Introduction
Gastric cancer (GC) is ranked as the fourth most
commonly diagnosed and the second most lethal type of malignancy
worldwide (1). A total of 989,600 new
cases of GC and 738,000 mortalities are estimated to occurglobally
per year, accounting for 8% of all cases of cancer and 10% of
cancer mortality (2). In the majority
of patients, GC is diagnosed at an advanced stage at which
malignant proliferation, extensive invasion, and lymphatic
metastasis have already occurred (3).
Despite the progress in chemotherapy, radiotherapy and surgical
techniques for GC in recent decades, the outcomes forpatients with
GC remain poor, and 5-year overall survival rates are <25%
(4,5).
Therefore, an improved understanding of the pathogenesis and
identification of the molecular alterations is essential for the
development of diagnostic markers and therapeutic targets to allow
the sensitive diagnosis and effective therapy of GC (3,6).
Long non-coding RNAs (lncRNAs), measuring >200
nucleotides in length, are a class of non-coding RNA molecule with
limited or no protein-coding capacity (7). They are associated with a variety of
biological processes, including cell proliferation, cell cycle,
differentiation and apoptosis, primarily by the regulation of gene
expression through chromatin remodeling, RNA maturation, transport
and protein synthesis (8). Advances
in the depth and quality of transcript me sequencing have revealed
a range of lncRNAs classes dysregulated in various human diseases,
particularly cancer (9–11). H19 is a member of a highly-conserved
cluster of imprinted genes, and was the first lncRNA to be
associated with cancer (12).
Previous studies have indicated that H19 exhibits dysregulated
expression in GC (13,14), hepatocellular carcinoma (15), bladder cancer (16), breast cancer (17) and esophageal cancer (18), and that itmay cause tumor promoting
and suppressing effects (19). Niu
et al (20) screened lncRNA
expression profilesfrom colorectal cancer and adenoma samples, and
identified a novel lncRNA, AK027294, which was associated with cell
proliferation, migration and apoptosis in colorectal cancer cells.
Li et al (21) and Song et
al (22) investigated the lncRNA
expression patterns in GC with a microarray, and identified 135
differentially expressed lncRNAs in GC tissues compared with
non-tumor adjacent tissues (NATs). Furthermore, the expression
level of gastric cancer-associated transcript 1 (GACAT1) was
associated with the lymph node metastasis, distant metastasis,
tumor node metastasis stage and differentiation of GC (23). By analyzing RNA-sequencing (seq) and
public microarray data, Park et al (24) identified 31 differently expressed
transcripts in GC, including BM742401, which was verified as a
potential lncRNA marker and therapeutic target in subsequent
experiments.
In the present study, the differentially expressed
genes (DEGs) and lncRNAs in GC samples compared with normal tissue
samples were screened using bioinformatics analysis. lncRNA-DEG
pairs were identified, and pathway enrichment analysis was
performed for the DEGs co-expressed with each lncRNA. Based on our
previous data (25), the present
study focused on the lncRNA TCONS_00068220, which was suspected to
be associated with GC pathogenesis. TCONS_00068220 encodes a
1,281-nt RNA that contains 3 exons at the human genome locus
8q11.21. Further functional studies of TCONS_00068220 in the
present study indicated that the down regulation of TCONS_00068220
may promote apoptosis in GC cells.
Materials and methods
Bioinformatics analysis
The GSE41476 RNA-seq data was obtained from the Gene
Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/). The expression
profilesof 3 primary cell culture samples from GC tissue and 2
normal tissue samples from GSE41476 were included in the dataset.
The RNA-seq data were compared with human genome 19 from the
University of California Santa Cruz (UCSC) database (http://genome.ucsc.edu) and aligned to achieve
transcript tome assembly. Subsequently, comparative genomics
methods were utilized to predict the novel lncRNAs. DEGs and
differentially expressed known and predicted lncRNAs were screened
usingan adjusted P-value of <0.05 and |log fold-change
(FC)|>1 as the cut-off criteria. The association between DEGs
and differentially expressed lncRNAs was calculated; a Pearson's
correlation coefficient >0.99 was used as the cut-off criterion
for DEG-lncRNA pairs. Subsequently, Kyoto Encyclopedia of Genes and
Genomes (KEGG; www.genome.jp/kegg/) pathway enrichment analysis was
conducted for the DEGs associated with each lncRNA. The detailed
procedures for bioinformatics analysis have been described in a
previous study (25).
Tumor tissues and cell lines
GC tissues and matched NATs were collected from 15
patients who had undergone the surgical resection of GC at the
First Affiliated Hospital of Harbin Medical University (Harbin,
China) and the People's Hospital of Heilongjiang Province (Harbin,
China) between January 2015 and March 2015. The mean age of the
patients was 59.5 years (range 47–74 years) and the ratio of males
to females was 9:6. No local or systemic treatment had been
administered to the patients prior to surgery. The NAT samples were
obtained 5 cm away from the tumor margin. All tissue samples were
snap-frozen in liquid nitrogen immediately following resection and
stored at −80°C until RNA extraction. The study was approved by the
Ethics Committee of First Affiliated Hospital of Harbin Medical
University and was performed in compliance with the Declaration of
Helsinki. Written, informed consent was obtained from the patients
involved in the study.
Human GC cell lines (including MKN-45, SNU-484 and
NCI-N87) and the human immortalized gastric epithelial cell line
GES-1 were purchased from the Biochemistry and Cell Biology at the
Chinese Academy of Sciences (Shanghai, China). All cell lines were
cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10%
fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific, Inc.),
100 U/ml penicillin, and 100 mg/ml streptomycin (Gibco; Thermo
Fisher Scientific, Inc.). The cells were incubated at 37°C in a
humidified atmosphere with 5% CO2.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analyses
According to the manufacturers' protocols, total RNA
was extracted from tissues and cultured cells with RNAiso Plus
(Takara Biotechnology Co., Ltd., Dalian, China), and reverse
transcription was performed using PrimeScript™ RT Master
Mix (Takara Biotechnology Co., Ltd.). qPCR was performed using
Power SYBR Green PCR Master mix (Applied Biosystems, Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The PCR
primers for TCONS_00068220 and GAPDH (Synbio Technologies LLC,
Suzhou, China) were as follows: TCONS_00068220 forwards,
5′-AATACCAGAGGCTCCAAGAACAG-3′, and reverse,
5′-CGACTACATAAGGGCTTAGAAACG-3′; GAPDH forwards,
5′-TGACAACTTTGGTATCGTGGAAGG-3′, and reverse,
5′-AGGCAGGGATGATGTTCTGGAGAG-3′. qPCR was performed in a total
reaction volume of 20 µl, including 2 µl RT products, 0.4 µl of
forward primers, 0.4 µl of reverse primers, 10 µl of 2X SYBR Green
Mix and 7.2 µl RNase-free dH2O. The reactions were
performed with the following protocol: An initial denaturation of
10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 60
sec. Relative gene expression was calculated using the comparative
cycle threshold method (value of 2−ΔΔCq) (26) with GAPDH as an endogenous control. The
RT-qPCR assays and data collection were performed using an ABI
ViiA7 instrument (Applied Biosystems; Thermo Fisher Scientific,
Inc.). Each sample was analyzed in triplicate.
Downregulation of TCONS_00068220
Small interfering RNA (siRNA) for TCONS_00068220
(si-TCONS_00068220, 5′-CCUGUAAUAAACGUCACAATT-3′) and negative
control (si-NC, 5′-UUCUCCGAACGUGUCACGUTT-3′) were synthesized by
Shanghai GenePharma Co., Ltd. (Shanghai, China). NCI-N87 cells were
transfected with si-TCONS_00068220 or si-NC using Lipofectamine
2000® transfection reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Cell viability assay
NCI-N87 cells transfected with si-NC or
si-TCONS_00068220 (1.5×105 cells/well) were seeded in
12-well plates. Trypan blue exclusion assays were performed to
assess cell viability at 1, 2, 3, 4 and 5 days. A mixture of
adherent and suspended cells was collected, mixed with 0.4% trypan
blue solution (Beyotime Institute of Biotechnology, Haimen, China),
added into a hemocytometer and observed under a light microscope
(magnification, ×100; BX43; Olympus Corporation, Tokyo, Japan).
Blue-stained cells were counted as dead, and cell viability (%) was
expressed as [1-(dead cell number/the total cell number)] ×100%.
The experiments were performed in triplicate.
Apoptosis analysis by flow
cytometry
NCI-N87 cells (1×105) were harvested with
Trypsin and washed with ice-cold PBS 3 days subsequent
totransfection. Apoptotic cell death was determined using a
fluorescein isothiocyanate-Annexin V Apoptosis Detection kit I (BD
Pharmingen; BD Biosciences, Franklin Lakes, NY, USA) according to
the manufacturer's protocol. Samples were stained for 30 min at
room temperature in the dark. The stained specimens were then
analyzed by FACSC alibur flow cytometry with Cell Quest software
version 3.0 (BD Biosciences). Experiments were performed in
triplicate.
Nuclear staining with DAPI
NCI-N87 cells (1×105 cells/well) were
seeded in a 12-well plate with a glass slide at the bottom of each
well, and then cultured and transfected with siRNAs as previously
described. Following 3 days of incubation, the cells were washed in
PBS and fixed with 1 ml 4% paraformaldehyde solution at room
temperature for 20 min. The fixed cells were subsequently washed
with PBS and stained with VECTASHIELD® Antifade Mounting
Medium with 1.5 µg/ml DAPI (Vector Laboratories, Inc., Burlingame,
CA, USA) at room temperature in the dark for 5 min. Nuclear
morphology of cells was examined by a confocal laser scanning
platform (TCS SP8; Leica Microsystems GmbH, Wetzlar, Germany).
Western blot analysis
NCI-N87 cells were seeded at a density of
3×105 cells per well in a 6-well plate and transfected
with si-NC or si-TCONS_00068220. The cells were collected at 3 days
after transfection and resuspended in RIPA Lysis Buffer III (Sangon
Biotech Co., Ltd., Shanghai, China). The concentration of total
protein was quantified by bicinchoninic acid (BCA) method. The
protein extracts (20 µg/well) were resolved by 10% SDS-PAGE and
transferred to a polyvinylidene difluoride membrane, followed by
blocking with PBS and Tween-20 solution supplemented with 5%
non-fat milk at room temperature for 1.5 h. The membrane was
incubated with primary antibodies against p17-specific cleaved
caspase-3 (cat. no., 25546-1-AP; dilution, 1:500) and GAPDH (cat.
no., 60004-1-Ig; dilution, 1:2,000; both ProteinTech Group, Inc.,
Chicago, IL, USA) at 4°C overnight, and then incubated with
horseradish peroxidase-conjugated goat anti-rabbit IgGsecondary
antibodies (cat. no., 111-035-045; dilution, 1:5,000; Jackson
ImmunoResearch Laboratories, Inc., West Grove, PA, USA) at room
temperature for 1.5 h. The ChemiDoc™ XRS+ system
(Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to
visualize the bands.
Statistical analysis
Data were expressed as the mean ± standard deviation
(SD) from three separate experiments. The western blot analysis
results are presented from a representative experiment. Statistical
significance was determined using the two-tailed Student's t-test.
Multiple groups were analyzed using a one-way analysis of variance
followed by a Newman-Keuls test. All statistical analyses were
performed using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA).
P<0.05 was considered to indicate a statically significant
difference.
Results
TCONS_00068220 is differentially
expressed in GC compared with NATs, and co-expressed DEGs of
TCONS_00068220 are enriched in cancer-associated pathways
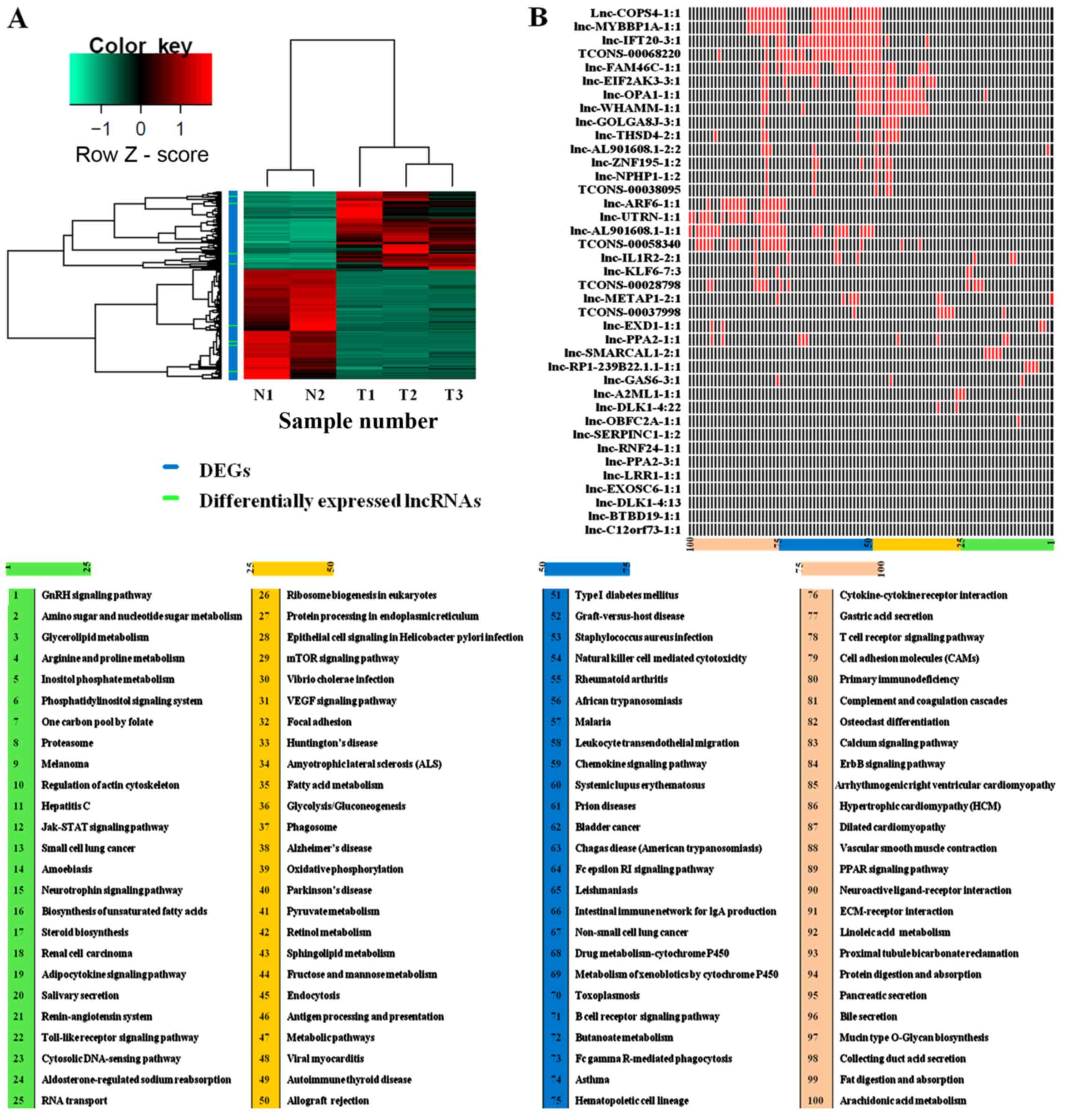
Compared with NATs, a total of 2,625 differentially
expressed transcripts' including 34 known lncRNAs, 5 predicted
lncRNAs and 2,586 mRNAs) were identified in the GC samples
(Fig. 1A; Table I). Following the identification of
lncRNA-DEG pairs, KEGG pathway enrichment analysis was conducted
for the DEGs co-expressed with each lncRNA (Fig. 1B). Based on this analysis, it was
revealed that the DEGs co-expressed with the predicted lncRNA
TCONS_00068220 were enriched in cancer-associated processes,
including ‘bladder cancer’, ‘cell adhesion molecules’ (CAMs),
‘chemokine signaling pathway’ and ‘natural killer cell-mediated
cytotoxicity’ (Table II) suggesting
that TCONS_00068220 may serve a crucial biological function in GC
cells.
 | Table I.Differentially expressed long
non-coding RNAs in gastric cancer tissue compared with normal
tissue samples. |
Table I.
Differentially expressed long
non-coding RNAs in gastric cancer tissue compared with normal
tissue samples.
| Name | |log FC| | P-value | Q-value |
|---|
| TCONS_00028798 | 1.79769E+308 |
2.74×10−4 |
5.26×10−3 |
| TCONS_00038095 | 2.79182 |
5.24×10−3 |
4.69×10−2 |
| TCONS_00058340 | 1.79769E+308 |
2.98×10−3 |
3.13×10−2 |
| TCONS_00068220 | 4.92069 |
1.57×10−3 |
1.94×10−2 |
| TCONS_00037998 | 4.74522 |
4.84×10−4 |
8.05×10−3 |
 | Table II.Top 10 enriched Kyoto Encyclopedia of
Genes and Genomes pathways for the differentially expressed genes
co-expressed with TCONS_00068220. |
Table II.
Top 10 enriched Kyoto Encyclopedia of
Genes and Genomes pathways for the differentially expressed genes
co-expressed with TCONS_00068220.
| Name | Genes, n | Associated
genes | P-value |
|---|
| Cell adhesion
molecules | 15 | CD2, CD28 |
3.10×10−6 |
| Chemokine signaling
pathway | 15 | CCL5, CCR1 |
2.02×10−4 |
| Bladder cancer | 5 | MMP9, MYC |
5.61×10−3 |
| Natural killer cell
mediated cytotoxicity | 8 | BID, ICAM2 |
3.38×10−2 |
| Intestinal immune
network for IgA production | 8 | CD28, ICOSLG |
4.17×10−5 |
| Staphylococcus
aureus infection | 8 | C1QB, C1QC |
1.14×10−4 |
| Leishmaniasis | 9 | ITGB1, MAPK12 |
1.43×10−4 |
| Type I diabetes
mellitus | 7 | CD28, PRF1 |
1.49×10−4 |
| Allograft
rejection | 6 | CD28, HLA-DMB |
4.61×10−4 |
| Viral
myocarditis | 8 | BID, CD28 |
6.24×10−4 |
TCONS_00068220 expression is
upregulated in human GC tissues and cell lines
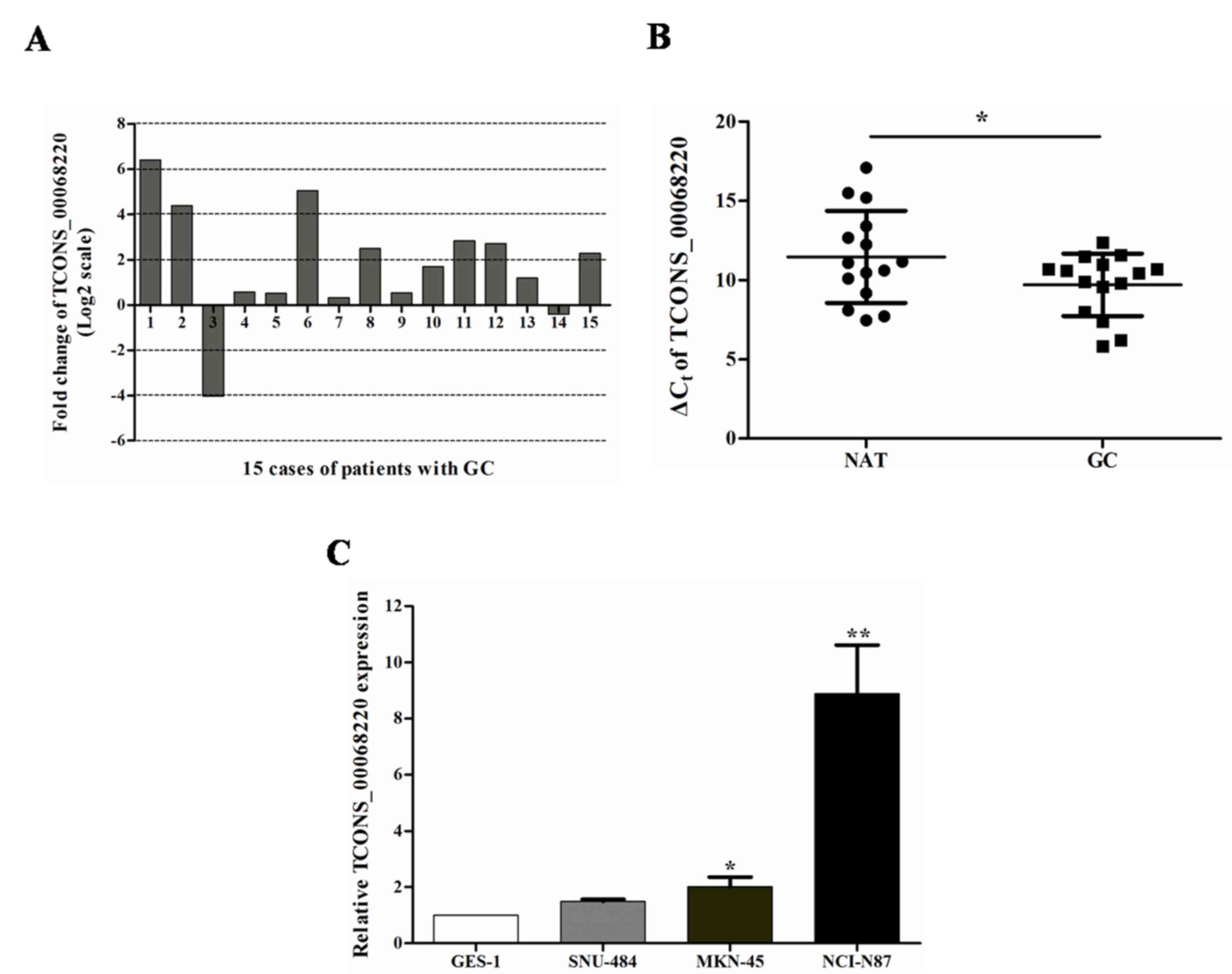
To assess the role of TCONS_00068220 in GC genesis
and progression, the TCONS_00068220 expression level was examined
in GC tissues and cell lines with RT-qPCR. The TCONS_00068220
levels were upregulated in 86.7% of GC tissues compared with NATs
(Fig. 2A). The median fold change was
3.16. The mean ± SD ΔCq value for TCONS_00068220 in GC
tissues was 9.70±1.97, whereas it was 11.46±2.91 for NATs (Fig. 2B).
It was then verified that the expression of
TCONS_00068220 was increased in two GC cell lines (MKN-45 and
NCI-N87) compared with the expression in the non-cancer GES-1 cells
(Fig. 2C; P<0.05). These data
indicated that abnormal TCONS_00068220 expression may be associated
with the genesis and progression of GC.
TCONS_00068220 downregulation inhibits
the viability of GC cells
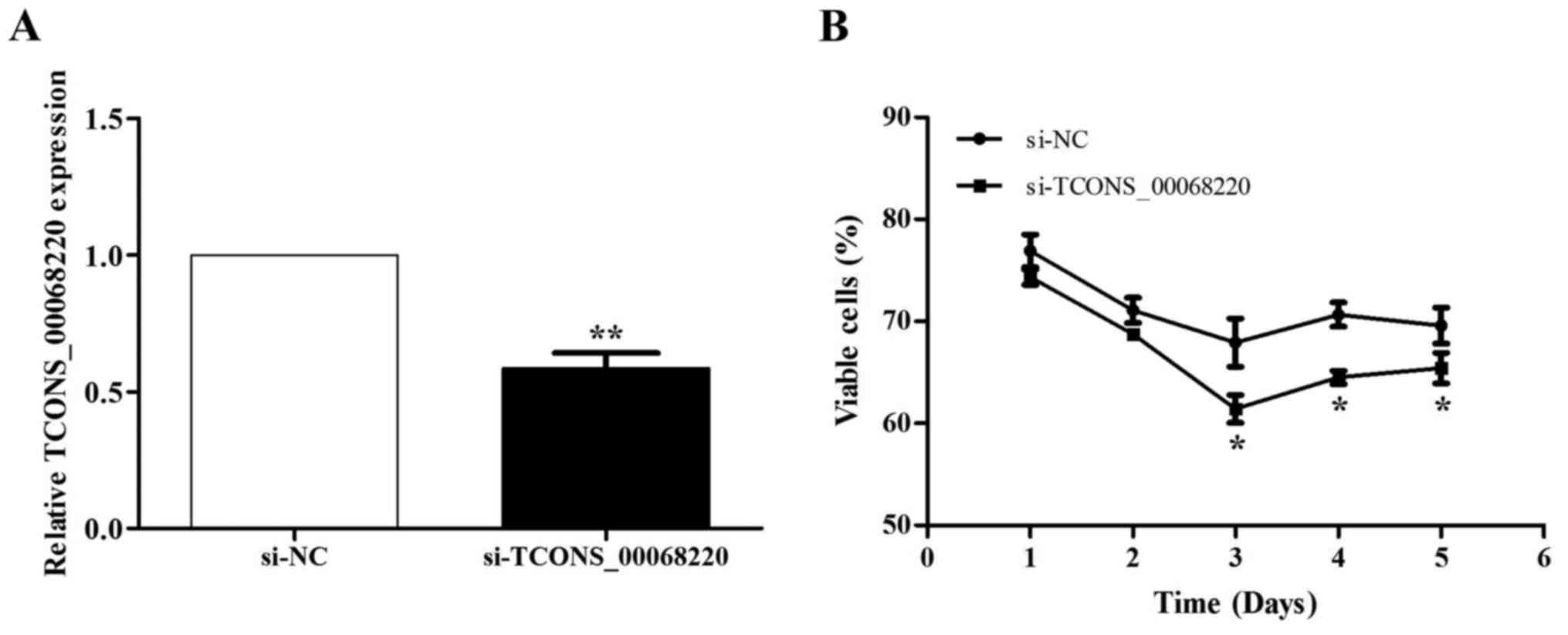
RNA interference is an effective strategy for
inhibiting gene expression in cultured cells (27). To verify whether TCONS_00068220
expression was associated with GC progression, siRNAs were used to
downregulate TCONS_00068220 expression in the NCI-N87 cell line.
The knockdown effect was confirmed by RT-qPCR analysis (Fig. 3A). Atrypan blue exclusion assay
indicated that the rate of viable cells was reduced compared with
cells transfected with the si-NC, and widespread cell death was
observed at 3 days after transfection with si-TCONS_00068220
(Fig. 3B).
TCONS_00068220 downregulation promotes
the apoptosis of GC cells
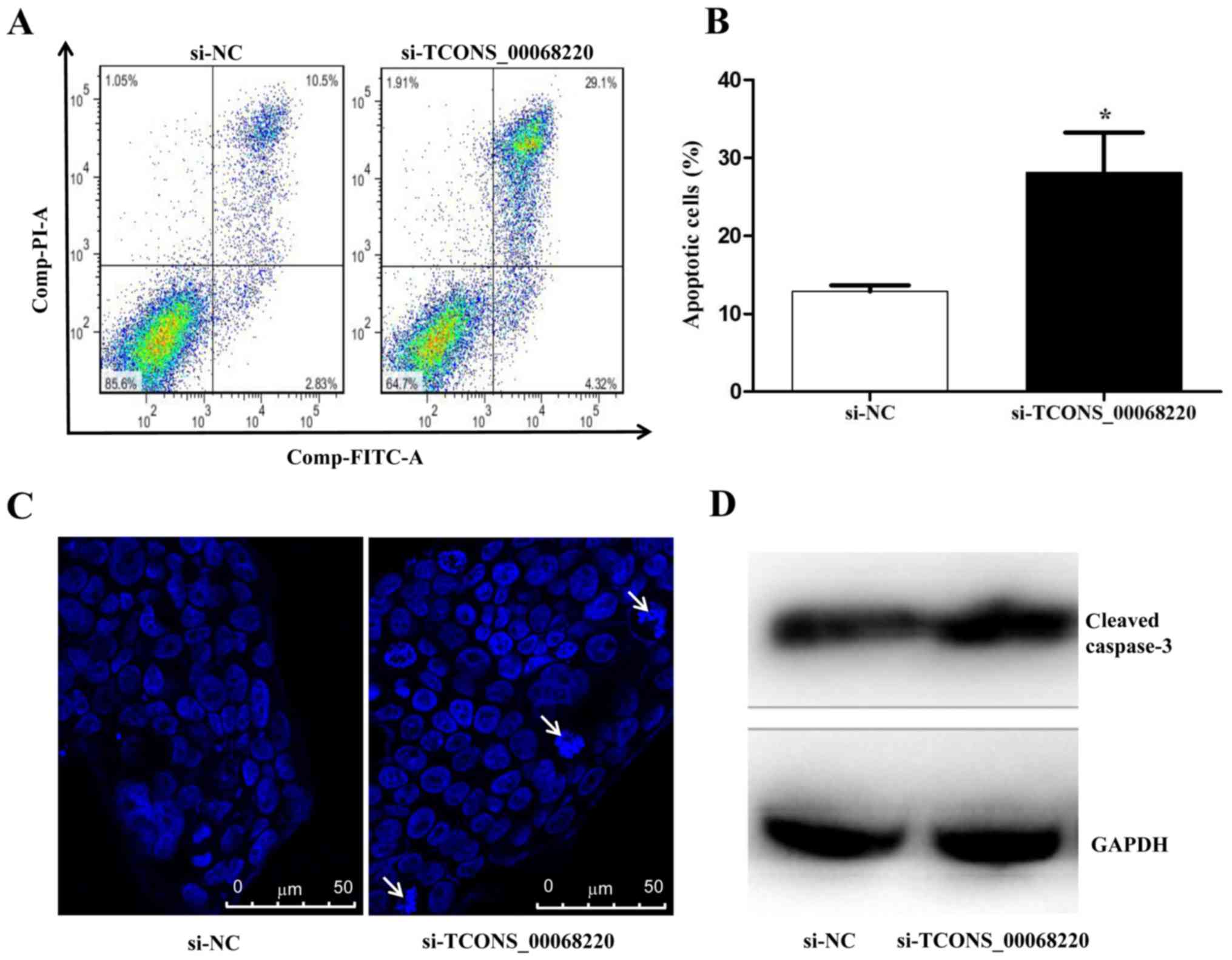
To confirm that TCONS_00068220 downregulation
promoted GC cell apoptosis, flow cytometry analysis was performed.
As demonstrated in Fig. 4A and B,
when TCONS_00068220 expression was downregulated with siRNA, the
rate of apoptosis was markedly increased. Based on observations of
nuclear morphology, apoptotic cells were increased in
si-TCONS_00068220 group compared with si-NC group (Fig. 4C). Additionally, the expression of
caspase-3, a key mediator of apoptosis (28), was detected by western blot analysis.
The results revealed that the expression of cleaved caspase-3 was
enhanced in NCI-N87 cells treated with TCONS_00068220 siRNA
compared with the control group (Fig.
4D).
Discussion
lncRNAs exhibit diverse functions in the
pathogenesis of cancer. The dysregulation of lncRNA is considered
to be a critical contributor to thetumorigenesis of various types
of cancer, as it mayupregulatetumor cell proliferation, allow the
evasion of growth suppressors, enable replicative immortality,
induce angiogenesis and increase apoptosis resistance (1). For example, an antisense lncRNA
transcribed from the p15 tumor suppressor locus induces alterations
to local heterochromatin and DNA methylation status, thereby
regulating p15 expression, and may be associated with leukemia
oncogenesis (29). TP53-regulated
lncRNA (LOC401317) is directly regulated by p53 and has been
demonstrated to exhibit antitumor effects in nasopharyngeal
carcinoma cells (30). LOC401317 may
inhibit cell cycle progression by upregulating p21, and decreasing
cyclin D1 and cyclin E1 expression; it may also promote apoptosis
through the induction of poly (ADP-ribose) polymerase (PARP) and
caspase-3 cleavage (30). Long
stress-induced non-coding transcript 5 (LSINCT5) is a 2.6 kb
polyadenylated intergenic nuclear lncRNA that is potentially
transcribed by RNA polymerase III. LSINCT5 expression is
significantly upregulated in gastrointestinal cancer tissues and
cell lines, and is associated with certain clinical pathologies,
such as tumor size, tumor invasion depth, lymphatic metastasis and
tumor node metastasis stages (31).
Due to advances in microarray production and novel
sequencing technologies, the identification and characterization of
lncRNAs is now possible. Huang et al (32) investigated the alteration in lncRNA
expression induced by hepatitis B virus X protein (HBx), which
wasimplicated as an oncogene in hepatocarcinogenesis, viaepigenetic
modification and genetic regulation, based on microarray studies.
They identified an lncRNA downregulated by HBx, designated as
lncRNA-Dreh, which inhibited hepatocellular carcinoma growth and
metastasis by targeting the intermediate filament protein vimentin.
Taurine up-regulated gene 1 (TUG1) was initially detected in a
genomic screen for genes upregulated in response to taurine
treatment in developing mouse retinal cells (33). A study demonstrated that the
dysregulation of TUG1 is associated with the progression of a
variety of tumors. The increased expression of TUG1 predicts a poor
prognosis of GC and regulates cell proliferation by epigenetically
silencing p57 (34).
In the present study, based on previous data, a DEGs
and differentially expressed lncRNAs expression profile of GC
tissue samples compared with NATswas screened from RNA-seq data
from GEO. A total of 39 lncRNAs were identified as differently
expressed. The lncRNA TCONS_00068220 was the focus of the present
study; DEGs associated with TCONS_00068220 wereidentified to be
enriched in cancer-associated processes. Therefore, TCONS_00068220
was predicted to function in the pathogenesis of GC. It was
identified that the expression of TCONS_00068220 was upregulated in
GC tissues compared with NATs. In addition, the significantly
increased expression of TCONS_00068220 was also identified in GC
cell lines. The knockdown of TCONS_00068220 reduced the viability
of NCI-N87 GC cells. To furtherclarify the role of TCONS_00068220
in GC cells, a flow cytometry assay was used to detect the
apoptosis rate in NCI-N87 cells following transfection with si-NC
or si-TCONS_00068220. The results indicated that the down
regulation of TCONS_00068220 upregulated the apoptosis of GC
cells.
Apoptosisis a genetically regulated ‘cellular
suicide’ mechanism that serves a crucial role in development and
homeostasis (35,36). Cancer cells adopt various strategies
to override apoptosis, including the upregulation of anti-apoptotic
machinery, the down regulation of pro-apoptotic factors or a
combination of these strategies (37). A number ofcancer-associated lncRNAs
have been identified that affect apoptosis via various pathways
(10,38,39). The
most prominent pathways for apoptosis are activated by the
mitochondria or death receptors from the tumor necrosis factor
(TNF) family through various cascade reactions (40). Part of these cascades is the initiator
caspases, which activatefurther executorcaspases by cleaving them
at aspartate residues. The activation of these executor caspases
leads to the activation of further caspases, and ultimately, to
cell death by initiating the degradation of DNA and other vital
cell components (41). Caspase-3, an
executor caspase that serves a central role in the execution of the
apoptotic program (42), is primarily
responsible for the cleavage of PARP during apoptosis (43). During apoptosis, PARP is cleaved by
caspase-3 into 89- and 24-kDa fragments that contain the active
site and the DNA-binding domain of the enzyme, respectively
(44–46). This cleavage inactivates PARP by
removing its ability to respond to DNA strand breakage (43). Caspase-3 also cleaves Bcl-2 and
Bcl-2-extra large, which removes the anti-apoptotic function of
these proteins and releases pro-apoptotic C-terminal fragments
(47). Caspase-3 also affects the
mitochondria; it induces the loss of mitochondrial membrane
potential and the release of apoptosis inducing factor (28). In the present study, following the
down regulation of TCONS_00068220, the level of cleaved caspase-3
was markedly increased, which implies TCONS_00068220 may serve a
role in preventing the apoptosis of cancer cells.
In summary, a DEGs and differentially expressed
lncRNAs expression profile of GC samples relative to normal tissue
samples was screened with bioinformatics methods. TCONS_00068220
was identified as a novel lncRNA associated with apoptosis
inhibition in GC cells. These data suggest that TCONS_00068220 may
serve a key functional role in GC occurrence and progression. To
the best of our knowledge, this is the first study to examine the
biological functions of TCONS_00068220. Therefore, additional
detailed study is required.
Acknowledgements
The authors wish to thank Ms. Qimeng Yuan (First
Affiliated Hospital of Harbin Medical University, Harbin, China)
and Dr Fengqi Jiang (People's Hospital of Heilongjiang Province,
Harbin, China) for gifting the tissue samples.
References
|
1
|
Fang XY, Pan HF, Leng RX and Ye DQ: Long
noncoding RNAs: Novel insights into gastric cancer. Cancer Lett.
356:357–366. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sun M, Xia R, Jin F, Xu T, Liu Z, De W and
Liu X: Downregulated long noncoding RNA MEG3 is associated with
poor prognosis and promotes cell proliferation in gastric cancer.
Tumor Biol. 35:1065–1073. 2014. View Article : Google Scholar
|
|
4
|
Chen X, Sun J, Song Y, Gao P, Zhao J,
Huang X, Liu B, Xu H and Wang Z: The novel long noncoding RNA
AC138128.1 may be a predictive biomarker in gastric cancer. Med
Oncol. 31:2622014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guo X, Xia J and Deng K: Long non-coding
RNAs: Emerging players in gastric cancer. Tumor Biol.
35:10591–10600. 2014. View Article : Google Scholar
|
|
6
|
Zhao JH, Sun JX, Song YX, Chen XW, Yang
YC, Ma B, Wang J, Gao P and Wang ZN: A novel long noncoding
RNA-LOWEG is low expressed in gastric cancer and acts as a tumor
suppressor by inhibiting cell invasion. J Cancer Res Clin Oncol.
142:601–609. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ding J, Lu B, Wang J, Shi Y, Lian Y, Zhu
Y, Wang J, Fan Y, Wang Z, De W and Wang K: Long non-coding RNA
Loc554202 induces apoptosis in colorectal cancer cells via the
caspase cleavage cascades. J Exp Clin Cancer Res. 34:1002015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Luo G, Wang M, Wu X, Tao D, Xiao X, Wang
L, Min F, Zeng F and Jiang G: Long non-coding RNA MEG3 inhibits
cell proliferation and induces apoptosis in prostate cancer. Cell
Physiol Biochem. 37:2209–2220. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhu H, Li X, Song Y, Zhang P, Xiao Y and
Xing Y: Long non-coding RNA ANRIL is up-regulated in bladder cancer
and regulates bladder cancer cell proliferation and apoptosis
through the intrinsic pathway. Biochem Biophys Res Commun.
467:223–228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zang W, Wang T, Wang Y, Chen X, Du Y, Sun
Q, Li M, Dong Z and Zhao G: Knockdown of long non-coding RNA
TP73-AS1 inhibits cell proliferation and induces apoptosis in
esophageal squamous cell carcinoma. Oncotarget. 7:19960–19974.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu S, Mao J, Shao Y, Chen F, Zhu X, Xu D,
Zhang X and Guo J: Reduced expression of the long non-coding RNA
AI364715 in gastric cancer and its clinical significance. Tumor
Biol. 36:8041–8045. 2015. View Article : Google Scholar
|
|
13
|
Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J
and Fang G: Up-regulated long non-coding RNA H19 contributes to
proliferation of gastric cancer cells. FEBS J. 279:3159–3165. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li H, Yu B, Li J, Su L, Yan M, Zhu Z and
Liu B: Overexpression of lncRNA H19 enhances carcinogenesis and
metastasis of gastric cancer. Oncotarget. 5:2318–2329. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim KS and Lee YI: Biallelic expression of
the H19 and IGF2 genes in hepatocellular carcinoma. Cancer Lett.
119:143–148. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Luo M, Li Z, Wang W, Zeng Y, Liu Z and Qiu
J: Upregulated H19 contributes to bladder cancer cell proliferation
by regulating ID2 expression. FEBS J. 280:1709–1716. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Berteaux N, Lottin S, Monté D, Pinte S,
Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T and
Adriaenssens E: H19 mRNA-like noncoding RNA promotes breast cancer
cell proliferation through positive control by E2F1. J Biol Chem.
280:29625–29636. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hibi K, Nakamura H, Hirai A, Fujikake Y,
Kasai Y, Akiyama S, Ito A and Takagi H: Loss of H19 imprinting in
esophageal cancer. Cancer Res. 56:480–482. 1996.PubMed/NCBI
|
|
19
|
Wang J, Song YX and Wang ZN: Non-coding
RNAs in gastric cancer. Gene. 560:1–8. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Niu H, Hu Z, Liu H, Hu G, Yang B, Wu S and
Li F: Long non-coding RNA AK027294 involves in the process of
proliferation, migration, and apoptosis of colorectal cancer cells.
Tumor Biol. 37:10097–10105. 2016. View Article : Google Scholar
|
|
21
|
Li PF, Chen SC, Xia T, Jiang XM, Shao YF,
Xiao BX and Guo JM: Non-coding RNAs and gastric cancer. World J
Gastroenterol. 20:5411–5419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Song H, Sun W, Ye G, Ding X, Liu Z, Zhang
S, Xia T, Xiao B, Xi Y and Guo J: Long non-coding RNA expression
profile in human gastric cancer and its clinical significances. J
Transl Med. 11:2252013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun W, Wu Y, Yu X, Liu Y, Song H, Xia T,
Xiao B and Guo J: Decreased expression of long noncoding RNA
AC096655.1–002 in gastric cancer and its clinical significance.
Tumor Biol. 34:2697–2701. 2013. View Article : Google Scholar
|
|
24
|
Park SM, Park SJ, Kim HJ, Kwon OH, Kang
TW, Sohn HA, Kim SK, Noh Moo S, Song KS, Jang SJ, et al: A known
expressed sequence tag, BM742401, is a potent lincRNA inhibiting
cancer metastasis. Exp Mol Med. 45:e312013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao Z, Song Y, Piao D, Liu T and Zhao L:
Identification of genes and long non-coding RNAs associated with
the pathogenesis of gastric cancer. Oncol Rep. 34:1301–1310. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liao X, Tang S, Thrasher B, Griebling TL
and Li B: Small-interfering RNA-induced androgen receptor silencing
leads to apoptotic cell death in prostate cancer. Mol Cancer Ther.
4:505–515. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lakhani SA, Masud A, Kuida K, Porter GA
Jr, Booth CJ, Mehal WZ, Inayat I and Flavell RA: Caspase 3 and 7:
Key mediators of mitochondrial events of apoptosis. Science.
311:847–851. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gong Z, Zhang S, Zeng Z, Wu H, Yang Q,
Xiong F, Shi L, Yang J, Zhang W, Zhou Y, et al: LOC401317, a
p53-regulated long non-coding RNA, inhibits cell proliferation and
induces apoptosis in the nasopharyngeal carcinoma cell line HNE2.
PLoS One. 9:e1106742014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu MD, Qi P, Weng WW, Shen XH, Ni SJ, Dong
L, Huang D, Tan C, Sheng WQ, Zhou XY and Du X: Long non-coding RAN
LSINCT5 predicts negative prognosis and exhibits oncogenic activity
in Gastric cancer. Medicine (Baltimore). 93:e3032014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang
Y, Tang GN, Zhou WP and Sun SH: Hepatitis B virus X protein
(HBx)-related long noncoding RNA (lncRNA) down-regulated expression
by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by
targeting the intermediate filament protein vimentin. Hepatology.
57:1882–1892. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Young TL, Matsuda T and Cepko CL: The
noncoding RNA taurine upregulated gene 1 is reguired for
differentiation of the murine retina. Curr Biol. 15:501–512. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang E, He X, Yin D, Han L, Qiu M, Xu T,
Xia R, Xu L, Yin R and De W: Increased expression of long noncoding
RNA TUG1 predicts a poor prognosis of gastric cancer and regulates
cell proliferation by epigenetically silencing of p57. Cell Death
Dis. 7:e21092016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cryns V and Yuan JY: Proteases to die for.
Gene & Development. 12:1551–1570. 1998. View Article : Google Scholar
|
|
36
|
Earnshaw WC, Martins LM and Kaufmann SH:
Mammalian caspases: Structure, activation, substrates, and
functions during apoptosis. Annu Rev Biochem. 68:383–424. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fernald K and Kurokawa M: Evading
apoptosis in cancer. Trends cell Biol. 23:620–633. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang M, Huang T, Luo G, Huang C, Xiao XY,
Wang L, Jiang GS and Zeng FQ: Long non-coding RNA MEG3 induces
renal cell carcinoma cells apoptosis by activating the
mitochondrial pathway. J Huazhong Univ Sci Technolog Med Sci.
35:541–545. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhu YP, Bian XJ, Ye DW, Yao XD, Zhang SL,
Dai B, Zhang HL and Shen YJ: Long noncoding RNA expression
signatures of bladder cancer revealed by microarray. Oncol Lett.
7:1197–1202. 2014.PubMed/NCBI
|
|
40
|
Daniel PT: Dissecting the pathways to
death. Leukemia. 14:2035–2044. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Juraver-Geslin HA and Durand BC: Early
development of the neural plate: New roles for apoptosis and for
one of its main effectors caspase-3. Genesis. 53:203–224. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Friedrich K, Wieder T, Von Haefen C,
Radetzki S, Jänicke R, Schulze-Osthoff K, Dörken B and Daniel PT:
Overexpression of caspase-3 restores sensitivity for drug-induced
apoptosis in breast cancer cell lines with acquired drug
resistance. Oncogene. 20:2749–2760. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Boulares AH, Yakovlev AG, Ivanova V,
Stoica BA, Wang G, Iyer S and Smulson M: Role of poly(ADP-ribose)
polymerase (PARP) cleavage in apoptosis. Caspase 3-resistant PARP
mutant increases rates of apoptosis in transfected cells. J Biol
Chem. 274:22932–22940. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tewari M, Quan LT, O'Rourke K, Desnoyers
S, Zeng Z, Beidler DR, Poirier GG, Salvesen GS and Dixit VM:
Yama/CPP32beta, a mammalian homolog of CED-3, is a CrmA-inhibitable
protease that cleaves the death substrate poly(ADP-Ribose)
polymerase. Cell. 81:801–809. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu J, Wu Y, Wang B, Yuan X and Fang B:
High levels of glucose induced the caspase-3/PARP signaling
pathway, leading to apoptosis in human periodontal ligament
fibroblasts. Cell Biochem Biophys. 66:229–237. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Krishnakumar R and Kraus WL: The PARP side
of the nucleus: Molecular actions, physiological outcomes, and
clinical targets. Mol Cell. 39:8–24. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wolf BB and Green DR: Suicidal tendencies:
Apoptotic cell death by caspase family proteinases. J Biol Chem.
274:20049–20052. 1999. View Article : Google Scholar : PubMed/NCBI
|