Introduction
Cancer stem cells, a small number of cells with the
potential for unlimited self-renewal, have been recently theorized
to play a substantial role in tumor growth and the promotion of
tumor formation (1). Cancer stem
cells have already been identified in numerous types of carcinoma,
including brain neoplasms, leukemia and cancer of the breast, colon
and lung (2–10). In malignant head and neck tumor stem
cells, cluster of differentiation (CD)133 has been reported to be a
biological marker of cancer stem cells in laryngeal cancer
(11) and nasopharyngeal carcinoma
(12); CD133, CD44 and aldehyde
dehydrogenase 1 are all cancer stem cell markers in oral carcinoma
(13,14). However, there have been few studies
investigating the role of cancer stem cells in hypopharyngeal
carcinoma. CD44+ cells from the hypopharyngeal cancer
FaDu cell line were previously found to have a higher proliferative
capacity and tumorigenic potential, which suggests that
CD44+ hypopharyngeal tumor cells may be cancer stem
cells, or that cancer stem cells in hypopharyngeal cancer exist in
the CD44+ tumor cell population (15). To determine whether the CD44 gene
serves a notable role in the cancer stem cell properties of
CD44+ cells, the tumorigenicity of CD44+
cells was assessed in the FaDu cell line following the silencing of
CD44 gene expression using recombinant lentiviral vectors that
specifically knocked down the expression of CD44. The viability of
FaDu cells in vitro and their tumorigenicity in vivo
were subsequently examined to determine whether CD44 confers the
biological characteristics of cancer stem cells in CD44+
cells in hypopharyngeal cancer.
Materials and methods
Design and construction of CD44 short
hairpin RNA (shRNA) -expressing lentivirus
The hU6-MCS-CMV-EGFP-iRNA plasmid (Shanghai Genechem
Co., Ltd., Shanghai, China) was used to generate the CD44
shRNA-expressing lentivirus. According to The National Center for
Biotechnology Information GenBank (https://www.ncbi.nlm.nih.gov/genbank/) published human
CD44 mRNA (NM_00100139) sequence information and RNA interference
sequence design principles, four interfering target sequences were
designed (Table I). Subsequently,
target sequences were used to carried out BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) comparison
in human genome data to exclude the other similar nucleotide
sequences. For each targeting sequence, two oligonucleotides
containing the sense and antisense of the targeting sequences (with
a loop sequence in between) were synthesized (Table II). The two oligonucleotides were
resolved together in water at 90°C for 15 min and then placed at
room temperature to cool down and form double-stranded DNA. The
double-stranded DNA was ligated into the hU6-MCS-CMV-EGFP-iRNA
plasmid digested with AgeI and EcoRI restriction
endonucleases (New England Biolabs, Inc., Ipswich, MA, USA) to
generate the CD44 shRNA-expressing lentiviral vectors:
LV-GFP-CD44-shRNA-1, LV-GFP-CD44-shRNA-2, LV-GFP-CD44-shRNA-3 and
LV-GFP-CD44-shRNA-4. The ligated vectors were transformed into
competent Escherichia coli DH5α cells (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), and a single colony was
selected for PCR amplification. The reaction mixture volume of 20
µl, comprising 0.4 µl upstream and downstream primers (upstream:
5′-CCATGATTCCTTCATATTTGC-3′; downstream:
5′-CGCGTGGATAACCGTATTAC-3′), 15.2 µl ddH2O, 2 µl 10X
buffer (Takara Bio Inc., Japan), 0.8 µl 2.5 mM dNTP Mixture (Takara
Bio Inc., Otsu, Japan), 0.2 µl Taq DNA polymerase (Takara Bio Inc.,
Japan), and 1 µl 10 ng/µl template. The PCR reaction thermocycler
conditions were as follows: Pre-denaturing at 94°C for 30 sec,
denaturing at 94°C for 30 sec, annealing at 55°C for 30 sec,
extending at 72°C for 40 sec, then 72°C for a final 6 min. The
total PCR process was 30 cycles. Reactions were performed with the
Applied Biosystems Veriti Thermal Cycler (Thermo Fisher Scientific,
Inc.). The QIAGEN Plasmid Midi Kit (Qiagen GbmH, Hilden, Germany)
was used to extract the plasmid, which was then sent to Shanghai
Genechem Co., Ltd., (Shanghai, China) for nucleotide sequencing to
verify the correct constructs (16).
293T cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.). Recombinant lentiviruses were produced by
co-transfecting 293T cells with the recombinant lentiviral vector
and packaging plasmids (Shanghai Genechem Co., Ltd.) (pHelper 1.0,
including gag/pol, and pHelper 2.0, including vesicular stomatitis
virus G) using the cationic lipid complex method (Lipofectamine
2000; Invitrogen; Thermo Fisher Scientific, Inc.). The culture
supernatants containing the produced viruses were harvested 48 h
after transfection and concentrated by centrifugation at 4,000 × g
at 4°C for 10–15 min. Aliquots of the concentrated viruses were
stored at 80°C for subsequent use. The infectious titer was
measured using a proportional dilution method with 293T cells and
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR).
 | Table I.Sequence of CD44 siRNA group. |
Table I.
Sequence of CD44 siRNA group.
| CD44-knockdown siRNA
group | Sequence | GC content, % |
|---|
| KD1 |
CCTCTGCAAGGCTTTCAAT | 47.37 |
| KD2 |
GCTCTGAGCATCGGATTTG | 52.63 |
| KD3 |
GCATCGGATTTGAGACCTG | 52.63 |
| KD4 |
GGCTTTCAATAGCACCTTG | 47.37 |
 | Table II.Sequences of oligonucleotides for
constructing recombinant lentiviral vectors expressing shRNAs. |
Table II.
Sequences of oligonucleotides for
constructing recombinant lentiviral vectors expressing shRNAs.
| shRNA group | 5′ | STEMP | Loop | STEMP | 3′ |
|---|
| KD1 sense | CCGG | GACCTCTGCAAGGC | CTCGAG |
ATTGAAAGCCTTGCAG | TTTTTG |
|
|
| TTTCAAT |
| AGGTC |
|
| KD1 antisense | AATTCAAAAA | GACCTCTGCAAGGC | CTCGAG |
ATTGAAAGCCTTGCAG |
|
|
|
| TTTCAAT |
| AGGTC |
|
| KD2 sense | CCGG | AAGCTCTGAGCATC | CTCGAG |
CAAATCCGATGCTCAG | TTTTTG |
|
|
| GGATTTG |
| AGCTT |
|
| KD2 antisense | AATTCAAAAA | AAGCTCTGAGCATC | CTCGAG |
CAAATCCGATGCTCAG |
|
|
|
| GGATTTG |
| AGCTT |
|
| KD3 sense | CCGG | GAGCATCGGATTTG | CTCGAG |
CAGGTCTCAAATCCGA | TTTTTG |
|
|
| AGACCTG |
| TGCTC |
|
| KD3 antisense | AATTCAAAAA | GAGCATCGGATTTG | CTCGAG |
CAGGTCTCAAATCCGA |
|
|
|
| AGACCTG |
| TGCTC |
|
| KD4 sense | CCGG | AAGGCTTTCAATAG | CTCGAG |
CAAGGTGCTATTGAAA | TTTTTG |
|
|
| CACCTTG |
| GCCTT |
|
| KD4 antisense | AATTCAAAAA | AAGGCTTTCAATAG | CTCGAG |
CAAGGTGCTATTGAAA |
|
|
|
| CACCTTG |
| GCCTT |
|
RT-qPCR
Total RNA from 293T or FaDu cells was extracted
using TRIzol® (Takara Bio Inc.), and reversed
transcribed according to the manufacturer's protocol. cDNA samples
were amplified using SYBR® Premix Ex Taq™ (Takara Bio
Inc.). The primer sequences were used as follows: CD44:
5′-AGGCTGAGACAGGAGGTTA-3′ (forward); 5′-CCTCCCTTATTTCTATCGTG-3′
(reverse); GAPDH: 5′-GGTGAAGGTCGGTGGAACG-3′ (forward);
5′-CTCGCTCCTGGAAGATGGTG-3′ (reverse). The PCR reaction conditions
were as follows: 95°C pre-denaturing for 30 sec, 95°C denaturing
for 5 sec, 60°C annealing for 34 sec for 40 cycles, then 95°C
denaturing for 15 sec, 60°C annealing for 60 sec, and 95°C for a
final 15 sec. Reactions were performed with the ABI PRISM 7500
System (Thermo Fisher Scientific, Inc.). Gene expression in each
sample was normalized to GADPH expression. Statistical analyses
were performed on qPCR results obtained through the
2−ΔΔCq method (17). The
experiment was repeated in triplicate.
Screening of shRNA-expressing
lentiviruses
FaDu cells were subcultured at a density of
1×106 cells per well into 6-well tissue culture plates.
The experiment was divided into a low multiplicity of infection
(MOI) group and a high MOI group. There were also six subgroups: A
blank control group [CON, infected with PBS (0.01 mol/l) alone]; a
negative virus control group [NC, infected with the negative
control lentiviral vector (8E + 8 TU/ml)]; and four RNAi groups
(KD1, infected with the shRNA1-CD44 lentiviral vector; KD2,
infected with the shRNA2-CD44 lentiviral vector; KD3, infected with
the shRNA3-CD44 lentiviral vector; and KD4, infected with the
shRNA4-CD44 lentiviral vector; all shRNA sequences are shown in
Table II). Following incubation at
37°C for 48 h of culture, cells were infected with specific or
negative control lentiviral vectors, at the aforementioned MOI.
Cells were examined by fluorescence microscopy 72 h after
lentiviral transduction. On day 5 after transduction, the cells
were harvested to determine the efficiency of CD44 silencing by
RT-qPCR. The cells were firstly washed with PBS and digested with
0.25% trypsin at 37°C for 3 min. RPMI-1640 medium (1 ml) with 10%
FBS to stop digestion was then added, followed by centrifugation at
500 × g for 5 min at 28°C. In the RNAi group, the most efficient
shRNA-CD44 lentiviral vector (hereafter termed KD) was used in the
following in vitro and in vivo experiments.
Cell culture
The FaDu and 293T cells, obtained from the Institute
of Biochemistry and Cell Biology, Shanghai Institute for Biological
Sciences, Chinese Academy of Sciences (Shanghai, China), were
recovered from frozen storage and cultured to 70–80% confluence in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.),
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.) in an incubator maintained with 5% CO2
at a temperature of 37°C. The cells were then washed with PBS
(Gibco; Thermo Fisher Scientific, Inc.) and digested with 0.25%
trypsin (Gibco; Thermo Fisher Scientific, Inc.).
FaDu cell viability assay
The FaDu cells from the CON, NC and KD groups were
seeded at a density of 5,000 cells/well in 96-well plates. Cell
viability assays were performed at days 1, 3, 5 and 7 using an MTT
assay (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany). Viable cells
were quantified by measuring the absorption spectra at 490 nm with
a Versamax microplate reader (Molecular Devices, LLC, Sunnyvale,
CA, USA). The wells were observed daily using an Olympus CKX41
light microscope (Olympus Corporation, Tokyo, Japan). A total of 6
duplicate wells were used for each group. The samples were loaded
into 96-well plates and the optical density values were measured at
490 nm with the spectrophotometer every 48 h. Growth curves were
prepared using the mean values.
In vivo tumorigenicity assay of FaDu
cells
A total of 20 non-obese diabetic/severe combined
immunodeficient (NOD/SCID) mice (6 weeks of age; weight, 20–25 g)
were obtained from Shanghai Si Laike Experimental Animals
(Shanghai, China). NOD/SCID mice consisted of equal numbers of
males and females. Food and water were available ad libitum
and the mice were kept in standard laboratory conditions. Mice were
maintained at a temperature of 23±2°C, 65% humidity and 12-h
light/dark cycles. Animals were randomly divided into three groups:
A blank control group (CON, injected with FaDu cells infected with
PBS alone; n=8), a negative virus control group (NC, injected with
FaDu cells infected with the negative control lentiviral vector;
n=4) and an RNAi group (KD, injected with FaDu cells infected with
the RNAi-CD44 lentiviral vector; n=8). Cells (1×106)
were suspended in 0.1 ml PBS and injected into the hypodermis of
the right armpit of the NOD/SCID mice. Twice a week, the injection
site of the NOD/SCID mice was palpated to check for tumor formation
and growth. A total of 7 weeks after the injection (when the tumor
size was ~1 cm in diameter), the mice were sacrificed using carbon
dioxide euthanasia. The flow rate of CO2 was 28% of the
chamber volume/minute, and the volume of the cage was 294×190×130
mm. The final concentration of CO2 was 80%. Euthanasia
was confirmed through the disappearance of physiological responses
such as corneal reflexes, and the lack of detectable heart beat for
1 min. Tumor nodules were immediately extracted from the animals.
The long axis (a) and short axis (b) of all tumors were measured
and the tumor volume (V) was calculated as follows: V =
ab2 / 2. Finally, all tumors were histopathologically
examined and diagnosed by two senior pathologists. The animal
experiments were approved by the Animal Ethics Committee of Xinhua
Hospital, Shanghai Jiao Tong University School of Medicine
(Shanghai, China).
Statistical analysis
Data are expressed as the mean ± standard deviation,
and were analyzed with SAS version 9.13 software (SAS Institute
Inc., Cary, NC, USA). Statistical analyses were performed on
RT-qPCR results obtained through the 2−ΔΔCq method. The
distribution of tumor volumes was analyzed for normality using the
Shapiro-Wilk test. The comparison of mean tumor volumes was
conducted using one-way analysis of variance followed by LSD
post-hoc test. Fisher's exact test was used to determine the
difference in the number of tumorigenic mice between two groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Verification of recombinant lentiviral
vectors targeting CD44
The four plasmids containing the four CD44-targeting
shRNA clones, with the correct insertion and orientation of the
shRNA fragment of the hU6-MCS-CMV-EGFP expression vector, were
detected by PCR (Fig. 1). These
recombinant lentiviral vectors (Lv-shRNA-CD44) were used to
specifically knockdown CD44 expression.
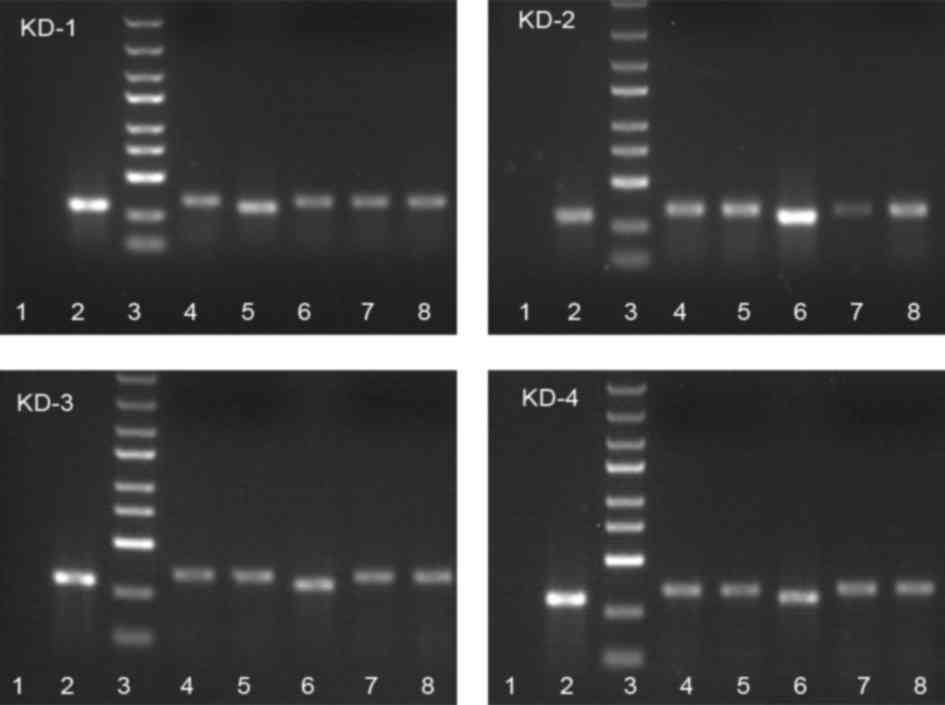 | Figure 1.Analysis of recombinant cluster of
differentiation 44 short hairpin RNA by polymerase chain reaction.
Lane 1, negative control (double-distilled H2O); lane 2,
negative control (shRNA fragment not inserted, 307 bp); lane 3,
markers from top to bottom: 5, 3, 2, 1.5, 1 kb, 750, 500, 250, 100
bp; lanes 4–8, 341-bp-long positive clones. |
Screening of lentiviral vectors
targeting CD44
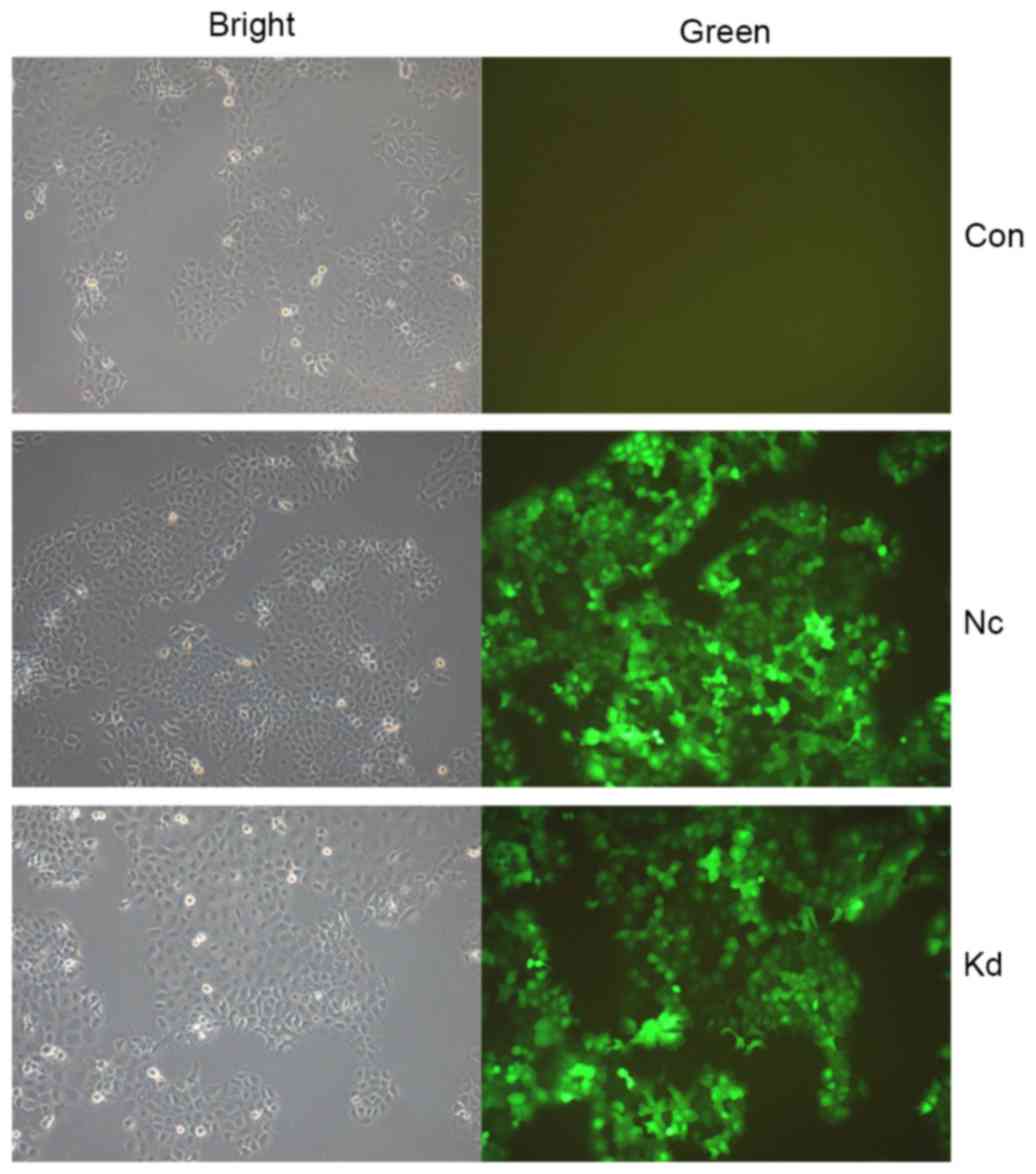
FaDu cells were transfected with Lv-shRNA-CD44
vectors (KD1, KD2, KD3 and KD4). Cells were examined by
fluorescence microscopy 72 h after lentivirus transduction
(Fig. 2). On day 5 after
transduction, cells were harvested to determine the efficiency of
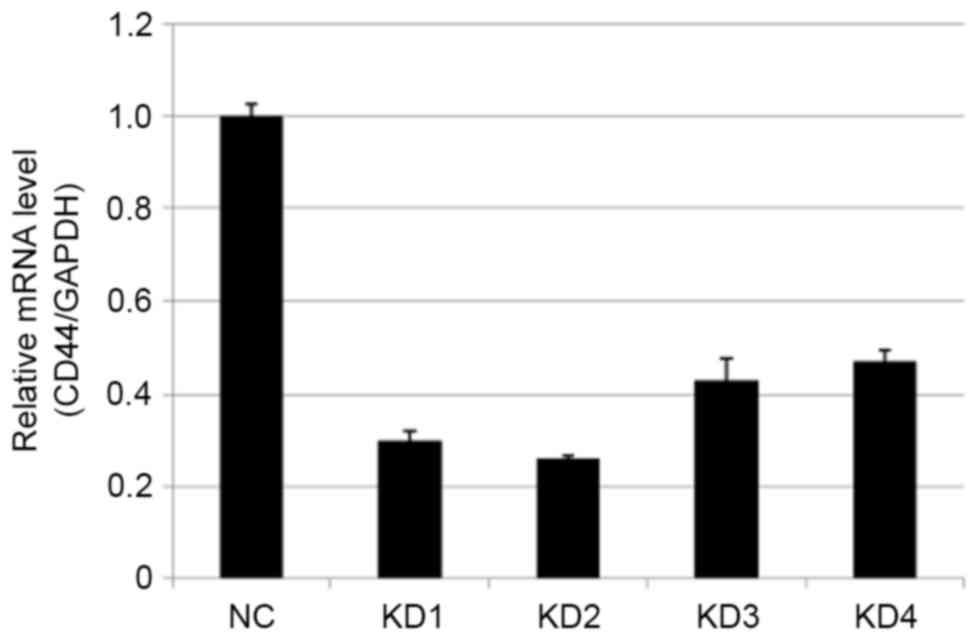
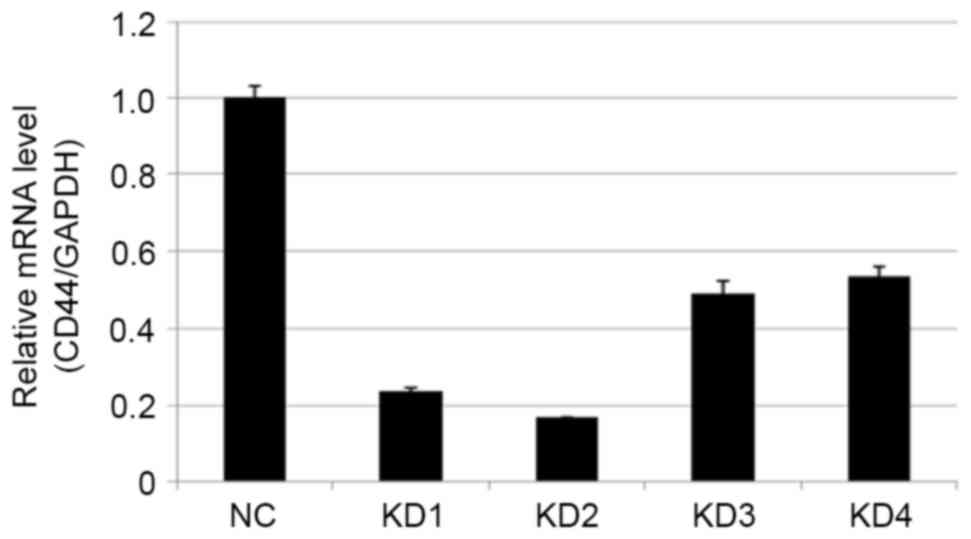
CD44 silencing by RT-qPCR (Figs. 3
and 4; Tables III and IV). Compared with the NC group, the most
effective knockdown efficiency for the CD44 gene was >70%, using
KD2 at a titer of 8E+8 TU/ml (P<0.01). Thus KD2, the most
efficient recombinant vector, was used in subsequent experiments
in vitro and in vivo.
 | Table III.Expression level of CD44 mRNA 5 days
after target cell transfection with the low multiplicity of
infection. |
Table III.
Expression level of CD44 mRNA 5 days
after target cell transfection with the low multiplicity of
infection.
| Groups | CD44/GAPDH |
P-valuea |
|---|
| NC | 1.000±0.028 |
|
| KD1 | 0.300±0.019 |
8.4211×10−6 |
| KD2 | 0.261±0.008 |
2.1910×10−4 |
| KD3 | 0.430±0.047 |
2.6567×10−4 |
| KD4 | 0.472±0.025 |
1.5357×10−5 |
 | Table IV.Expression level of CD44 mRNA 5 days
after target cell transfection with the high multiplicity of
infection. |
Table IV.
Expression level of CD44 mRNA 5 days
after target cell transfection with the high multiplicity of
infection.
| Groups | CD44/GAPDH |
P-valuea |
|---|
| NC | 1.000±0.031 |
|
| KD1 | 0.236±0.010 |
1.9202×10−4 |
| KD2 | 0.168±0.001 |
4.6399×10−4 |
| KD3 | 0.493±0.034 |
4.7761×10−5 |
| KD4 | 0.539±0.020 |
9.5098×10−5 |
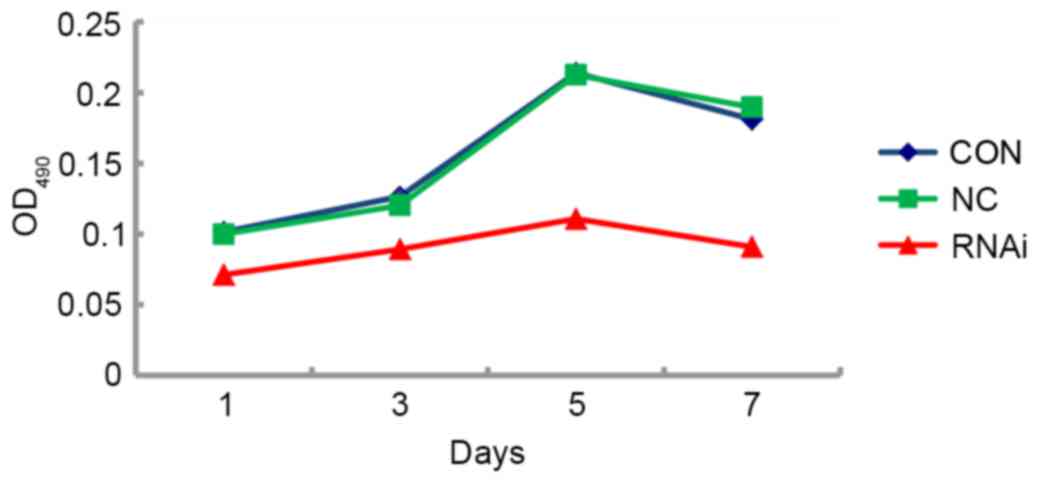
Proliferative capabilities of FaDu
cells following CD44 gene-knockdown in vitro
To evaluate the proliferative capabilities of FaDu
cells in vitro following CD44 gene silencing, FaDu cells
from the CON, NC and RNAi groups were seeded at the same density in
96-well plates, and the numbers of viable cells were quantified at
days 1, 3, 5 and 7 using the MTT assay. This assay revealed
distinct differences in cell growth curves in vitro between
the RNAi group and the control groups. Although the growth rate was
similar between CON and NC groups, the RNAi group had a
significantly slower proliferation rate (P<0.05; Fig. 5).
Tumorigenicity of FaDu cells following
CD44 gene knockdown in vivo
To assess the tumorigenic ability of FaDu cells
in vivo, 1×106 cells from each of the three
groups were injected into the right armpit hypodermis of NOD/SCID
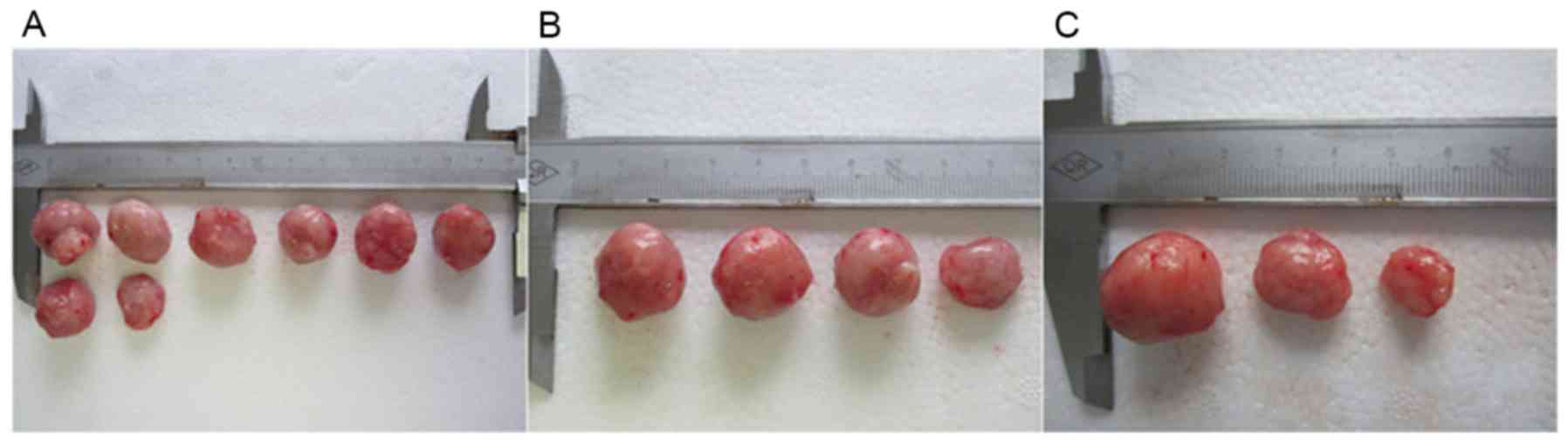
mice. All mice were monitored for 7 weeks (Fig. 6; Table
V). There was no significant difference in tumorigenesis
between the CON and NC groups (Fisher's exact test,
χ2=1.726, P>0.05), although there was a significant
difference in tumorigenesis between the CON and RNAi groups
(Fisher's exact test, χ2=7.809, P<0.05; Table VI). Mice from the RNAi group required
a longer incubation period to develop tumors than mice from the CON
group. In addition, the mean tumor volume in the CON group
(1,554.56±216.83 mm3) was not significantly different to
the mean tumor volume in the NC group (1,512.38±239.86
mm3) (t=0.31, P>0.05), while the mean tumor volume in
the RNAi group (1,061.00±185.76 mm3) was significantly
smaller than the CON group (t=3.47, P<0.05) (Table VII). The tumorigenic ability of FaDu
hypopharyngeal cancer cells was therefore significantly weaker
following knockdown of the CD44 gene. These observations suggest
that the CD44 gene may serve an important role in the cancer stem
cell properties of CD44+ cells in the FaDu
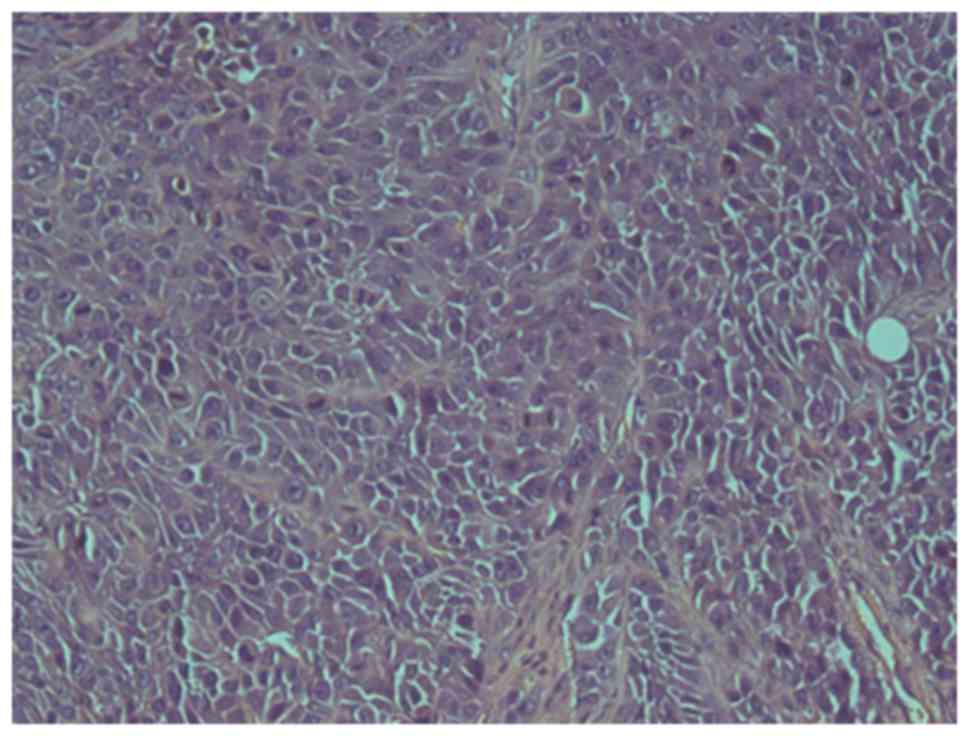
hypopharyngeal cancer cell line. A pathological study confirmed
that all tumors were poorly differentiated squamous cell carcinomas
with cellular heterogeneity (Fig.
7).
 | Table V.Tumorigenic rate of FaDu cells in
vivo following CD44 RNAi. |
Table V.
Tumorigenic rate of FaDu cells in
vivo following CD44 RNAi.
| Groups | Tumor incidence
(incubation period, days) | Tumorigenicity
rate, % |
|---|
| CON | 8/8a (14, 13, 16, 17, 14, 16, 19,
18) | 100 |
| RNAi | 3/8 (27, 24,
25) | 37.5b |
| NC | 4/4 (16, 17, 19,
14) | 100 |
 | Table VI.Number of nude mice that formed or
failed to form tumors. |
Table VI.
Number of nude mice that formed or
failed to form tumors.
| Groups | No. of mice with
tumor failure (n=5) | No. of mice with
tumor formation (n=15) |
|---|
| Control | 0 | 8 |
| RNAi-CD44 | 5 | 3 |
| NC-RNAi | 0 | 4 |
 | Table VII.Tumor volumes in mice injected with
FaDu cells following transfection with CD44 RNAi. |
Table VII.
Tumor volumes in mice injected with
FaDu cells following transfection with CD44 RNAi.
| Groups | Long axis, mm | Short axis, mm | Tumor volume,
mm3 | Mean tumor volume,
mm3 |
|---|
| CON | 17 | 15 | 1912.5 | 1554.56±216.83 |
|
| 15 | 13 | 1267.5 |
|
|
| 16 | 14 | 1568.0 |
|
|
| 15 | 14 | 1470.0 |
|
|
| 17 | 13 | 1436.5 |
|
|
| 17 | 14 | 1666.0 |
|
|
| 18 | 14 | 1764.0 |
|
|
| 16 | 13 | 1352.0 |
|
| RNAi | 14 | 12 | 1008.0 |
1061.00±185.76a |
|
| 15 | 13 | 1267.5 |
|
|
| 15 | 11 |
907.5 |
|
| NC | 18 | 14 | 1764.0 |
1512.38±239.86b |
|
| 17 | 14 | 1666.0 |
|
|
| 16 | 13 | 1352.0 |
|
|
| 15 | 13 | 1267.5 |
|
Discussion
Recent data have indicated that tumor stem cells
exist in numerous types of malignant tumors (2–10),
including head and neck malignant tumors (11–14), and
are closely associated with the occurrence, development and
metastasis of malignant tumors (14,18–21).
Hypopharyngeal carcinoma is a head and neck malignant tumor with
one of the poorest prognoses, and the mechanisms of its tumor
development and progression are unclear (22). Results from a previous study
demonstrated that CD44+ cells in hypopharyngeal cancer
had a stronger proliferative capability than CD44− cells
and a higher tumorigenic potential, indicating that cancer stem
cells in hypopharyngeal cancer may exist in the CD44+
tumor cell population, or that CD44+ cells may be
hypopharyngeal cancer stem cells (15). To investigate whether the CD44 gene
confers the biological characteristics of CD44+ cancer
stem cells in hypopharyngeal cancer, the tumorigenicity of
CD44+ cells required the knockout or knockdown of the
CD44 gene. Previous studies have demonstrated that RNAi is an
economical, fast and highly efficient technique for knocking down
gene expression (23–26). Thus, RNAi technology was used to
suppress the expression of CD44 in the present study.
To confirm the role of CD44 in cancer stem cells in
hypopharyngeal cancer further, RNAi technology was used to
knockdown CD44 gene expression in FaDu hypopharyngeal cancer cells
and any changes in their tumorigenicity were assessed. Following
RNAi of the CD44 gene, the expression level of CD44 mRNA was
significantly decreased, with a silencing efficiency >70%
(reflective of successful gene silencing). The proliferative
capacities of FaDu cells in vitro following CD44-silencing
differed between the KD group and the CON or NC groups, which
indicated that FaDu cells have a slower growth rate following CD44
gene-knockdown. Subsequent to the injection of FaDu cells into
NOD/SCID mice, the tumorigenic rate in the KD group was
significantly lower than the CON or NC group. Mice from the KD
group also required a longer incubation period to develop tumors
compared with mice in the CON or NC group. Furthermore, the tumor
volumes in the KD group were markedly smaller than those in the CON
or NC groups. In conclusion, the tumorigenic potential of FaDu
cells became significantly weaker once CD44 expression was knocked
down by RNAi. These observations suggest that the CD44 gene may
play an important role in the cancer stem cell properties of
CD44+ cells in hypopharyngeal cancer.
The results from the present study and a previous
study (15) indicate that CD44 is an
important biological marker of hypopharyngeal cancer stem cells.
The results of the present study also indicate that FaDu cells
retained proliferative abilities in vitro and tumorigenicity
in vivo following CD44 gene-knockdown; thus, it cannot be
considered that CD44 is a completely unique molecular biomarker for
hypopharyngeal cancer or that all biological characteristics of
stem cells in hypopharyngeal cancers are conferred by the CD44
gene. The results of the current study continue to suggest that
CD44+ cells are notable cancer stem cells in
hypopharyngeal cancer, and that certain biological characteristics
of cancer stem cells in hypopharyngeal cancer are conferred by the
CD44 gene. Other molecular markers of hypopharyngeal cancer stem
cells, and the precise role of CD44 amongst all molecular markers
of hypopharyngeal cancer stem cells, require further study.
Acknowledgements
The study was supported by grants from the National
Natural Science Foundation of China (no. 81271088), the Natural
Science Foundation of Shanghai (no. 11ZR1423600) and Shanghai Key
Laboratory of Translational Medicine on Ear and Nose diseases
Foundation (no. 14DZ2260300).
References
|
1
|
Han J, Fujisawa T, Husain SR and Puri RK:
Identification and characterization of cancer stem cells in human
head and neck squamous cell carcinoma. BMC Cancer. 14:1732014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Singh SK, Hawkins C, Clarke ID, Squire JA,
Bayani J, Hide T, Henkelman RM, Cusimano MD and Dirks PB:
Identification of human brain tumour initiating cells. Nature.
432:396–401. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Barreyro L, Will B, Bartholdy B, Zhou L,
Todorova TI, Stanley RF, Ben-Neriah S, Montagna C, Parekh S,
Pellagatti A, et al: Overexpression of IL-1 receptor accessory
protein in stem and progenitor cells and outcome correlation in AML
and MDS. Blood. 120:1290–1298. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hwang-Verslues WW, Kuo WH, Chang PH, Pan
CC, Wang HH, Tsai ST, Jeng YM, Shew JY, Kung JT, Chen CH, et al:
Multiple lineages of human breast cancer stem/progenitor cells
identified by profiling with stem cell markers. PLoS One.
4:e83772009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Haraguchi N, Ohkuma M, Sakashita H,
Matsuzaki S, Tanaka F, Mimori K, Kamohara Y, Inoue H and Mori M:
CD133+CD44+ population efficiently enriches
colon cancer initiating cells. Ann Surg Oncol. 15:2927–2933. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Karimi-Busheri F, Zadorozhny V, Li T, Lin
H, Shawler DL and Fakhrai H: Pivotal role of CD38 biomarker in
combination with CD24, EpCAM, and ALDH for identification of H460
derived lung cancer stem cells. J Stem Cells. 6:9–20.
2011.PubMed/NCBI
|
|
7
|
Curley MD, Therrien VA, Cummings CL,
Sergent PA, Koulouris CR, Friel AM, Roberts DJ, Seiden MV, Scadden
DT, Rueda BR and Foster R: CD133 expression defines a tumor
initiating cell population in primary human ovarian cancer. Stem
Cells. 27:2875–2883. 2009.PubMed/NCBI
|
|
8
|
Zimmerer RM, Korn P, Demougin P, Kampmann
A, Kokemüller H, Eckardt AM, Gellrich NC and Tavassol F: Functional
features of cancer stem cells in melanoma cell lines. Cancer Cell
Int. 13:782013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li C, Lee CJ and Simeone DM:
Identification of human pancreatic cancer stem cells. Methods Mol
Biol. 568:161–173. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Feng D, Peng C, Li C, Zhou Y, Li M, Ling
B, Wei H and Tian Z: Identification and characterization of cancer
stem-like cells from primary carcinoma of the cervix uteri. Oncol
Rep. 22:1129–1134. 2009.PubMed/NCBI
|
|
11
|
Wei XD, Zhou L, Cheng L, Tian J, Jiang JJ
and Maccallum J: In vivo investigation of CD133 as a putative
marker of cancer stem cells in Hep-2 cell line. Head Neck.
31:94–101. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhuang HW, Mo TT, Hou WJ, Xiong GX, Zhu
XL, Fu QL and Wen WP: Biological characteristics of CD133(+) cells
in nasopharyngeal carcinoma. Oncol Rep. 30:57–63. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Oliveira LR, Castilho-Fernandes A,
Oliveira-Costa JP, Soares FA, Zucoloto S and Ribeiro-Silva A:
CD44+/CD133+ immunophenotype and matrix
metalloproteinase-9: Influence on prognosis in early-stage oral
squamous cell carcinoma. Head Neck. 36:1718–1726. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu W, Wu L, Shen XM, Shi LJ, Zhang CP, Xu
LQ and Zhou ZT: Expression patterns of cancer stem cell markers
ALDH1 and CD133 correlate with a high risk of malignant
transformation of oral leukoplakia. Int J Cancer. 132:868–874.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shen C, Xiang M, Nie C, Hu H, Ma Y and Wu
H: CD44 as a molecular marker to screen cancer stem cells in
hypopharyngeal cancer. Acta Otolaryngol. 133:1219–1226. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang YP, Wang MZ, Luo YR, Shen Y and Wei
ZX: Lentivirus-mediated shRNA interference targeting SLUG inhibits
lung cancer growth and metastasis. Asian Pac J Cancer Prev.
13:4947–4951. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lin ZX: Patterns in the occurrence and
development of tumors. Chin Med J (Engl). 124:1097–1104.
2011.PubMed/NCBI
|
|
19
|
Miyoshi N, Ishii H, Sekimoto M, Haraguchi
N, Doki Y and Mori M: Properties and identification of cancer stem
cells: A changing insight into intractable cancer. Surg Today.
40:608–613. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Charafe-Jauffret E, Ginestier C, Iovino F,
Tarpin C, Diebel M, Esterni B, Houvenaeghel G, Extra JM, Bertucci
F, Jacquemier J, et al: Aldehyde dehydrogenase 1-positive cancer
stem cells mediate metastasis and poor clinical outcome in
inflammatory breast cancer. Clin Cancer Res. 16:45–55. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Koukourakis MI, Giatromanolaki A, Tsakmaki
V, Danielidis V and Sivridis E: Cancer stem cell phenotype relates
to radio-chemotherapy outcome in locally advanced squamous cell
head-neck cancer. Br J Cancer. 106:846–853. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hall SF, Groome PA, Irish J and O'Sullivan
B: The natural history of patients with squamous cell carcinoma of
the hypopharynx. Laryngoscope. 118:1362–1371. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fire A, Xu S, Montgomery MK, Kostas SA,
Driver SE and Mello CC: Potent and specific genetic interference by
double-stranded RNA in Caenorhabditis elegans. Nature. 391:806–811.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rubinson DA, Dillon CP, Kwiatkowski AV,
Sievers C, Yang L, Kopinja J, Rooney DL, Zhang M, Ihrig MM, McManus
MT, et al: A lentivirus-based system to functionally silence genes
in primary mammalian cells, stem cells and transgenic mice by RNA
interference. Nat Genet. 33:401–406. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liau SS, Ashley SW and Whang EE:
Lentivirus-mediated RNA interference of HMGA1 promotes
chemosensitivity to gemcitabine in pancreatic adenocarcinoma. J
Gastrointest Surg. 10:1254–1262. 2006.discussion 1263. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Couto LB and High KA: Viral
vector-mediated RNA interference. Curr Opin Pharmacol. 10:534–542.
2010. View Article : Google Scholar : PubMed/NCBI
|