared to be the positive internal control. Each set of
amplifications included a positive control, a negative control, and
a non-template control.
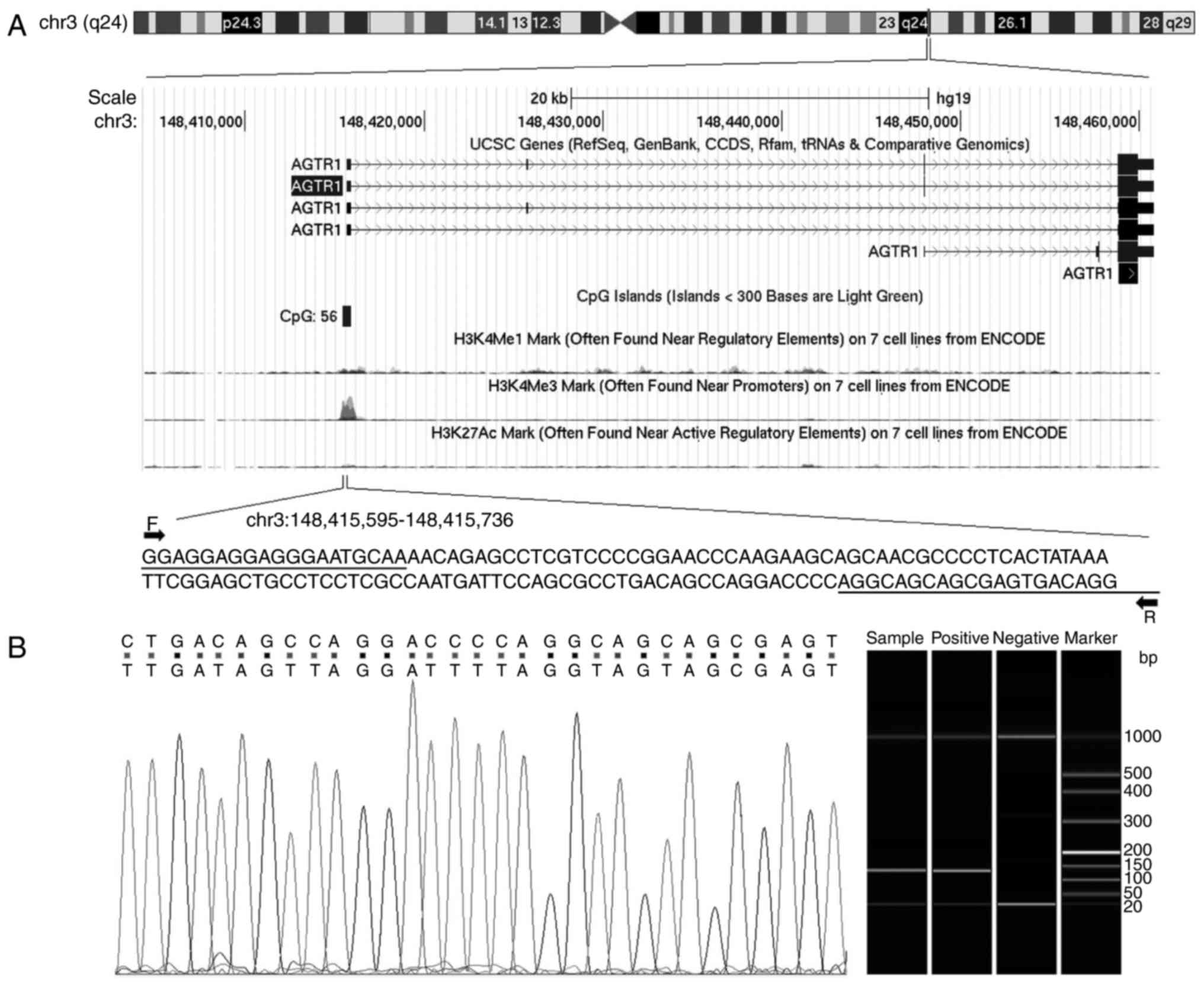
The sequences of primers for target and internal
reference genes were summarized in Table
I, and the genomic location of AGTR1 gene was shown in
Fig. 1A. Some of the PCR products
were analysed using to the Qsep100 DNA Analyzer (Bioptic Inc,
Taiwan, China) to validate the methylation status, and ultimately
the visible peaks were exported by the Q-analyzer software
(Fig. 1B). Besides, some of the
products were sequenced randomly using the Applied Bio
systems® 3730 DNA Analyzer (Applied Biosystems,
Warrington, UK) to confirm a complete bisulphite conversion
(Fig. 1B).
 | Table I.Primer sequences for quantitative
methylation-specific polymerase chain reaction. |
Table I.
Primer sequences for quantitative
methylation-specific polymerase chain reaction.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) | Product (bp) | Tm (°C) |
|---|
| AGTR1 |
GGAGGAGGAGGGAATGTAA |
CCTATCACTCGCTACTACCT | 142 | 58 |
| ACTB |
TGGTGATGGAGGAGGTTTAGTAAGT |
AACCAATAAAACCTACTCCTCCCTTAA | 133 | 58 |
Statistical analysis
For each sample the comparative Ct (ΔΔCt) method was
used to determine the relative methylation values. And the
percentage of methylated reference (PMR) was shown by using the
following formula (substituted the actual methylation value here):
[(gene/AGTR1) sample/(gene/AGTR1) positive] ×100%.
The median of PMR (97.4) was set as the cutoff value and defined
methylation as hypermethylation (positive) and hypomethylation
(negative) subgroups. Statistical analyses were performed using
SPSS 18.0 version (SPSS Inc., Chicago, IL, USA). The wilcoxon
signed ranks test was used to determine the difference of the
methylation index between tumor tissues and non-tumor tissues.
χ2 test was used to evaluate the association between
promoter methylation and clinical parameters. Overall survival in
relation to methylation status was calculated by Kaplan-Meier
survival curves, and survival differences were assessed in the
log-rank test. P<0.05 was considered to indicate statistical
significance.
Results
The result of sequenced PCR products showed that all
the non-CpG cytosines were converted to thymine, whereas the
cytosines of CG dinucleotide remained unchanged (Fig. 1). And the results of capillary
electrophoresis experiments showed the length of products were
correct.
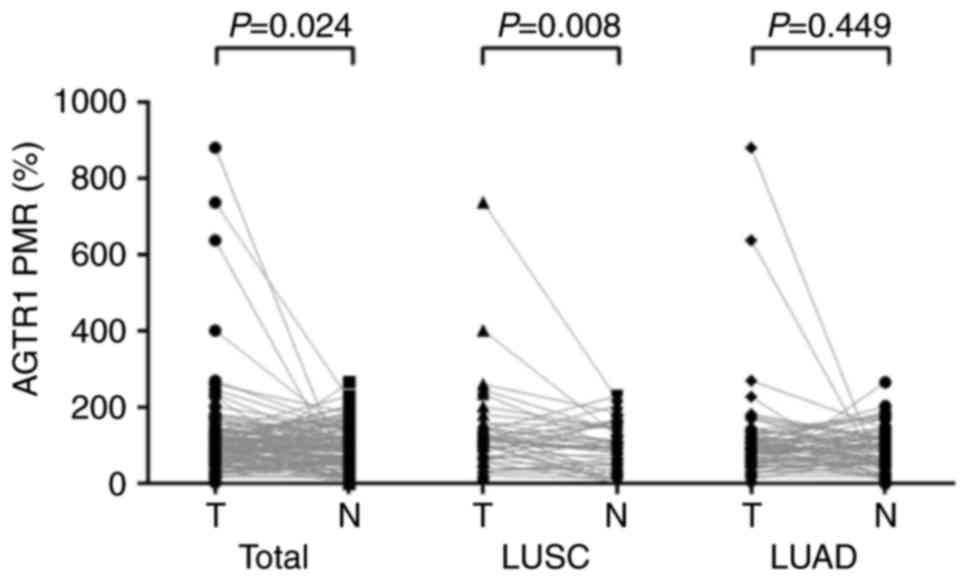
As the results revealed, there existed statistical
differences in AGTR1 methylation between tumor tissues and
the adjacent non-tumor tissues (PMR: 97.4 vs. 85, P=0.024). A
subgroup analysis indicated that there was a significantly induced
hypermethylation of AGTR1 in LUSC tumors (P=0.008) but not
in LUAD tumors (P=0.449, Fig. 2).
Further correlation analysis of clinicopathologic
characteristics in patients with promoter methylation status was
also performed (Table II). There was
higher AGTR1 promoter methylation in LUSC patients than LUAD
(Odds ratio=2.483, 95% CI=1.125–5.480, P=0.023). In addition,
AGTR1 promoter methylation was not associated with gender,
age, smoking history, clinical stage and lesion location in NSCLC
(Table II).
 | Table II.Association between gene methylation
and clinicopathological characteristics in patients with lung
cancer. |
Table II.
Association between gene methylation
and clinicopathological characteristics in patients with lung
cancer.
| Variables | n | AGTR1
hypermethylation | AGTR1
hypomethylation | OR (95% CI) | P-value |
|---|
| Gender |
|
|
|
|
|
|
Male | 73 | 38 | 35 | 1.206
(0.550–2.645) | 0.639 |
|
Female | 38 | 18 | 20 | 1 |
|
| Age (years) |
|
|
|
|
|
|
≤65 | 62 | 31 | 31 | 0.960
(0.454–2.031) | 0.915 |
|
>65 | 49 | 25 | 24 | 1 |
|
| Smoking
history |
|
|
|
|
|
|
Nonsmoker | 50 | 26 | 24 | 1.119
(0.530–2.366) | 0.768 |
|
Smoker | 61 | 30 | 31 | 1 |
|
| Histological
type |
|
|
|
|
|
|
LUSC | 42 | 27 | 15 | 2.483
(1.125–5.480) | 0.023 |
|
LUAD | 69 | 29 | 40 | 1 |
|
| Clinical stage |
|
|
|
|
|
|
I+II | 88 | 41 | 47 | 0.465
(0.179–1.209) | 0.112 |
|
III+IV | 23 | 15 | 8 | 1 |
|
| Tumor location |
|
|
|
|
|
| Left
lung | 46 | 25 | 21 | 1.306
(0.612–2.784) | 0.490 |
| Right
lung | 65 | 31 | 34 |
|
|
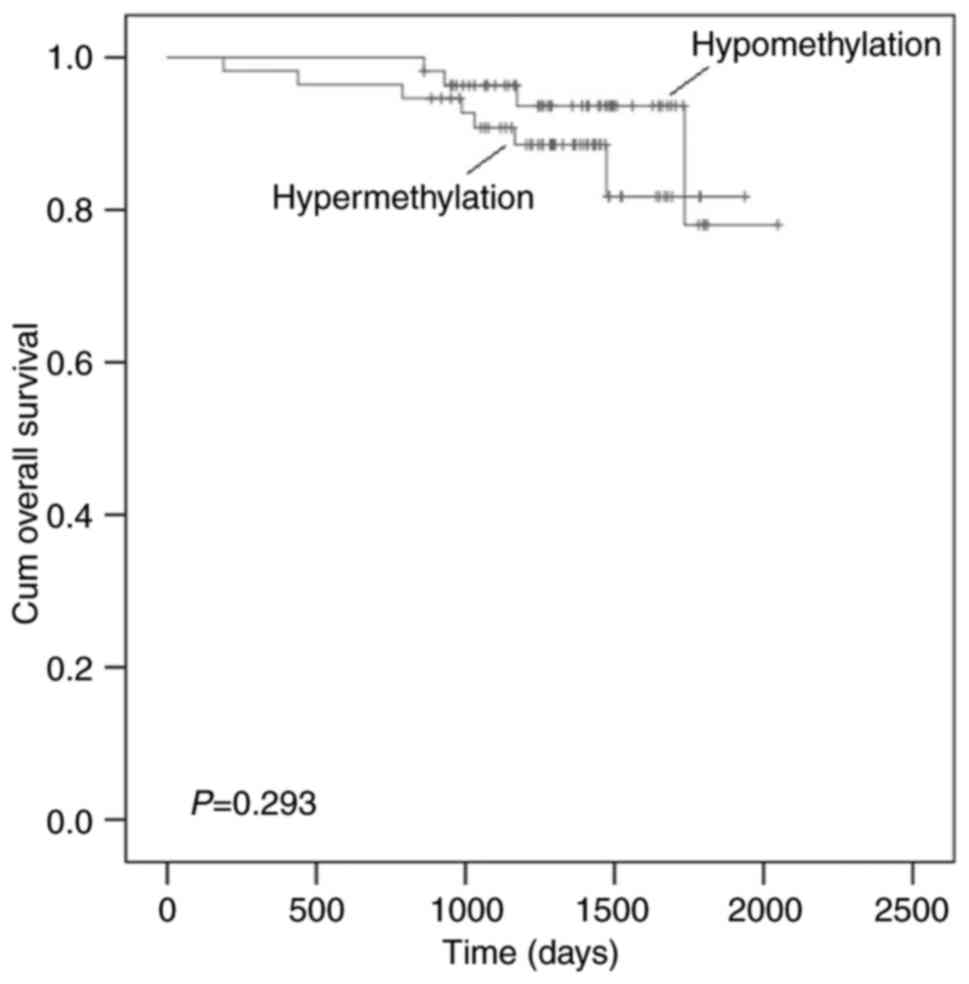
Previous study showed that AGTR1 is predictor
of progression-free survival (PFS) and response to
advanced/metastatic breast cancer (20). However, the correlation of
AGTR1 methylation with overall survival of cancer was
unknown. In this study, we examined the overall survival of
individuals according to AGTR1 PMR value, and no significant
difference was observed between LUSC and the LUAD patients
(P=0.293, Fig. 3).
To further reveal the correlation between expression
and methylation status of AGTR1 in LUSC and LUAD, the
MEXPRESS system (21) (http://mexpress.be) was used. And the Cancer Genome
Atlas (TCGA) data of 378 LUSC and 477 LUAD patients (21) indicated a lower expression in cancer
tissues, supporting an inverse correlation between gene expression
and AGTR1 methylation.
Discussion
NSCLC is a huge threat to human health, and the
study of the methylation and its relation to NSCLC is a field of
growing interest. AGTR1 acts as a novel component of the
renin-angiotensin system (RAS) to affect the blood pressure and
heart hypertrophy (15), targeting of
renin-angiotensin system (RAS) through AGTR1 blocker is
associated with the resistance phenomenon in treatment of resistant
breast cancers. And now the AGTR1 blocker has been applied
in the clinical oncology, such as the losartan for breast cancer
(22) and candesartan for prostate
cancer (23). And now emerging
evidences showed that AGTR1 regulates the cell proliferation
during cancer development, and promotes tumor invasion, migration,
metastasis and angiogenesis (24). In
consideration of the unfavorable treatment status of NSCLC,
detection of biomarkers like the AGTR1 is really necessary
and urgent.
In the present study, DNA methylation differences of
AGTR1 between 111 paired tumor sample and adjacent non-tumor
tissues were assessed using quantitative methylation-specific PCR.
We identified significant differences in AGTR1 methylation
between tumor tissues and the adjacent non-tumor tissues, this
observation lead us to suggest that CpG island methylation
phenotype in AGTR1 may be an early event during NSCLC
development. Besides, AGTR1 were hypermethylated in LUSC but
not in LUAD, and there were significant differences in AGTR1
promoter methylation between LUAD and LUSC patients, suggesting a
potential usage of this epigenetic biomarker in the diagnosis of
LUSC. In addition, we found that AGTR1 promoter methylation
was not associated with gender, age, smoking history, clinical
stage and lesion location in NSCLC. And research on the data of
TCGA suggested a negative correlation between gene expression and
AGTR1 methylation.
Our finding of AGTR1 promoter methylation as
a diagnostic marker of NSCLC was consistent with a study of
high-throughput DNA methylation microarray dataset for Chinese Han
NSCLC retrospective cohort which showed the significant association
between methylation and NSCLC (17).
This was also consistent with the result of TCGA for LUSC and LUAD
patients, suggesting that AGTR1 might be associated with
increased risk of NSCLC.
Our results showed that there were significant
differences in AGTR1 promoter methylation between LUAD and
LUSC patients, and there was a significantly induced
hypermethylation of AGTR1 in LUSC tumors compared with the
normal tissues but not in LUAD. Previous study declared
differentially methylated genes were specially differential
methylation in unique cancer, and the DNA methylation correlation
network had been built based on the methylation correlation, such
as seven biomarkers (PCDHB15, IGF1, PRRT1, CYGB, WBSCR17,
ACTG2 and GYPC) can distinguish the breast cancer into
high-risk group and low-risk group. Likewise, different methylation
status in eight biomarkers (ZBTB32, GPSM1, SALL1, OR51B4,
MAGEA8, SALL3, CCL8 and TMEFF2) in colon cancer showed
the different risk level (25). So
the methylated AGTR1 might be identified as the specific
driver gene for LUSC, which might be implicated in cancer type
specific pathway and be expected to be used to explain part of the
heterogeneity between these two NSCLC types.
NSCLC was regarded as a complex diseases, and some
clinicopathologic characteristics were involved in cancer, such as
the smoking history, which was identified as the main risk factor
of lung cancer (26), and association
between tobacco smoking and promoter DNA hypermethylation had been
demonstrated for several genes, such as the transcription factor 21
(TCF21) and MicroRNA Let-7a-3 (27). The previous study on primary lung
cancer cases in China (N=40,022) showed that males were 1.5 times
more likely to have lung cancer than females (28), but the incidence of lung cancer among
women was rising exponentially as a consequence of recent changes
in gender-specific smoking patterns. Besides, women with lung
cancer were diagnosed at younger ages than men, suggesting the
different gender and age susceptibility existed in patients
(29). Notablely, the clinical stages
were important prognostic factors in NSCLC. The advanced disease
stages affected survival negatively (30). Besides, even though apparently
symmetric, previous study showed that higher incidence of lung
cancer was found on the right side (31), just as the breast cancer, which was
about 5% more likely to be diagnosed in the left breast than the
right (32). Breast cancer that
arising on different sides of the body presented different cancer
traits inferred from methylation and expression profiles (31), and this result contributed to serve as
proof of principle for other bilateral cancers like the lung
cancer. In our study, AGTR1 promoter methylation was shown
likely to be not linked with gender, age, smoking history, clinical
stage and tumor location. But the further verification is required
to ensure the results considering of the limitation factors existed
in our experimental design.
In the present study, we enrolled the relatively
large cohorts that contained 111 patients, and we collected the
precious surgery tissues to perform the survey by general
methodology that easily to be applied to different cancer subtypes.
However, there existed several limitations here. Firstly, because
of the limited time, the large cell carcinoma was not included in
our study, the study design may still be too simple to uncover the
complicated trait of NSCLC. Secondly, because of the limited amount
of sample, we did not detect the expression of different methylated
AGTR1 and replaced by a database analysis of TCGA, which
showed the negative relevance between methylation and expression of
AGTR1. However, one previous studies showed there was no
correlation between the degree of methylation and mRNA abundance of
AGTR1 in early gestation amnion and placenta (33), so the further studies are needed.
Thirdly, the mechanisms by which AGTR1 methylation affect
the LUSC remained largely elusive. Fourthly, we just detect the
part of the content area, it could be possible that some more CpG
sites are required for gene function, besides, additional
epigenetic alteration, such as the histone modifications that
closely connected with the gene methylation are needed for study on
NSCLC progression. In addition, only the AGTR1 gene was
investigated in the present study, and additional relevant genes
should be explored in the future.
In conclusion, our findings of the association
between AGTR1 methylation and LUSC provided a potential
biomarker for detection, diagnosis and risk prediction for
LUSC.
Acknowledgements
The research and publication was supported by the
grants from the National Natural Science Foundation of China
(31100919, 81371469), K. C. Wong Magna Fund of the Ningbo
University, National Natural Science Foundation for the Youth of
China (81402220) and Suzhou Planning Project of Science and
Technology (SYS201301).
References
|
1
|
Barlési F, Giaccone G, Gallegos-Ruiz MI,
Loundou A, Span SW, Lefesvre P, Kruyt FA and Rodriguez JA: Global
histone modifications predict prognosis of resected non small-cell
lung cancer. J Clin Oncol. 25:4358–4364. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McGuire S: World cancer report 2014.
Geneva, Switzerland: World health organization, international
agency for research on cancer, WHO press, 2015. Adv Nutr.
7:418–419. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Collins LG, Haines C, Perkel R and Enck
RE: Lung cancer: Diagnosis and management. Am Fam Physician.
75:56–63. 2007.PubMed/NCBI
|
|
4
|
NSCLC Meta-analysis Collaborative Group:
Preoperative chemotherapy for non-small-cell lung cancer: A
systematic review and meta-analysis of individual participant data.
Lancet. 383:1561–1571. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhan C, Yan L, Wang L, Sun Y, Wang X, Lin
Z, Zhang Y, Shi Y, Jiang W and Wang Q: Identification of
immunohistochemical markers for distinguishing lung adenocarcinoma
from squamous cell carcinoma. J Thorac Dis. 7:1398–1405.
2015.PubMed/NCBI
|
|
6
|
Hong Q, Li Y, Chen X, Ye H, Tang L, Zhou
A, Hu Y, Gao Y, Chen R, Xia Y and Duan S: CDKN2B, SLC19A3 and DLEC1
promoter methylation alterations in the bone marrow of patients
with acute myeloid leukemia during chemotherapy. Exp Ther Med.
11:1901–1907. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yin D, Jia Y, Yu Y, Brock MV, Herman JG,
Han C, Su X, Liu Y and Guo M: SOX17 methylation inhibits its
antagonism of Wnt signaling pathway in lung cancer. Discov Med.
14:33–40. 2012.PubMed/NCBI
|
|
8
|
Haas U, Sczakiel G and Laufer SD:
MicroRNA-mediated regulation of gene expression is affected by
disease-associated SNPs within the 3′-UTR via altered RNA
structure. RNA Biol. 9:924–937. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsygankova OM, Peng M, Maloney JA, Hopkins
N and Williamson JR: Angiotensin II induces diverse signal
transduction pathways via both Gq and Gi proteins in liver
epithelial cells. J Cell Biochem. 69:63–71. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Briet M, Barhoumi T, Mian MOR, Coelho SC,
Ouerd S, Rautureau Y, Coffman TM, Paradis P and Schiffrin EL:
Aldosterone-induced vascular remodeling and endothelial dysfunction
require functional angiotensin type 1a receptors. Hypertension.
67:897–905. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fan R, Mao S, Zhong F, Gong M, Yin F, Hao
L and Zhang L: Association of AGTR1 promoter methylation levels
with essential hypertension risk: A matched case-control study.
Cytogenet Genome Res. 147:95–1029. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Foy JP, Pickering CR, Papadimitrakopoulou
VA, Jelinek J, Lin SH, William WN Jr, Frederick MJ, Wang J, Lang W,
Feng L, et al: New DNA methylation markers and global DNA
hypomethylation are associated with oral cancer development. Cancer
Prev Res (Phila). 8:1027–1035. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Carmona FJ, Azuara D, Berenguer-Llergo A,
Fernández AF, Biondo S, de Oca J, Rodriguez-Moranta F, Salazar R,
Villanueva A, Fraga MF, et al: DNA methylation biomarkers for
noninvasive diagnosis of colorectal cancer. Cancer Prev Res
(Phila). 6:656–665. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rhodes DR, Ateeq B, Cao Q, Tomlins SA,
Mehra R, Laxman B, Kalyana-Sundaram S, Lonigro RJ, Helgeson BE,
Bhojani MS, et al: AGTR1 overexpression defines a subset of breast
cancer and confers sensitivity to losartan, an AGTR1 antagonist.
Proc Natl Acad Sci USA. 106:10284–10289. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bi FF, Li D, Cao C, Li CY and Yang Q:
Regulation of angiotensin II type 1 receptor expression in ovarian
cancer: A potential role for BRCA1. J Ovarian Res. 6:892013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Du N, Feng J, Hu LJ, Sun X, Sun HB, Zhao
Y, Yang YP and Ren H: Angiotensin II receptor type 1 blockers
suppress the cell proliferation effects of angiotensin II in breast
cancer cells by inhibiting AT1R signaling. Oncol Rep. 27:1893–1903.
2012.PubMed/NCBI
|
|
17
|
Guo S, Yan F, Xu J, Bao Y, Zhu J, Wang X,
Wu J, Li Y, Pu W, Liu Y, et al: Identification and validation of
the methylation biomarkers of non-small cell lung cancer (NSCLC).
Clin Epigenetics. 7:32015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lewis SZ, Diekemper R and Addrizzo-Harris
DJ: Methodology for development of guidelines for lung cancer:
Diagnosis and management of lung cancer, 3rd ed: American college
of chest Physicians evidence-based clinical practice guidelines.
Chest. 143 (5 Suppl):41S–50S. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hughes S and Jones JL: The use of multiple
displacement amplified DNA as a control for methylation specific
PCR, pyrosequencing, bisulfite sequencing and methylation-sensitive
restriction enzyme PCR. BMC Mol Biol. 8:912007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Salvador J, Manso L, de la Haba J, Jaen A,
Ciruelos E, de Villena MC, Gil M, Murias A, Galan A, Jara C, et al:
Final results of a phase II study of paclitaxel, bevacizumab, and
gemcitabine as first-line therapy for patients with HER2-negative
metastatic breast cancer. Clin Transl Oncol. 17:160–166. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Koch A, De Meyer T, Jeschke J and Van
Criekinge W: MEXPRESS: Visualizing expression, DNA methylation and
clinical TCGA data. BMC Genomics. 16:6362015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Namazi S, Sahebi E, Rostami-Yalmeh J,
Jaberipour M, Razmkhah M, Hosseini A and Arabsolghar R: Effect of
angiotensin receptor blockade on prevention and reversion of
tamoxifen-resistant phenotype in MCF-7 cells. Tumour Biol.
36:893–900. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Alhusban A, Al-Azayzih A, Goc A, Gao F,
Fagan SC and Somanath PR: Clinically relevant doses of candesartan
inhibit growth of prostate tumor xenografts in vivo through
modulation of tumor angiogenesis. J Pharmacol Exp Ther.
350:635–645. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen X, Meng Q, Zhao Y, Liu M, Li D, Yang
Y, Sun L, Sui G, Cai L and Dong X: Angiotensin II type 1 receptor
antagonists inhibit cell proliferation and angiogenesis in breast
cancer. Cancer Lett. 328:318–324. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang C, Zhao H, Li J, Liu H, Wang F, Wei
Y, Su J, Zhang D, Liu T and Zhang Y: The identification of specific
methylation patterns across different cancers. PLoS One.
10:e01203612015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Word B, Lyn-Cook LE Jr, Mwamba B, Wang H,
Lyn-Cook B and Hammons G: Cigarette smoke condensate induces
differential expression and promoter methylation profiles of
critical genes involved in lung cancer in NL-20 lung cells in
vitro: Short-term and chronic exposure. Int J Toxicol. 32:23–31.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lyn-Cook L, Word B, George N, Lyn-Cook B
and Hammons G: Effect of cigarette smoke condensate on gene
promoter methylation in human lung cells. Tob Induc Dis. 12:152014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen K, Wang PP, Sun B, Li Q, Perruccio A,
Power D, Wang C, He MMH, Shibei Y, et al: Twenty-year secular
changes in sex specific lung cancer incidence rates in an urban
Chinese population. Lung Cancer. 51:13–19. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Isla D, Majem M, Viñolas N, Artal A,
Blasco A, Felip E, Garrido P, Remón J, Baquedano M, Borrás JM, et
al: A consensus statement on the gender perspective in lung cancer.
Clin Transl Oncol. 19:527–535. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Urvay SE, Yucel B, Erdis E and Turan N:
Prognostic factors in stage III non-small-cell lung cancer
patients. Asian Pac J Cancer Prev. 17:4693–4697. 2016.PubMed/NCBI
|
|
31
|
Campoy EM, Laurito SR, Branham MT, Urrutia
G, Mathison A, Gago F, Orozco J, Urrutia R, Mayorga LS and Roqué M:
Asymmetric cancer hallmarks in breast tumors on different sides of
the body. PLoS One. 11:e01574162016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Perkins CI, Hotes J, Kohler BA and Howe
HL: Association between breast cancer laterality and tumor
location, United States, 1994–1998. Cancer Causes Control.
15:637–645. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sykes SD, Mitchell C, Pringle KG, Wang Y,
Zakar T and Lumbers ER: Methylation of promoter regions of genes of
the human intrauterine Renin Angiotensin system and their
expression. Int J Endocrinol. 2015:4598182015. View Article : Google Scholar : PubMed/NCBI
|