Introduction
Adoptive immunotherapy is an effective and safe
anticancer therapy (1), and currently
used for various tumor treatments, including renal cell carcinoma
(2), lung cancer (3), breast cancer (4), melanoma (5), and hepatoma (6).
As a valuable cancer therapeutic scheme, numerous
studies focused on T cells in order to further improve the
antitumor activity of adoptive immunotherapy (1). Rettinger et al (7) and Meng et al (8) demonstrated that IL-15 and IL-21,
respectively were able to promote the proliferative, and cytotoxic
activity of cytokine-induced killer cells (CIKs). Rutella et
al (9) noted that thymoglobulin
efficiently expanded the CIK population, and that the CIKs
generated by thymoglobulin were feasible and safe for solid tumor
treatment. Tan et al (10)
verified that K-ras dendritic cells were able to enhance CIK
proliferation and increase the killing effect on pancreatic cancer
cells. Though the activity of CIKs may be enhanced by technological
strategies, maneuvers to improve the antitumor efficacy through the
use of T cells have been under exploration (11). However, the proteins associated with
CIK activity remain unclear.
In the present study, it was demonstrated that the
cytotoxicity of CIKs was significantly influenced following
gentamicin treatment. Further research verified that a protein
identified by proteomic and reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) strategies was associated with
CIK activity. The results of the current study have important
implications for the investigation of the CIK activity-enhancement
mechanism.
Materials and methods
CIK culture and protein
extraction
Peripheral blood was donated from a 31-year-old male
volunteer following written informed consent being obtained. The
present study was approved by the Animal Welfare and Research
Ethics Committee of the Institute of University of South China
(Hengyang, China).
Lymphocytes were separated and cultured as reported
by Pan et al (12), and Laport
et al (13) with certain
modifications. Equivalent volumes of peripheral blood and 0.9%
physiological saline were mixed followed with ficoll gradient
separation (LymphoPrep; PAA Laboratories; GE Healthcare, Chicago,
IL, USA). Following centrifugation at 800 × g for 20 min at room
temperature, the leukocyte layer was collected and washed twice
with 0.9% physiological saline. Following centrifugation at 500 × g
for 7 min at room temperature, the cells cultured with gentamicin
(80 U/ml; Yichang Humanwell Pharmaceutical Co., Ltd., Yichang,
China) were defined as the experiment group, and the cells cultured
with no gentamicin were defined as the control group. The
lymphocytes were cultured in GT-T551 medium (Takara Biotechnology
Co., Ltd., Dalian, China) with 1,000 U/ml γ-interferon (Beijing
Biocoen Biotechnology Co., Ltd., Beijing, China) and 10% autologous
plasma added on day 0. Then, 50 µg/ml CD3 monoclonal antibody
(catalogue no. Mab-37; Skoda Biotechnology Co., Ltd., Shanghai,
China) and 100 U/ml interleukin 1α PeproTech China, Suzhou, China)
were added on day 1. Subsequently, 1,000 U/ml recombinant human
interleukin 2 (SL Pharma Labs, Inc., Wilmington, DE, USA) and 2%
autologous plasma were added to the medium from day 1 onward. The
cells were cultured at 37°C with 5% CO2 until the 11th
day.
The CIK protein was extracted as reported by Gao
et al (14) with certain
modifications. The CIKs were centrifuged at 150 × g for 10 min at
4°C, and the precipitate was washed with 1 ml 0.9% physiological
saline three times. After resuspension in 0.1 ml 0.9% physiological
saline, the CIKs were lysed by freezing-thawing from −80°C to room
temperature five times, and then centrifuged at 13,000 × g for 10
min at 4°C. After determining the protein concentration using the
Bradford method (15), the
supernatant was collected and stored at −20°C with 1 Mm
phenylmethanesulfonyl fluoride (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) for subsequent analysis.
Flow cytometry analysis
A total of 1 ml CIK suspension was collected
following centrifugation at 150 × g for 10 min at room temperature,
the precipitate was resuspended in 1 ml of 0.9% physiological
saline and centrifuged at 150 × g for 10 min at room temperature.
The precipitate was then resuspended in 150 µl 0.9% physiological
saline and divided into two groups. One group as the isotype
control with FITC mouse IgG 2α (5 µl; catalogue no. 555573; BD
Biosciences, Franklin Lakes, NJ, USA) added, and the control or
experiment groups had FITC mouse anti-human CD3 (5 µl; catalogue
no. 555339; BD Biosciences) added. The three groups were all
incubated 15 min at room temperature, then resuspended in 1 ml 0.9%
physiological saline, and centrifuged at 150 × g for 10 min at room
temperature. Finally, the precipitate was resuspended in 0.2 ml
0.9% physiological saline, and prepared for analysis using a BD
Accuri C6 flow cytometer (BD Biosciences).
MTT analysis
The HepG2 hepatoma cell line, which was purchased
from the Advanced Research Center of Central South University
(Changsha, China), was cultured in Dulbecco's modified Eagle's
medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% FBS (Thermo Fisher Scientific, Inc.) at 37°C
and in 5% CO2. Hepatoma cells used as target cells were
obtained at the logarithmic growth phase and the concentration was
adjusted to 4×104 cells/ml. CIKs cultured for 11 days
were used as effector cells and mixed with target cells in the
ratio 40:1 (effector cell:target cell). A total of 10 ml CIK
culture medium was collected following centrifugation at 150 × g
for 10 min at room temperature. The precipitate was resuspended in
GT-T551 containing 2% autologous plasma and diluted to
1.6×106 cells/ml. The suspension was divided into three
groups: The effector-target group, 100 µl effector and target
cells; effector cell group, 100 µl effector cells and GT-T551
culture medium; target cell group, 100 µl target cells and GT-T551
culture medium. All groups were cultured at 37°C and 5%
CO2 for 24 h. A total of five parallel experiments were
used in each group. Then, 10 µl MTT (5 mg/ml) was added and
cultured at 37°C with 5% CO2 for 4 h. Following
centrifugation at 900 × g for 5 min at room temperature, the
precipitate was dissolved with 100 µl DMSO, agitated for 15 min at
37°C, and the optical density (OD) was detected at 490 nm. The
killing rate (%) was calculated as follows: [1 -
(ODeffector-target cell well - ODeffector cell
well) / ODtarget cell well × 100%.
One-dimensional gel electrophoresis
(1-DE) analysis
1-DE was performed using a 5% stacking gel (pH 6.8)
and an 8% separating gel (pH 8.9) in Tris-glycine buffer (pH 8.3),
and 10 µg protein was analyzed. The gels were run at 60 V for 45
min, then 120 V for 2–3 h per gel in an ice-bath until the
bromophenol blue dye had migrated to <1 cm from the bottom of
the gel. Subsequently, The gels were stained with coomassie
brilliant blue R-250 at room temperature for 15 min. The bands was
analyzed using Quantity One 4.6.2 (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA).
Mass spectrometry analysis
Differentially expressed proteins bands were excised
from 1-DE gels for LC-MS/MS mass spectrometry analysis. Briefly,
after the gel plug was digested with trypsin, 10 µl of the peptide
mixture was separated with a linear gradient of 5–80% buffer B
[100% acetonitrile, 0.1% formic acid (FA)] at a flow rate of 400
nl/min on a C18-reversed phase column packed in-house
with Eprogen-Pur C18-AQ 5 µm resin in buffer A (100%
H2O, 0.1% FA). A prominence nano 2D chromatography
system (Shimadzu Corp., Kyoto, Japan) was coupled online to the
micrOTOF-QII (Bruker Corporation, Billerica, MA, USA). The data was
collected using BrukerDaltonicsmicrOTOFcontrol software 3.2 (Bruker
Corporation) with the conditions 50–2,200 m/z scan range, 1,500 V
capillary voltages, and 150°C drying argon gas temperature.
Finally, the selected peptide masses were analyzed using
DataAnalysis software 4.1 (Bruker Corporation) and searched using
the Mascot search engine version 2.3.01 (http://www.bgi-proteomics.cn/tocsam/cgi/master_results_2.pl?file=20140317%2FF044726.dat;pr.eh=3%2C3p#tc:rf:hits:3).
Western blot analysis
Following 1-DE, the gel was separately transferred
to a polyvinylidene fluoride (PVDF) membrane for 90 min at 300 mA
in transfer buffer (25 mM Tris, 0.1 M glycine, and 20% methanol).
Subsequently, the membranes were blocked for 1 h in TBS-Tween 20
(TBST; 0.05% Tween-20, 20 mM Tris, 150 mM NaCl, pH 7.4) containing
5% skim milk at room temperature. After washing three times with
TBS for 5 min each, the PVDF membrane was incubated with rabbit
anti-human haptoglobin (catalogue no. BA3744; Wuhan Boster
Biological Technology, Ltd., Wuhan, China) at a dilution of 1:200
in TBST containing 5% skim milk at room temperature for 1 h. The
membrane was washed three times with TBS for 20 min and incubated
with goat anti-rabbit IgG-horseradish peroxidase antibody
(catalogue no. MBS856805; Sino-American Bio. Corp., Luoyang, China)
at a dilution of 1:1,000 in TBST containing 5% skim milk at room
temperature for 40 min. Next, the membrane was washed two times
with TBS for 20 min and analyzed using the ECL Plus (Beijing
Solarbio Science & Technology Co., Ltd., Beijing, China)
chemiluminescence detection method. Finally, the results were
analyzed using Quantity One 4.6.2 (Bio-Rad Laboratories, Inc.).
RNA isolation and RT-qPCR
analysis
Total RNA was extracted from CIKs using the
RNAsimple Total RNA Kit (Tiangen Biotech Co., Ltd., Beijing, China)
according to the manufacturer's protocol. A total of 1 µg RNA was
synthesized to cDNA using the RevertAid First Strand cDNA Synthesis
kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Following cDNA synthesis, a RT-qPCR assay
was performed as Livak and Schmittgen (16) reported with some modification. A
segment of 141 and 221 bp was amplified using primer sets
Haptoglobin-F/Haptoglobin-R (Table I)
and β-actin-F/β-actin-R (Table I),
respectively. RT-qPCR was performed in a volume of 20 µl containing
10 µl 2X Master mix (Maxima SYBR Green/ROX qPCR Master Mix kit;
Thermo Fisher Scientific, Inc.), 0.3 µl of 10 µM Forward Primer,
0.3 µl of 10 µM Reverse Primer, 0.8 µl cDNA and 8.6 µl
dH2O. Next, the 20-µl mixture was analyzed using ABI
Stepone Plus (Thermo Fisher Scientific, Inc.), under the following
thermocycling conditions: 1 cycle of 95°C for 10 min, followed by
40 cycles of 95°C for 15 sec, 60°C for 30 sec and 72°C for 15 sec
and a final cycle of 60°C for 1 min and 95°C for 15 sec. Data were
normalized according to the 2−ΔΔCq method (16). The assays were performed in three
independent experiments.
 | Table I.Nucleotide sequences of primers. |
Table I.
Nucleotide sequences of primers.
| Primera | Sequence
(5′-3′) |
|---|
| Haptoglobin-F |
CAGCCAGAAACATAACCC |
| Haptoglobin-R |
TCTACACCCTAACTACTCCC |
| β-actin-F |
ATCGTGCGTGACATTAAGGAG |
| β-actin-R |
TAGGTGCTTTGATGGAAGTTGAG |
Statistical analysis
The data were presented as the mean ± standard
deviation. Statistical analyses were performed using SPSS version
17.0 (SPSS, Inc., Chicago, IL, USA). Differences were determined
using the Student's t-test. P<0.05 or P<0.01 were
respectively considered to indicate significant or very significant
differences. All experiments were repeated at least three
times.
Results
Ratio and activity analyzed for
CIKs
In order to investigate the associated protein
activity of CIKs cultured in vitro, gentamicin was selected
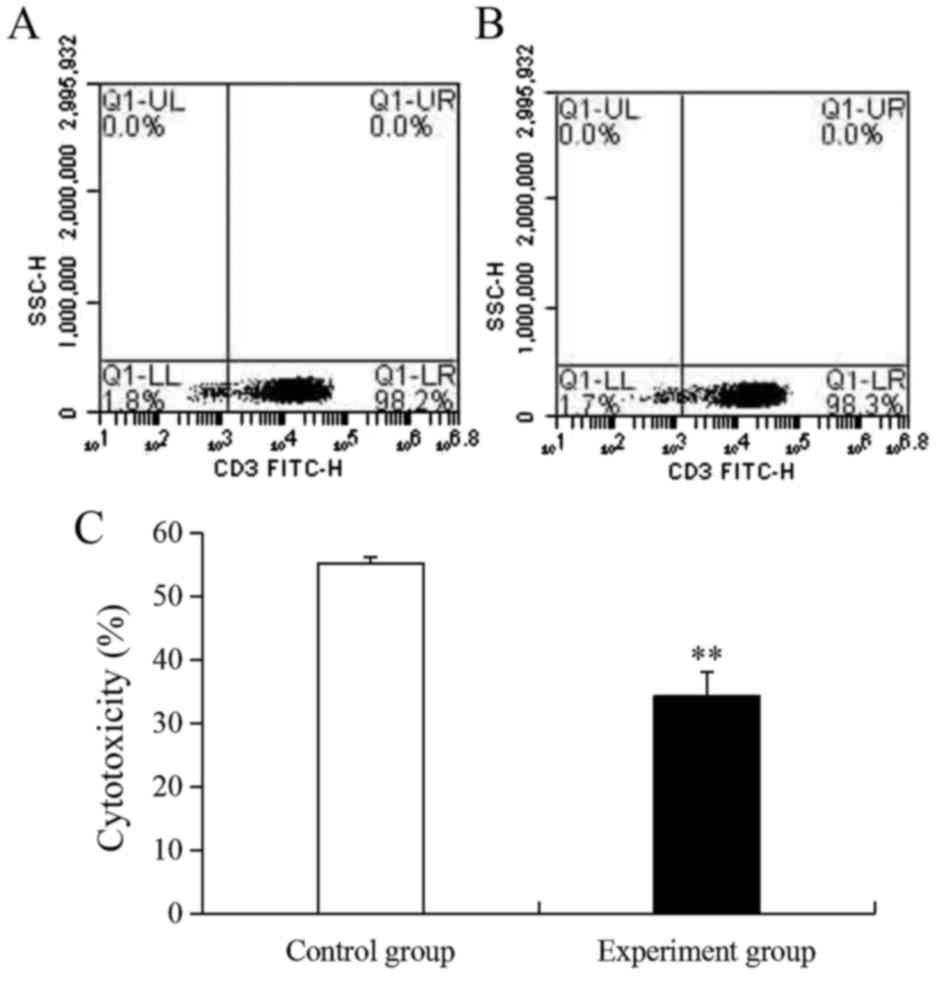
as an activity inhibitor. Following stimulation, the ratio of CIKs
in the control and experiment groups were 98.2 and 98.3%,
respectively (Fig. 1A and B).
However, the activity of CIKs in the experiment group (34.3%) was
very significantly lower compared with the control group (55.2%;
P<0.01; Fig. 1C) because of the
effect of gentamicin.
Differentially expressed protein
analyzed
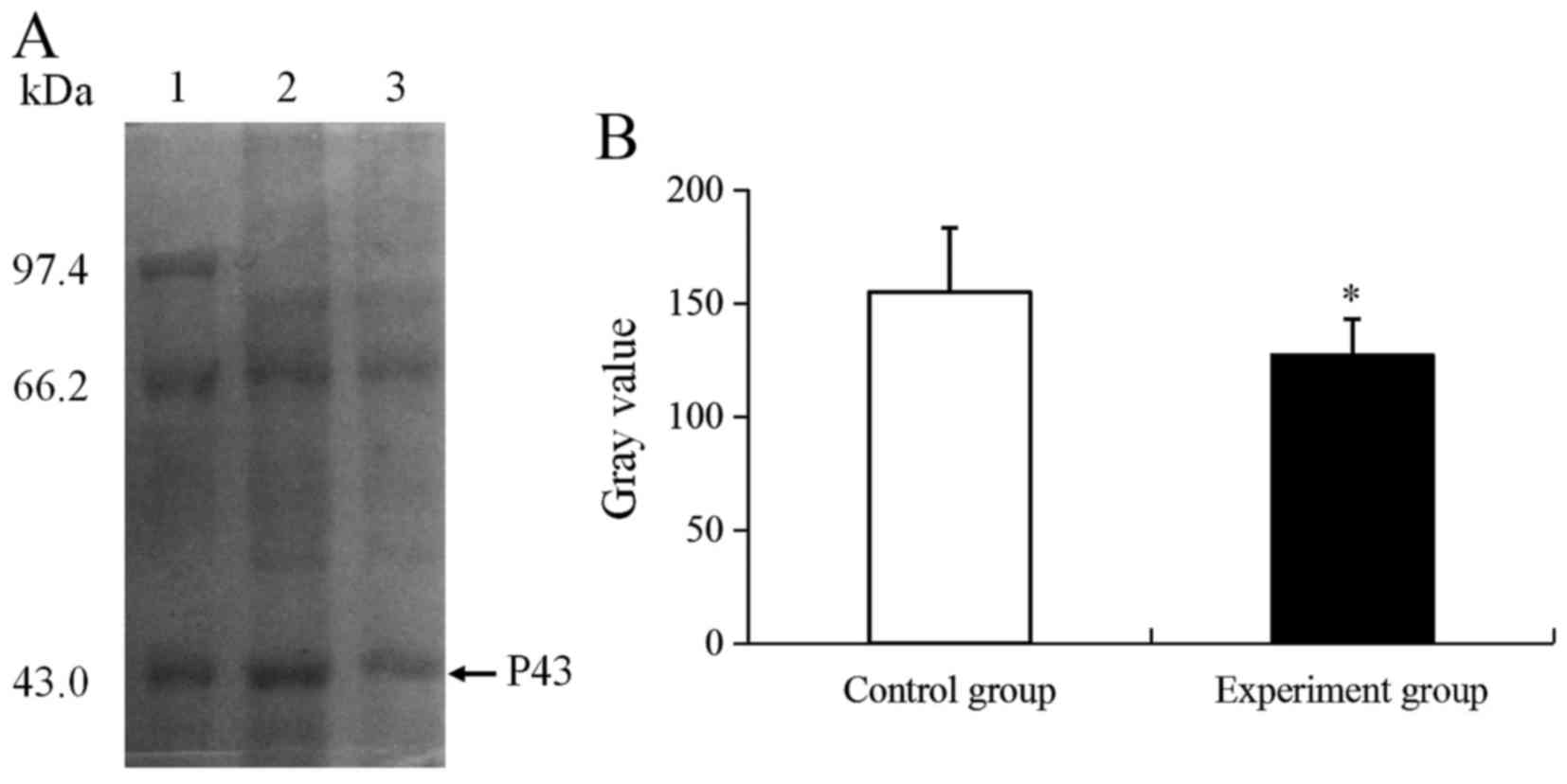
Based on the changes in CIK activity, an unknown
differential expression protein (P43) was identified using 1-DE
(Fig. 2A). As presented in Fig. 2A, the P43 molecular weight was ~43
kDa, and the expression of P43 was significantly downregulated by
1.22-fold compared with the control group (P<0.05; Fig. 2B).
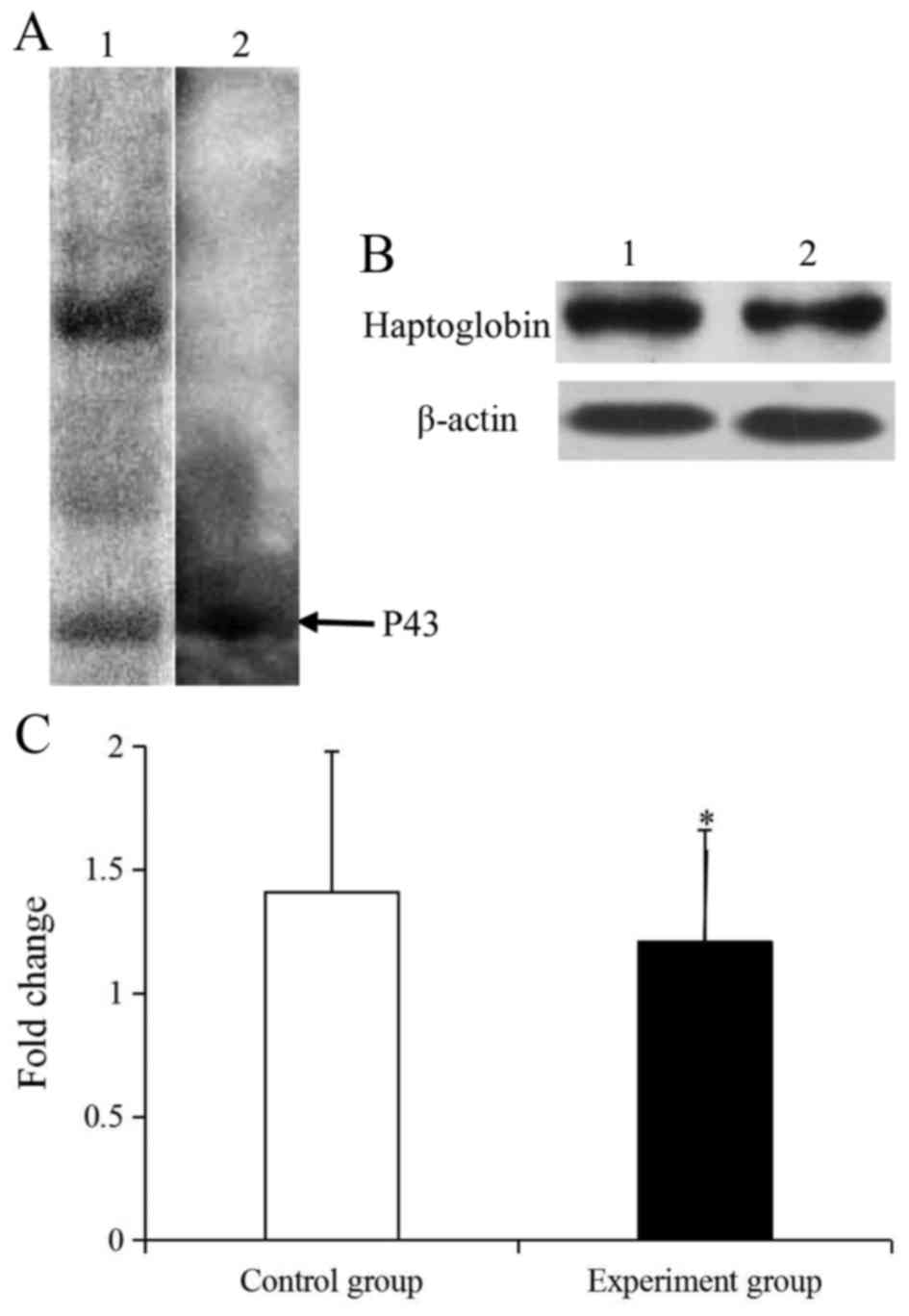
Identification of P43
For further investigation on the P43 protein in
CIKs, the P43 foci was detected as Homo sapien haptoglobin
protein (P00738) using LC-MS/MS (Tables
II and III). As Tables II and III demonstrates, the matched amino acid
sequences of haptoglobin attained an overall score of 159, and the
molecular and isoelectric point was 45861 Da, and 6.13,
respectively. Following a 1-DE separation and western blotting, the
P43 protein in CIKs lysis was demonstrated to specifically react
with the rabbit anti-human-haptoglobin (Fig. 3A). Together, these results suggested
that the P43 protein was haptoglobin.
 | Table II.Identification of the differentially
expressed protein in cytokine-induced killer cells using liquid
chromatography-mass spectrometry. |
Table II.
Identification of the differentially
expressed protein in cytokine-induced killer cells using liquid
chromatography-mass spectrometry.
| Protein | Accession name | Description | Species | Mr (Da) | pI | Trends in
expression |
|---|
| P43 | P00738 | Haptoglobin | Homo
sapiens | 45861 | 6.13 | ↓ |
 | Table III.Matched amino acid sequences of
haptoglobin (P00738) protein identified using liquid
chromatography-mass spectrometry. |
Table III.
Matched amino acid sequences of
haptoglobin (P00738) protein identified using liquid
chromatography-mass spectrometry.
| Observed | Mr (expt) | Mr (calc) | Delta | Miss | Score | Start-end | Sequence |
|---|
| 709.9410 | 1417.8673 | 1417.8181 | 0.0493 | 1 | 56 | 216–228 | DIAPTLTLYVGKK |
| 854.4398 | 1706.8649 | 1706.8120 | 0.0529 | 0 | 27 | 298–311 | YVMLPVADQDQCIR |
| 724.7164 | 2171.1274 | 2171.0504 | 0.0770 | 0 | 34 | 326–345 |
SPVGVQPILNEHTFCAGMSK |
| 602.3429 | 1202.6712 | 1202.6295 | 0.0417 | 0 | 42 | 392–401 | VTSIQDWVQK |
Haptoglobin expression analyzed
To analyze the expression of haptoglobin protein,
western blotting was performed (Fig.
3). In the CIK protein, only haptoglobin could specifically
reacted with rabbit anti-haptoglobin (Fig. 3A). Further investigation revealed that
the expression level of b-actin in the control and experiment
groups was similar and the haptoglobin was significantly
downregulated by 1.17-fold compared with the control group
(P<0.05; Fig. 3B and C).
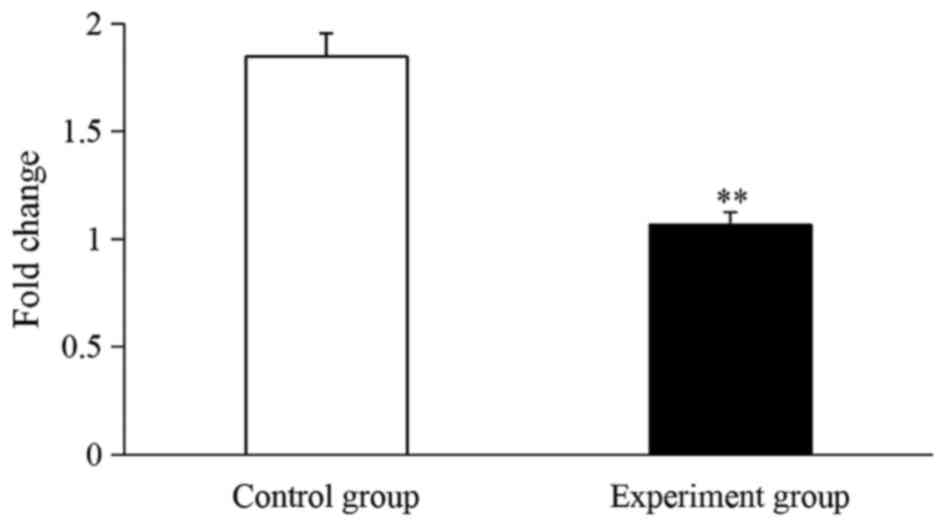
Haptoglobin mRNA analyzed
Based on the proteomic results, a RT-qPCR assay was
performed to determine the transcription levels of haptoglobin in
CIKs. As presented in Fig. 4, SpHMC
mRNA in the experiment group was significantly downregulated by
1.73-fold compared with the control group (P<0.01; Fig. 4).
Discussion
CIKs adoptive immunotherapy is used in tumor
treatment due to its efficiently antitumor activity (17–20).
Gentamicin is a broad-spectrum antibiotic used to preserve the
aseptic culture of cells (21–23). In
the present study, it was demonstrated that the CIK activity was
significant inhibited by gentamicin, and further research revealed
that a protein P43 was associated with CIK activity.
To determine the identity of the P43 protein, 1-DE
with LC-MS/MC was performed (24,25). The
P43 protein was successfully identified as human haptoglobin. This
was similar to the findings that an unknown protein spot identified
as haptoglobin in patient sera using the mass spectra strategy
(26–28). For further verification of the mass
spectra results, western blotting was used (29), and the results demonstrated that
haptoglobin protein was specificity connected with rabbit
anti-human-haptoglobin. Therefore, it was concluded that the P43
protein was haptoglobin.
Haptoglobin is a hemoglobin-binding protein
expressed by a genetic polymorphism for prevented oxidative damage
in plasma (30–32). However, research has identified that
haptoglobin was also involved in the immune response (33,34).
Delanghe et al (35)
demonstrated that haptoglobin phenotypes may influence T cells
activation and effect the interplay of lymphocytes. Shen et
al (36) reported that
haptoglobin activates the innate immunity. Kreisel and Goldstein
(37) noted that haptoglobin was a
novel protein activator of the innate immune system and involved in
immune modulatory as a known acute phase protein. These results
demonstrated that the concentration of haptoglobin in CIKs treated
with gentamicin was significantly downregulated by 1.17 using
western blotting. Thus, it was deduced that the haptoglobin was
associated with the enhancement of CIK activity.
To further illustrate the association of haptoglobin
with CIKs activity, a quantitative RT-qPCR assay was performed. As
a result, the mRNA level of haptoglobin in CIKs treated with
gentamicin was significantly downregulated by 1.73-fold compared
with the control group. This is similar to the finding that
haptoglobin expression signal was decreased 5.1-fold in peripheral
blood mononuclear cells from healthy participants compared with
methotrexate-resistant patients with rheumatoid arthritis (38). Herein, these results suggest that
haptoglobin was involved in the activity-enhancement of CIKs.
In conclusion, it was identified that haptoglobin in
CIKs was an activity-enhancement-associated protein. To the best of
our knowledge, this is the first study to demonstrate that
haptoglobin may be associated with the activity-enhancement of
CIKs. Further investigation is required to explore its exact role
of the signaling pathway associated with an increase in CIK
activity.
Acknowledgements
The present study was supported by The Foundation of
Hunan Provincial Key Laboratory for Special Pathogens Prevention
and Control Foundation (grant no. 2014-5), The Hunan Province
Cooperative Innovation Center for Molecular Target New Drug Study
(grant no. 2015-351), and National Natural Science Foundation of
China (grant no. 81101274).
References
|
1
|
Yang JC and Rosenberg SA: Adoptive T-cell
therapy for cancer. Adv Immunol. 130:279–294. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jäkel CE, Hauser S, Rogenhofer S, Müller
SC, Brossart P and Schmidt-Wolf IG: Clinical studies applying
cytokine-induced killer cells for the treatment of renal cell
carcinoma. Clin Dev Immunol. 2012:4732452012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li R, Wang C, Liu L, Du C, Cao S, Yu J,
Wang SE, Hao X, Ren X and Li H: Autologous cytokine-induced killer
cell immunotherapy in lung cancer: A phase II clinical study.
Cancer Immunol Immunother. 61:2125–2133. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pan K, Guan XX, Li YQ, Zhao JJ, Li JJ, Qiu
HJ, Weng DS, Wang QJ, Liu Q, Huang LX, et al: Clinical activity of
adjuvant cytokine-induced killer cell immunotherapy in patients
with post-mastectomy triple-negative cancer patients. Clin Cancer
Res. 20:3003–3011. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gammaitoni L, Giraudo L, Leuci V,
Todorovic M, Mesiano G, Picciotto F, Pisacane A, Zaccagna A, Volpe
MG, Gallo S, et al: Effective activity of cytokine-induced killer
cells against autologous metastatic melanoma including cells with
stemness features. Clin Cancer Res. 19:4347–4358. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pan QZ, Wang QJ, Dan JQ, Pan K, Li YQ,
Zhang YJ, Zhao JJ, Weng DS, Tang Y, Huang LX, et al: A nomogram for
predicting the benefit of adjuvant cytokine-induced killer cell
immunotherapy in patients with hepatocellular carcinoma. Sci Rep.
5:92022015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rettinger E, Kuçi S, Naumann I, Becker P,
Kreyenberg H, Anzaghe M, Willasch A, Koehl U, Bug G, Ruthardt M, et
al: The cytotoxic potential of interleukin-15-stimulated
cytokine-induced killer cells against leukemia cells. Cytotherapy.
14:91–103. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Meng JX, Zhao MF, Chai X, Xiao X, Mu J, Li
Q, Deng Q and Li YM: IL-21 enhances anti-leukemia effect by acting
on both CD3+CD56+ CIK cells and regulatory T cells derived from
umbilical cord blood in vitro. Blood. 122:10512013.
|
|
9
|
Rutella S, Iudicone P, Bonanno G,
Fioravanti D, Procoli A, Lavorino C, Foddai ML, Lorusso D,
Martinelli E, Vacca M, et al: Adoptive immunotherapy with
cytokine-induced killer cells generated with a new good
manufacturing practice-grade protocol. Cytotherapy. 14:841–850.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tan G, Zhang X, Feng H, Luo H and Wang Z:
The therapeutic effect of cytokine-induced killer cells on
pancreatic cancer enhanced by dendritic cells pulsed with K-Ras
mutant peptide. Clin Dev Immunol. 2011:6493592011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ji Y, Hocker JD and Gattinoni L: Enhancing
adoptive T cell immunotherapy with microRNA therapeutics. Semin
Immunol. 28:45–53. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pan K, Li YQ, Wang W, Xu L, Zhang YJ,
Zheng HX, Zhao JJ, Qiu HJ, Weng DS, Li JJ, et al: The efficacy of
cytokine-induced killer cell infusion as an adjuvant therapy for
postoperative hepatocellular carcinoma patients. Ann Surg Oncol.
20:4305–4311. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Laport GG, Sheehan K, Baker J, Armstrong
R, Wong RM, Lowsky R, Johnston LJ, Shizuru JA, Miklos D, Arai S, et
al: Adoptive immunotherapy with cytokine-induced killer cells for
patients with relapsed hematologic malignancies after allogeneic
hematopoietic cell transplantation. Biol Blood Marrow Transplant.
17:1679–1687. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao D, Li C, Xie X, Zhao P, Wei X, Sun W,
Liu HC, Alexandrou AT, Jones J, Zhao R and Li JJ: Autologous tumor
lysate-pulsed dendritic cell immunotherapy with cytokine-induced
killer cells improves survival in gastric and colorectal cancer
patients. PLoS One. 9:e938862014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bradford MM: A rapid and sensitive method
for the quantitation of microgram quantities of protein utilizing
the principle of protein-dye binding. Anal Biochem. 72:248–254.
1976. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Thorne SH, Negrin RS and Contag CH:
Synergistic antitumor effects of immune cell-viral biotherapy.
Science. 311:1780–1784. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen Y, Lin G, Guo ZQ, Zhou ZF, He ZY and
Ye YB: Effets of MICA expression on the prognosis of advanced
non-small cell lung cancer and the efficacy of CIK therapy. PLoS
One. 8:e690442013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Marin V, Pizzitola I, Agostoni V,
Attianese GM, Finney H, Lawson A, Pule M, Rousseau R, Biondi A and
Biagi E: Cytokine-induced killer cells for cell therapy of acute
myeloid leukemia: Improvement of their immune activity by
expression of CD33-specific chimeric receptors. Haematologica.
95:2144–2152. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zou Y, Li F, Hou W, Sampath P, Zhang Y and
Thorne SH: Manipulating the expression of chemokine receptors
enhances delivery and activity of cytokine-induced killer cells. Br
J Cancer. 110:1992–1999. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Batorov EV, Shevela EY, Tikhonova MA,
Batorova DS, Ushakova GY, Sizikova SA, Sergeevicheva VV, Gilevich
AV, Kryuchkova IV, Ostanin AA and Chernykh ER: Mesenchymal stromal
cells improve early lymphocyte recovery and T cell reconstitution
after autologous hematopoietic stem cell transplantation in
patients with malignant lymphomas. Cell Immunol. 297:80–86. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang J, Li C, Wang Y, Lv H, Guo Y, Dai H,
Wicha MS, Chang AE and Li Q: Cytokine-induced killer (CIK) cells
bound with anti-CD3/anti-CD133 bispecific antibodies target
CD133(high) cancer stem cells in vitro and in vivo. Clin Immunol.
149:156–168. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gargett T and Brown MP: Different cytokine
and stimulation conditions influence the expansion and immune
phenotype of third-generation chimeric antigen receptor T cells
specific for tumor antigen GD2. Cytotherapy. 17:487–495. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang J, Lou X, Shen H, Zellmer L, Sun Y,
Liu S, Xu N and Liao DJ: Isoforms of wild type proteins often
appear as low molecular weight bands on SDS-PAGE. Biotechnol J.
9:1044–1054. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chehayeb JF, Robertson AP, Martin RJ and
Geary TG: Proteomic analysis of adult ascaris suum fluid
compartments and secretory products. PLoS Negl Trop Dis.
8:e29392014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Othman N, Zainudin NS, Mohamed Z, Yahya
MM, Leow VM and Noordin R: Protein expression in sera of patients
with amoebic liver abscess (ALA): Potential use of haptoglobin as a
surrogate disease marker. Trop Biomed. 30:257–266. 2013.PubMed/NCBI
|
|
27
|
Jiang S, Lu Q, Hu S, Chen Y, Liu XL, Yang
Y and Ding MP: Proteomics comparison of the sera from multiple
sclerosis patients and neuromyelitis optica patients. Genet Mol
Res. 13:9292–9299. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Felix K, Hauck O, Fritz S, Hinz U,
Schnölzer M, Kempf T, Warnken U, Michel A, Pawlita M and Werner J:
Serum protein signatures differentiating autoimmune pancreatitis
versus pancreatic cancer. PLoS One. 8:e827552013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wen Q, Huang LT, Luo N, Wang YT, Li XY,
Mao HP, Zhang L, Dong XQ and Yu XQ: Proteomic profiling identifies
haptoglobin as a potential serum biomarker for steroid-resistant
nephrotic syndrome. Am J Nephrol. 36:105–113. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Langlois MR and Delanghe JR: Biological
and clinical significance of haptoglobin polymorphism in humans.
Clin Chem. 42:1589–1600. 1996.PubMed/NCBI
|
|
31
|
Flanagan JJ, Arjomandi A, Delanoy ML, Du
Paty E, Galea P, Laune D, Rieunier F, Walker RP and Binder SR:
Development of monoclonal antibodies to pre-haptoglobin 2 and their
use in an enzyme-linked immunosorbent assay (ELISA). J Immunol
Methods. 406:34–42. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bertaggia E, Scabia G, Dalise S, Lo Verso
F, Santini F, Vitti P, Chisari C, Sandri M and Maffei M:
Haptoglobin is required to prevent oxidative stress and muscle
atrophy. PLoS One. 9:e1007452014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vanuytsel T, Vermeire S and Cleynen I: The
role of haptoglobin and its related protein, zonulin, in
inflammatory bowel disease. Tissue Barriers. 1:e273212013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Asleh R, Ward J, Levy NS, Safuri S,
Aronson D and Levy AP: Haptoglobin genotype-dependent differences
in macrophage lysosomal oxidative injury. J Biol Chem.
289:16313–16325. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Delanghe JR, Langlois MR and De Buyzere
ML: Haptoglobin polymorphism: A key factor in the proatherogenic
role of B cells? Atherosclerosis. 217:80–82. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shen H, Song Y, Christopher CM, Wu T,
Bruce C, Scabia G, Galan A, Maffei M and Daniel RG: Haptoglobin
activates innate immunity to enhance acute transplant rejection in
mice. J Clin Invest. 122:383–387. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kreisel D and Goldstein DR: Innate
immunity and organ transplantation: Focus on lung transplantation.
Transpl Int. 26:2–10. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tan W, Wang F, Guo D, Ke Y, Shen Y, Lv C
and Zhang M: High serum level of haptoglobin is associated with the
response of 12 weeks methotrexate therapy in recent-onset
rheumatoid arthritis patients. Int J Rheum Dis. 19:482–489. 2016.
View Article : Google Scholar : PubMed/NCBI
|