Introduction
Colon cancer is the most common type of
gastrointestinal cancer. The risk of the colon cancer is associated
with lifestyle, inherent cause and colorectal adenoma. Diagnosis of
colon cancer includes laboratory endoscopy, biopsy, exfoliative
cytopathology and carcinoembryonic antigen (CEA) testing. The CEA
test measures the amount of this protein that may appear in the
blood of certain people who have certain types of cancer,
particularly cancer of the large intestine (colon and rectal
cancer). However, high CEA levels do not indicate cancer cell
metastasis, as low CEA levels were also detected in patients with
metastatic colon cancer. Exfoliative cytopathology is not
frequently used in clinics, because an ideal exfoliative sample was
difficult to obtain. Specific and sensitive biomarkers or a
distinct biomarker assemblage may facilitate the diagnosis and
monitoring of patients with colon cancer.
Fusion genes are often identified in cancer and
numerous newly identified fusion genes have oncogenic properties
(1). Therefore, fusion genes are
potential biomarkers or therapeutic targets in cancer (2). Fusion genes are a novel type of gene
that are a full or partial fusion of two genes and usually result
from chromosomal rearrangements (1).
Recently, RNA sequencing (RNA-seq) was applied in the study of
transcriptomes and novel fusion genes were identified, including
SLC45A3-ELK4 and PAX3-FOXO1, which were ascertained
to promote cancer progression (3–5). Using
transcriptome sequencing, Seshagiri et al (6) identified IF3E-RSPO2 and
PTPRK-RSPO3 in clinical colon cancer samples and predicted
that these fusion genes are involved in colon cancer
progression.
CCCTC-binding factor (CTCF) was previously revealed
to suppress expression of the fusion transcript SLC45A3-ELK4
in prostate cancer by binding to the insulators on the genome
between SLC45A3 and ELK4 (7). CTCF binds to enhances or insulators on
chromosomes to inhibit the spread of heterochromatin and regulate
gene expression (8,9). As there are ~15,000 binding sites for
CTCF in the human genome (10), CTCF
may regulate the expression of numerous other fusion genes. Qin
et al (11) identified that
CTCF regulates the expression of fusion transcripts that are not
unique to cancer cells. CTCF is widely expressed in normal tissues,
so may not drive the onset and progression of cancer (10). Brother of the Regulator of Imprinted
Sites (BORIS) is a paralog of CTCF and is expressed in normal
testis, ovary and skin cells; however, it is abnormally expressed
in breast, prostate and colon cancer cells (12–17). As
BORIS has the same zinc-finger domains as those of CTCF (13), BORIS may bind to CTCF-binding sites
and regulate fusion transcripts. Considering the potential effect
of BORIS on fusion genes and its carcinogenicity, the regulation of
fusion genes identified using RNA-seq by Seshagiri et al
(6) was evaluated using BORIS in the
HCT116 colon cancer cell line. The copy number of BORIS increased
as the colorectal carcinoma progressed from the M0 to the M1a stage
(www.oncomine.org) (18). The HCT116 colon cancer cell line
possesses EIF3E(e1)-RSPO2(e2), EIF3E(e1)-RSPO2(e3),
PTPRK(e1)-RSPO3(e2), PTPRK(e7)-RSPO3(e2),
TADA2A-MEF2B and MED13L-CD4 fusion transcripts within
the transcriptome. CDC42SE2-KIAAO146 is a genomic fusion
transcript originating from DNA arrangement in HCT116 cells. BORIS
suppresses the expression of EIF3E, RSPO2,
PTPRK, RSPO3, TADA2A and CD4 to inhibit
the expression of fusion transcripts in HCT116 cells. It was
hypothesized that the fusion transcripts investigated here may not
have oncogenic functions in HCT116 cells. As BORIS is not
colorectal carcinoma-specific, the fusion genes investigated may
constitute a biomarker assemblage for monitoring the progression of
colorectal carcinoma.
Materials and methods
Cell culture
The human HCT116 colon carcinoma cell line and the
K562 chronic myelogenous leukemia cell linewere cultivated in
Dulbecco's modified Eagle's medium supplemented with 10% fetal
bovine serum. Cells were seeded at a density of 1×105
into 6-well plates prior to drug treatment. When cells reached 70%
confluence, 5 µM 5-aza-2′-deoxycytidine (5-Aza-dC; Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) was applied to treat the cells for
48 h, following which an equal volume was added to re-treat the
cells for 48 h. Acetic acid (50%) in water was used as the negative
control. RNA from the cells was extracted using TRIzol®
reagent (Thermo Fisher Scientific, Inc. Waltham, MA, USA),
according to the manufacturer's protocol.
siRNA silencing and transfection
Negative siRNA and BORIS siRNA were synthesized by
Invitrogen; Thermo Fisher Scientific, Inc. (Waltham, MA, USA).
BORIS siRNA targeting; forward, 5′-AACGACGAUGCCGAACCAAUU-3′
was used for silencing BORIS. The target of negative siRNA
is not homologous with any sequence of the human genome. The BORIS
overexpression plasmid (p-BORIS) was obtained from OriGene
Technologies, Inc. (Rockville, MD, USA). Cells were seeded onto a
6-well plate for transfection. Lipofectamine® RNAiMAX
reagent and Lipofectamine® 3000 reagent (Thermo Fisher
Scientific, Inc.) were applied to, respectively, transfect the
siRNA and the plasmid, and to silence and induce the expression of
BORIS in HCT116 cells. RNA was extracted 3 days after
transfection.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RNA was extracted using TRIzol® (Thermo
Fisher Scientific, Inc.) for subsequent RT using
TransScript®One-Step gDNA Removal and cDNA Synthesis
SuperMix (Beijing Transgen Biotech Co., Ltd., Beijing, China). The
transcript amount was quantified using RT-qPCR and calculated using
the 2−ΔΔCq method (19).
The UltraSYBR Mixture was purchased from cwbiotech (Beijing CWBIO
Biotech Co., Ltd., Beijing, China) for RT-qPCR reaction. Primers
are listed in Table I. RT-qPCR was
conducted using the Applied Biosystems® 7500 Real-Time
PCR System (Thermo Fisher Scientific, Inc.).
 | Table I.Primers used in the present
study. |
Table I.
Primers used in the present
study.
| Fusion gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) | Transcript length,
bp |
|---|
|
EIF3E(e1)-RSPO2(e2) |
ACTACTCGCATCGCGCACT |
GGGAGGACTCAGAGGGAGAC | 155 |
|
EIF3E(e1)-RSPO2(e3) |
ACTACTCGCATCGCGCACT |
TGCAGGCACTCTCCATACTG | 205 |
|
PTPRK(e1)-RSPO3(e2) |
AAACTCGGCATGGATACGAC |
GCTTCATGCCAATTCTTTCC | 226 |
|
PTPRK(e7)-RSPO3(e2) |
TGCAGTCAATGCTCCAACTT |
GCCAATTCTTTCCAGAGCAA | 250 |
|
ETV6-NTRK3 |
AAGCCCATCAACCTCTCTCA |
GGGCTGAGGTTGTAGCACTC | 206 |
|
CDC42SE2-KIAA0146 |
AGGGCCAGATTTGAGTGTGT |
AAACTGAAAATCCCCGCTGT | 188 |
|
TADA2A-MEF2B |
GCTCTTTGGCGCGGATTA |
GGAGCTACCTGTGGCCCT | 152 |
|
MED13L-CD4 |
GTGTATGGCGTCGTGATGTC |
TCCCAAAGGCTTCTTCTTGA | 151 |
Genomic DNA extraction and genomic
fusion detection
HCT116 and K562 cell genomic DNA was extracted using
the phenol-chloroform extraction method (7). Primers used to test fusion genes are
listed in Table I. Taq DNA
polymerase purchased from Takara Biotechnology Co., Ltd., Dalian,
China was utilized for PCR. HCT116 and K562 genomic DNA were
applied as templates to detect the putative fusions. The PCR
amplicons were separated by 1.5% agarose gel electrophoresis and
visualized by BIORAD ChemiDoc imaging system (Bio-Rad Laboratories,
Inc., Hercules, CA, USA).
Clinical data
The copy number amplification frequency of BORIS in
two TCGA colorectal carcinoma datasets was summarized by the cBio
Cancer Genomics Portal (cBioPorta, the date of access is September
24th, 2015) (20,21). Associations between the copy number of
BORIS and the progression of colorectal carcinoma were analyzed
using Oncomine 4.4 (© 2015 Thermo Fisher Scientific Inc.).
Statistical analysis
All experimental data are presented as the mean ±
standard deviation. Data in the TCGA Colorectal 2 dataset exported
from the Oncomine database were replotted and the significance was
calculated using analysis of variance by the software of SPSS
Statistics 17.0.0 (SPSS Inc., Chicago, IL, USA). The significance
of up- or downregulation was calculated using Student' t-tests type
2 analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
BORIS copy number alteration in
colorectal carcinoma
BORIS is expressed in the testis, ovary and skin
cells of healthy individuals (13,22). Prior
studies have identified the abnormal expression of BORIS in a
number of somatic cancer types, including breast cancer and
prostate cancer (15–17,23).
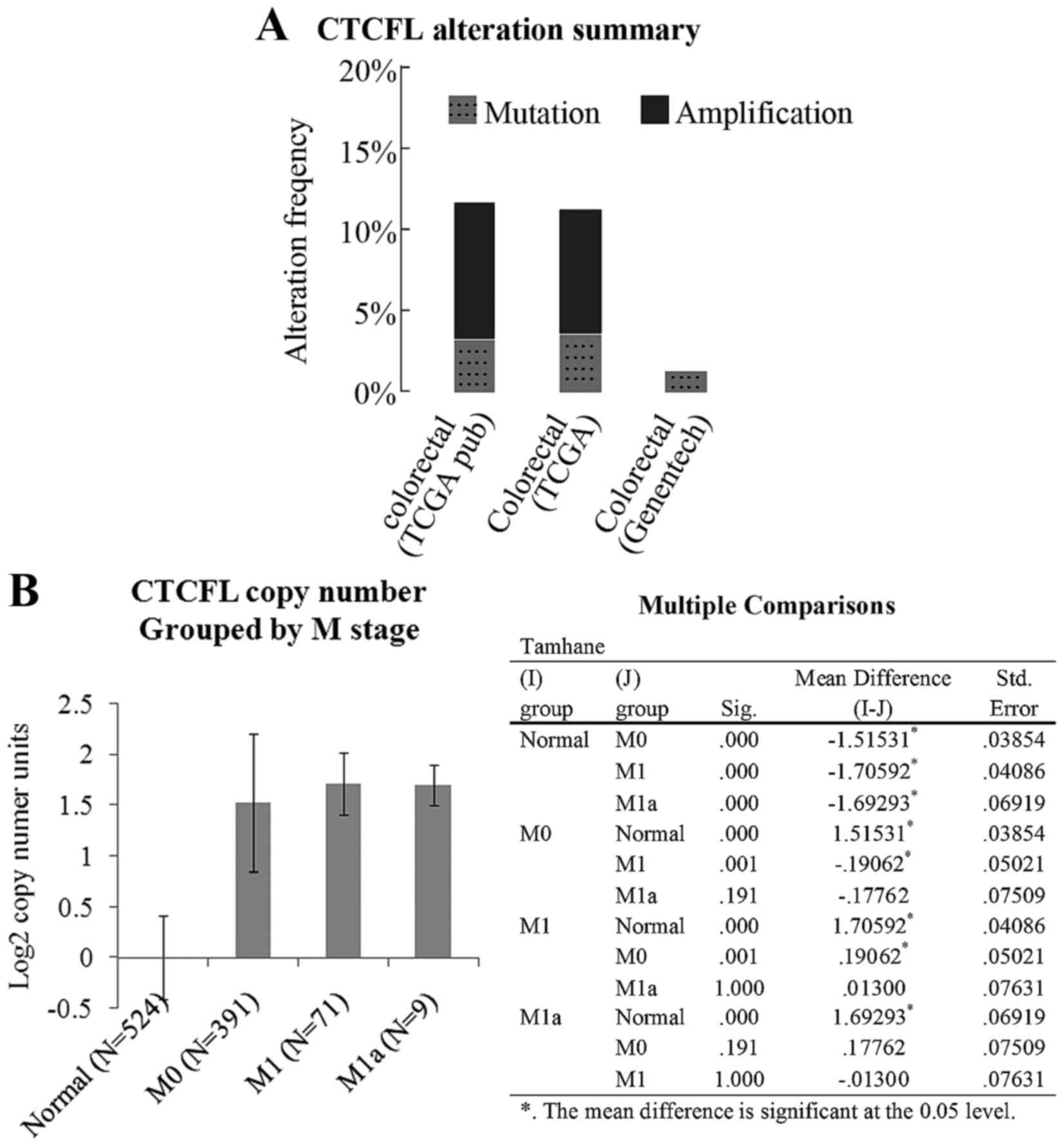
Fig. 1A indicates a copy number
amplification frequency of ~10% for BORIS in two TCGA colorectal
carcinoma datasets, as summarized by cBioPortal (20,21).
Fig. 1B indicates that the copy
number of BORIS is increased according to M stage progression. The
data were analyzed using Oncomine. It was considered that the
increased expression of BORIS may be associated with colorectal
carcinoma progression.
Genomic fusion gene examination
Seshagiri et al (6) analyzed 70 pairs of colon cancer and
adjacent non-cancerous tissues using RNA-seq. This study identified
multiple fusion transcripts, including recurrent gene fusions,
involving the R-spondin family members RSPO2 and
RSPO3 that together occur in ~10% of patients with colon
cancer. By fusing with exon one of EIF3E or PTPRK,
the expression of RSPO2 or RSPO3 was increased, and
the Wnt signaling pathway was correspondingly activated. To
investigate the fusion genes in colon cancer, the existence of
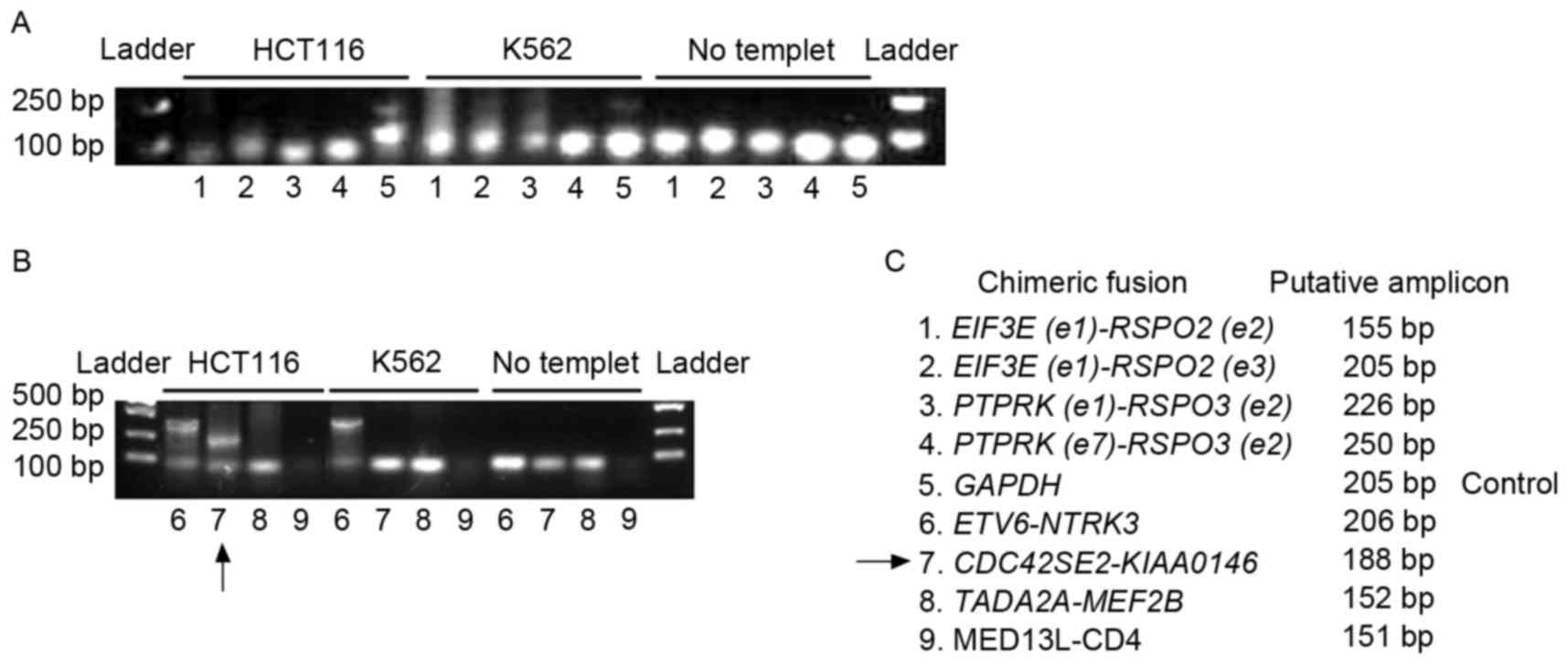
fusion genes in the genome of HCT116 cells was initially evaluated.
Genomic DNA was extracted from HCT116 and K562 cells. The primers
listed in Table I were used to
determine the presence of the fusion genes. Putative lengths of the
amplicons are listed in Table I and
Fig. 2. As the K562 cell line is
BCR-ABL fusion gene-positive and its genome is unstable
(24), it was investigated whether
K562 cells possess the same genomic fusion genes that were
identified in colon cancer, in order to validate the specificity of
the investigated fusion genes in colon cancer. ‘No template’ in
Fig. 2 indicates the negative control
of PCR, which did not contain template to preclude the
contamination of the PCR system. GAPDH was used as a positive
control for PCR amplification. The arrow in Fig. 2B indicates the amplicon of
CDC42SE2-KIAAO146 from the HCT116 genome. Fig. 2B indicates that a larger band of the
ETV6-NTRK3 amplicon was detected in the HCT116 and the K562
genome. The lowest bands on the gels were primer dimers (Fig. 2A and B). The fusion between
ETV6 and NTRK3 is beyond the range of prediction.
Furthermore, the ETV6-NTRK3 amplicon is not specific for
colon cancer (Fig. 2B). Therefore,
ETV6-NTRK3 may not be worthy of further study. Although
Seshagiri et al (6)
demonstrated that fusion genes involving RSPO2 and
RSPO3 existed in the genome of a limited number of patients
with colon cancer, genomic fusion of EIF3E(e1)-RSPO2(e2),
EIF3E(e1)-RSPO2(e3), PTPRK(e1)-RSPO3(e2),
PTPRK(e7)-RSPO3(e2), TADA2A-MEF2B and
MED13L-CD4 was not identified in the genome of HCT116 or
K562 (Fig. 2A and B). However, the
expression of these fusion genes was detected in the transcriptome
of HCT116 (Figs. 3–6).
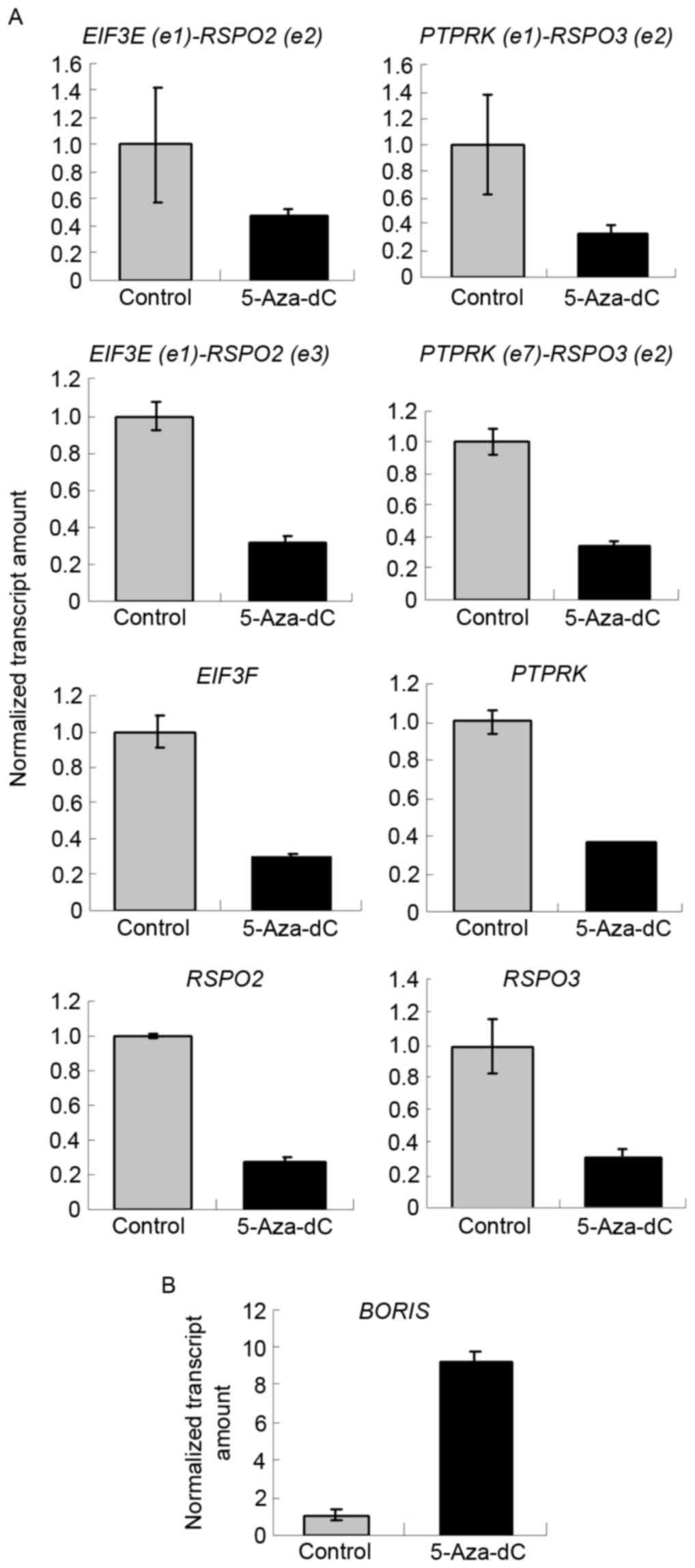
5-Aza-dC induces the expression of
BORIS to downregulate the expression of fusion transcripts
BORIS is the homolog of CTCF and is expressed
abnormally in colorectal carcinoma (Fig.
1A). As CTCF regulates the expression of SLC45A3-ELK4
and other fusion transcriptsin prostate cancer (11), BORIS may also regulate fusion
transcripts in colon cancer. In the present study, 5-Aza-dC was
used to induce the expression of BORIS (14). BORIS was upregulated by
demethylation, which is induced by 5-Aza-dC treatment (Fig. 3B). The results demonstrated that
EIF3E(e1)-RSPO2(e2), EIF3E(e1)-RSPO2(e3),
PTPRK(e1)-RSPO3(e2) and PTPRK(e7)-RSPO3(e2) were
suppressed by 5-Aza-dC treatment (Fig.
3A). The expression of BORIS was upregulated, and that
of EIF3E, RSPO2, PTPRK and RSPO3 was
downregulated. This suggests that 5-Aza-dC promotes the expression
of BORIS to suppress the expression of the investigated
fusion transcriptsand their parent genes (Fig. 3).
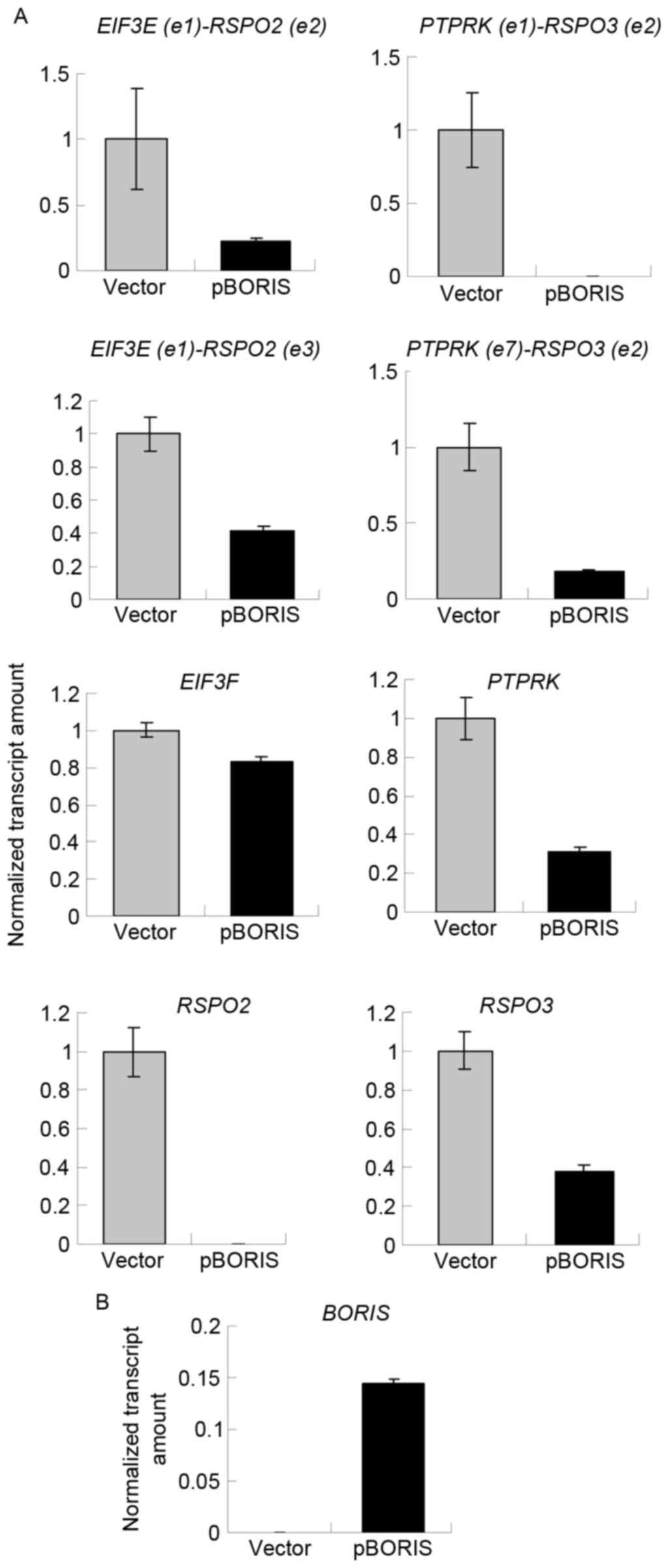
Overexpression of BORIS inhibits the
expression of fusion transcripts
Although 5-Aza-dC treatment induced the expression
of BORIS, 5-Aza-dC induced apoptosis (25). 5-Aza-dC may regulate other genes, in
addition to BORIS, to suppress the expression of fusion
transcripts. To exclude this possibility, BORIS was
overexpressed by transfecting the pBORIS plasmid into HCT116 cells.
The empty vector was set as the negative control. BORIS
overexpression efficiency is indicated in Fig. 4B. The results revealed that the fusion
transcripts and fusion parent genes were all downregulated
(Fig. 4A). These results were in
agreement with those of 5-Aza-dC treatment, and suggested that
BORIS suppresses the expression of fusion transcripts by
downregulating the parent genes.
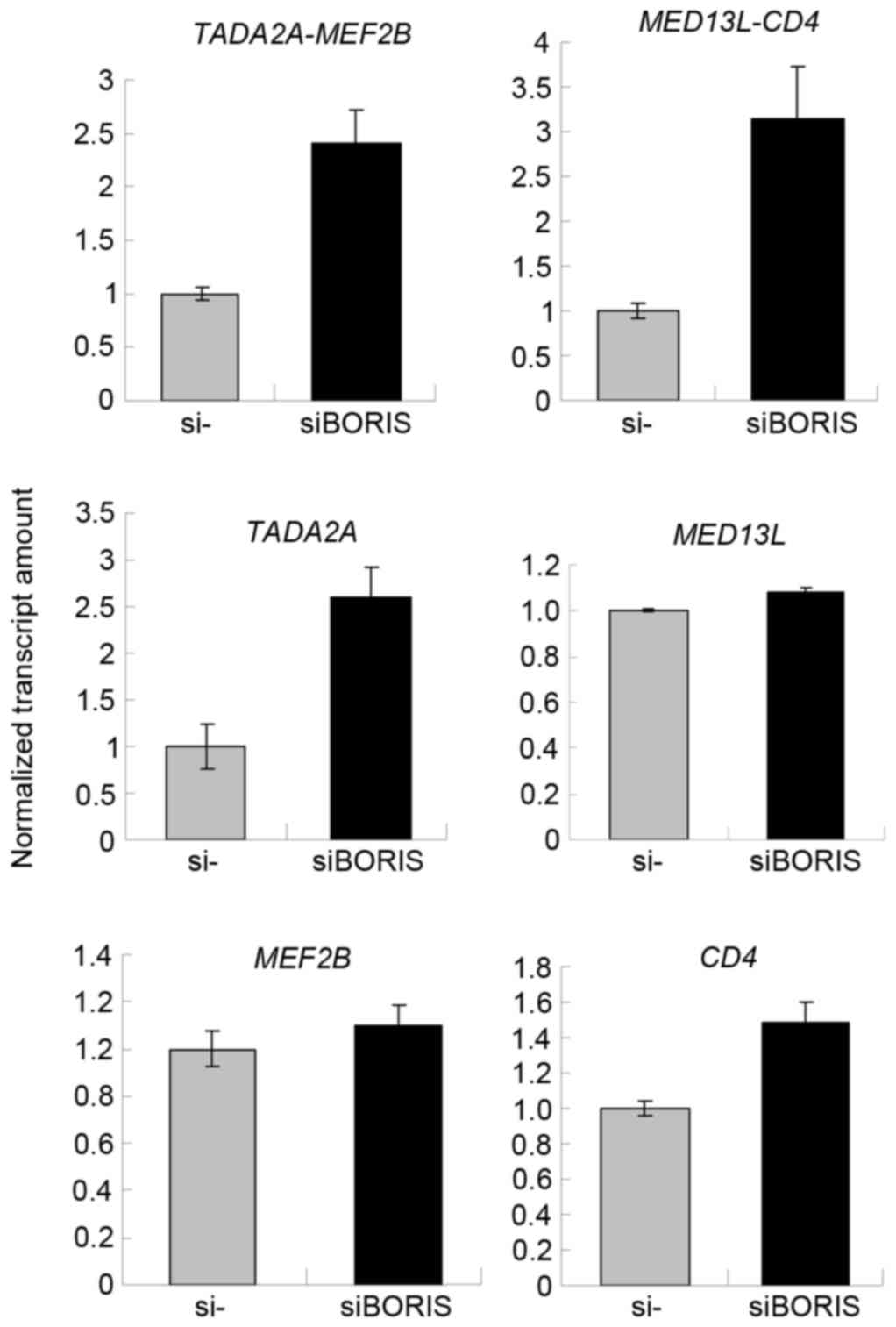
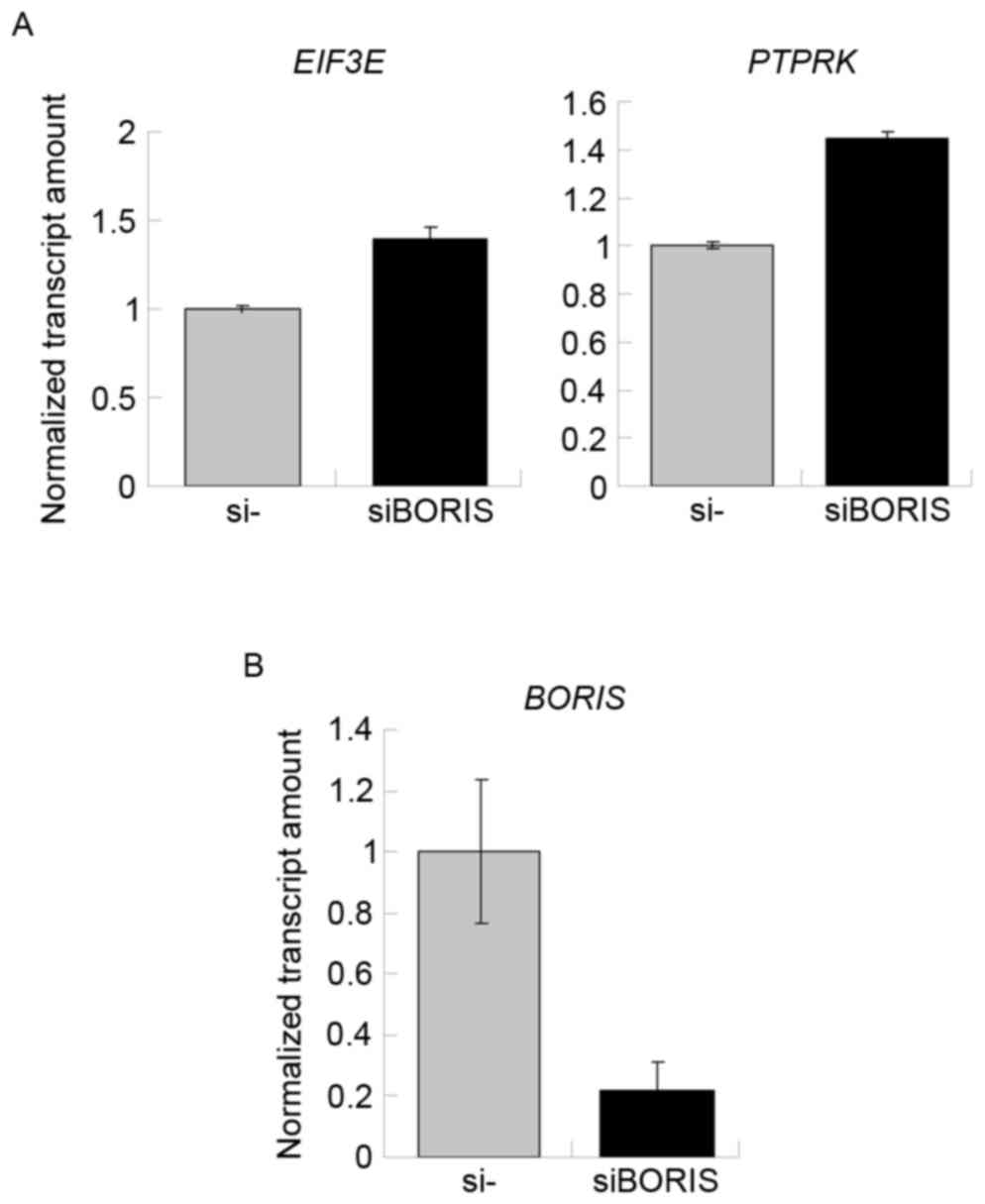
Silencing of BORIS upregulates fusion
transcripts by promoting the parent genes
To confirm the function of BORIS in the regulation
of fusion transcripts, BORIS was silenced in HCT116 cells.
Silencing efficiency was ~70% following transfecting siRNA of
BORIS into HCT116 cells (Fig.
5B). The expression of EIF3E and PTPRK was
upregulated (Fig. 5A). Owing to the
potential toxicity of transfection, the expression of RSPO2,
RSPO3 and the fusion transcripts were undetected. Therefore,
the expression of another two fusion transcripts
TADA2A-MEF2Band MED13L-CD4 identified by Seshagiri
et al (6) in colon cancer was
investigated. As the fusions of TADA2A-MEF2B and
MED13L-CD4 was not detected in the genome of HCT116
(Fig. 2B), TADA2A-MEF2B and
MED13L-CD4 may be regulated by BORIS in the transcriptome of
HCT116 cells. The RNA of siBORIS-silenced HCT116 cells was
extracted and reversed-transcribed into cDNA. RT-qPCR was applied
to determine the expression of certain transcripts. The results
revealed that the fusion parent genes TADA2A and CD4
were upregulated and the fusion transcripts of TADA2A-MEF2B
and MED13L-CD4 were upregulated (Fig. 6). These results suggested that the
downregulation of BORIS promotes the expression of the
investigated fusion transcripts via upregulating the expression of
the parent genes.
Discussion
Chimeric fused nucleic acids were classified as
fusion DNA in the genome and as fusion RNA transcripts in the
transcriptome. As a rule, fusion products in the genome are formed
through DNA rearrangement (26–28).
Fusion RNA transcripts in the transcriptome comprised of
cis-splicing of adjacent genes and trans-splicing
(fusion between genes on different strands of the same chromosome
or different chromosomes) (4,7,11,29). Our study revealed that the existence
of fusion RNA transcripts was not always accompanied by DNA
rearrangements.
As genomic fusion genes are not typically regulated,
the type of fusion investigated in the present study was first
confirmed to be DNA rearrangement or RNA fusion.
CDC42SE2-KIAAO146 was detected in the HCT116 genome
(Fig. 2B). A larger band, in
comparison with the putative ETV6-NTRK3 amplicon, was
detected in HCT116 and K562 cell lines (Fig. 2B). The fusion between ETV6 and
NTRK3 is out of the prediction range. In addition,
ETV6-NTRK3 is not specific for colon cancer, therefore the
regulation of ETV6-NTRK3 was not examined in HCT116 by
BORIS. The other fusion candidates investigated were not detected
in the genome of HCT116. The appearance of the fusions at the RNA
level, whilst a lack of existence at the genomic DNA level,
suggests that they are fusion transcripts on transcriptome.
Two parent transcripts joined to be one fusion
transcript. Therefore, the expression of fusion transcripts may be
affected by the expression of the parent genes. The regulation of
the parent genes was examined in the present study and BORIS was
identified to suppress the parent gene expression to inhibit the
expression of the fusion transcripts (Figs. 3–6).
The statistical data analyzed using cBioPortal
indicatedthat the copy number of BORIS was amplified
significantly in colorectal cancer. However, a rare mutation of
BORIS was detected (Fig. 1A)
that suggests that high expression levels of BORIS may be
associated with colon cancer.
BORIS is the homolog of CTCF. CTCF regulates the
expression of fusion RNA transcripts, including SLC45A3-ELK4
and ADCK4-NUMBL chimeric RNA transcripts (7,11). The
cis-splicing of adjacent genes was identified to be
regulated by the binding events of CTCF to genome. BORIS and CTCF
share the same zinc-finger DNA binding domains (13), therefore BORIS may regulate fusion
transcripts by binding to similar genomic regions to CTCF.
Furthermore, BORIS has a role in chromatin organization and gene
expression; it demethylates chromatin to regulate the gene
expression of cancer testis antigens, and recruits H3K4
methyltransferase to promote the expression of MYC and
BRCA1 (30,31). Thus, BORIS may also regulate fusion
transcripts by affecting the expression of the parent genes. In the
present study, BORIS was identified to inhibit the expression of
EIF3E, RSPO2, PTPRK, RSPO3,
TADA2A and CD4 (Figs.
3–6). The underlying molecular
mechanism requires further study.
The colon cancer genomic fusion gene
CDC42SE2-KIAAO146 was detected in the present study (Fig. 2B). Whether it may be used as target
for diagnosis, prognosis and therapy requires further examination
in other colon cancer cell lines and clinic samples. The expression
of EIF3E (e1)-RSPO2(e2), EIF3E(e1)-RSPO2(e3),
PTPRK(e1)-RSPO3(e2), PTPRK(e7)-RSPO3(e2),
TADA2A-MEF2B and MED13L-CD4 was detected in the
HCT116 transcriptome. The expression of the fusion transcripts was
affected by the parent genes, which were inhibited by BORIS
(Figs. 3–6). High expression of BORIS frequently
associated with cancer and BORIS promotes cancer cell
proliferation. On the other hand, chimeric fusion nuclear acids
were traditionally considered abnormal products of cancer. In
addition, it was predicted that BORIS promotes the expression of
abnormal chimeric fusions. However, BORIS did not induce or
suppress the expression of the tested chimeric fusions in our
study. Therefore, the association between cancer and chimeric
fusion RNA transcripts needs further determination. Clinical
investigation of the copy number alteration of BORIS in colorectal
carcinoma patients suggested that the BORIS copy number is
increased according to M stage progression (Fig. 1B). As BORIS is not colorectal
carcinoma-specific, the fusion genes investigated may constitute a
biomarker assemblage for monitoring colorectal carcinoma
progression.
Acknowledgements
The present study was supported by National Natural
Science Foundation of China (grant no. 81301782) and the Project
supported for Returned Overseas Chinese Scholars. The authors wish
to thank Mr. Xiaoliang Zheng and Mrs. Dongmei Yan of the Zhejiang
Academy of Medical Science (Hangzhou, China) for their discussion
and support.
References
|
1
|
Tomlins SA, Rhodes DR, Perner S,
Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J,
Kuefer R, et al: Recurrent fusion of TMPRSS2 and ETS transcription
factor genes in prostate cancer. Science. 310:644–648. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Druker BJ, Talpaz M, Resta DJ, Peng B,
Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R,
Ohno-Jones S and Sawyers CL: Efficacy and Safety of a Specific
Inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid
leukemia. N Engl J Med. 344:1031–1037. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kumar-Sinha C, Kalyana-Sundaram S and
Chinnaiyan AM: SLC45A3-ELK4 chimera in prostate cancer: Spotlight
on cis-splicing. Cancer Discov. 2:582–585. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rickman DS, Pflueger D, Moss B, Van Doren
VE, Chen CX, de la Taille A, Kuefer R, Tewari AK, Setlur SR,
Demichelis F and Rubin MA: SLC45A3-ELK4 is a novel and frequent
erythroblast transformation-specific fusion transcript in prostate
cancer. Cancer Res. 69:2734–2738. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maher CA, Kumar-Sinha C, Cao X,
Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N
and Chinnaiyan AM: Transcriptome sequencing to detect gene fusions
in cancer. Nature. 458:97–101. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Seshagiri S, Stawiski EW, Durinck S,
Modrusan Z, Storm EE, Conboy CB, Chaudhuri S, Guan Y, Janakiraman
V, Jaiswal BS, et al: Recurrent R-spondin fusions in colon cancer.
Nature. 488:660–664. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Y, Gong M, Yuan H, Park HG, Frierson
HF and Li H: Chimeric transcript generated by cis-splicing of
adjacent genes regulates prostate cancer cell proliferation. Cancer
Discov. 2:598–607. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zlatanova J and Caiafa P: CTCF and its
protein partners: Divide and rule? J Cell Sci. 122:1275–1284. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lobanenkov VV, Nicolas RH, Adler VV,
Paterson H, Klenova EM, Polotskaja AV and Goodwin GH: A novel
sequence-specific DNA binding protein which interacts with three
regularly spaced direct repeats of the CCCTC-motif in the
5′-flanking sequence of the chicken c-myc gene. Oncogene.
5:1743–1753. 1990.PubMed/NCBI
|
|
10
|
Kim TH, Abdullaev ZK, Smith AD, Ching KA,
Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV and Ren B: Analysis
of the vertebrate insulator protein CTCF-binding sites in the human
genome. Cell. 128:1231–1245. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qin F, Song Z, Babiceanu M, Song Y,
Facemire L, Singh R, Adli M and Li H: Discovery of CTCF-sensitive
Cis-spliced fusion RNAs between adjacent genes in human prostate
cells. PLoS Genet. 11:e10050012015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vatolin S, Abdullaev Z, Pack SD, Flanagan
PT, Custer M, Loukinov DI, Pugacheva E, Hong JA, Morse H III,
Schrump DS, et al: Conditional expression of the CTCF-paralogous
transcriptional factor BORIS in normal cells results in
demethylation and derepression of MAGE-A1 and reactivation of other
cancer-testis genes. Cancer Res. 65:7751–7762. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Loukinov DI, Pugacheva E, Vatolin S, Pack
SD, Moon H, Chernukhin I, Mannan P, Larsson E, Kanduri C, Vostrov
AA, et al: BORIS, a novel male germ-line-specific protein
associated with epigenetic reprogramming events, shares the same
11-zinc-finger domain with CTCF, the insulator protein involved in
reading imprinting marks in the soma. Proc Natl Acad Sci USA.
99:pp. 6806–6811. 2002; View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hoffmann MJ, Müller M, Engers R and Schulz
WA: Epigenetic control of CTCFL/BORIS and OCT4 expression in
urogenital malignancies. Biochem Pharmacol. 72:1577–1588. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
D'Arcy V, Abdullaev ZK, Pore N, Docquier
F, Torrano V, Chernukhin I, Smart M, Farrar D, Metodiev M,
Fernandez N, et al: The potential of BORIS detected in the
leukocytes of breast cancer patients as an early marker of
tumorigenesis. Clin Cancer Res. 12:5978–5986. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
D'Arcy V, Pore N, Docquier F, Abdullaev
ZK, Chernukhin I, Kita GX, Rai S, Smart M, Farrar D, Pack S, et al:
BORIS, a paralogue of the transcription factor, CTCF, is aberrantly
expressed in breast tumours. Br J Cancer. 98:571–579. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dougherty CJ, Ichim TE, Liu L, Reznik G,
Min WP, Ghochikyan A, Agadjanyan MG and Reznik BN: Selective
apoptosis of breast cancer cells by siRNA targeting of BORIS.
Biochem Biophys Res Commun. 370:109–112. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Method. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rosa-Garrido M, Ceballos L, Alonso-Lecue
P, Abraira C, Delgado MD and Gandarillas A: A cell cycle role for
the epigenetic factor CTCF-L/BORIS. PLoS One. 7:e393712012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Martin-Kleiner I: BORIS in human cancers-a
review. Eur J Cancer. 48:929–935. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Weber A, Borghouts C, Brendel C, Moriggl
R, Delis N, Brill B, Vafaizadeh V and Groner B: Stat5 exerts
distinct, vital functions in the cytoplasm and nucleus of Bcr-Abl+
K562 and Jak2(V617F)+ HEL leukemia cells. Cancers. 7:503–537. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schneider-Stock R, Diab-Assef M, Rohrbeck
A, Foltzer-Jourdainne C, Boltze C, Hartig R, Schönfeld P, Roessner
A and Gali-Muhtasib H: 5-Aza-cytidine is a potent inhibitor of DNA
methyltransferase 3a and induces apoptosis in HCT-116 colon cancer
cells via Gadd45- and p53-dependent mechanisms. J Pharmacol Exp
Ther. 312:525–536. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rabbitts TH: Chromosomal translocations in
human cancer. Nature. 372:143–149. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rowley JD: The role of chromosome
translocations in leukemogenesis. Semin Hematol. 36 4 Suppl
7:S59–S72. 1999.
|
|
28
|
Heim S and Mitelman F: Molecular screening
for new fusion genes in cancer. Nat Genet. 40:685–686. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yuan H, Qin F, Movassagh M, Park H, Golden
W, Xie Z, Zhang P, Sklar J and Li H: A chimeric RNA characteristic
of rhabdomyosarcoma in normal myogenesis process. Cancer Discov.
3:1394–1403. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bhan S, Negi SS, Shao C, Glazer CA, Chuang
A, Gaykalova DA, Sun W, Sidransky D, Ha PK and Califano JA: BORIS
binding to the promoters of cancer testis antigens, MAGEA2, MAGEA3,
and MAGEA4, is associated with their transcriptional activation in
lung cancer. Clin Cancer Res. 17:4267–4276. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nguyen P, Bar-Sela G, Sun L, Bisht KS, Cui
H, Kohn E, Feinberg AP and Gius D: BAT3 and SET1A form a complex
with CTCFL/BORIS to modulate H3K4 histone dimethylation and gene
expression. Mol Cell Biol. 28:6720–6729. 2008. View Article : Google Scholar : PubMed/NCBI
|