Introduction
Esophageal cancer cases are distributed worldwide;
its mortality rate ranks sixth among all types of cancer and it is
the fourth most commonly occurring cancer in China (1,2). China is
among the highest risk areas of esophageal cancer, where ~90% of
cases are squamous cell carcinoma (SCC) (3). The 5-year survival rate and quality of
life remain low (3). However, the
molecular mechanisms underlying the initiation and progression of
esophageal SCC (ESCC) remain unclear.
Special AT-rich sequence binding protein-1 (SATB1),
a tissue-specific nuclear matrix binding protein, was identified in
1992 (4). SATB1 is primarily
expressed in thymocytes, and also the expression of SATB1 in the
basal layer of the epidermis is regulated by p63 (5,6). SATB1 can
regulate gene expression by folding chromatin into loop domains and
tethering DNA domains to the SATB1 network structure (7,8). Under
normal conditions, SATB1 is expressed at low levels in cells and
tissues, but is overexpressed in a variety of malignant tumors,
including laryngeal squamous cell carcinoma, endometrial cancer,
hepatocellular carcinoma, rectal cancer, cutaneous malignant
melanoma, gastric cancer, prostate cancer, lung cancer and
cutaneous T-cell lymphoma (7,9–20). The
expression of SATB1 has been examined in esophageal adenocarcinoma
and is an independent prognostic factor (20). In several types of cancer, including
laryngeal squamous cell carcinoma, endometrial cancer,
hepatocellular cancer and lung cancer, high expression of SATB1
promotes tumor growth and metastasis, and is a negative prognostic
factor (7,9–20). SATB1
can regulate the expression of >1,000 genes involved in the
processes of DNA organization, proliferation and apoptosis
(8,9).
SATB1 has been demonstrated to serve roles in malignancies, in
addition to malignant transformation (7,20).
The present study aimed to explore the role of SATB1
in ESCC by investigating the association between SATB1 mRNA
expression and prognosis, and recurrence in 102 patients with ESCC
following surgery.
Materials and methods
Tissue samples, patient data and
follow-up
A total of 102 tissue samples of ESCC were obtained
from patients (78 male, 24 female) undergoing surgery at the
Department of Thoracic Oncology, West China Hospital, Sichuan
University (Chengdu, China) between March 2010 and July 2014.
Patient median age was 60.5 years old (range, 39–81 years old). All
specimens were immediately frozen in liquid nitrogen and stored at
−80°C until RNA extraction. All human ESCC tissue samples were
obtained and handled in accordance with an approved Institutional
Review Board application (Ethics Committee of Sichuan University).
Written informed consent was obtained from all patients.
All patients were diagnosed with primary ESCC based
on pathological assessment and had undergone a complete surgical
resection (R0). Patients who underwent non-curative resection (R1),
succumbed to postoperative complications, missed valid follow-up
conditions or lacked clinical data were all excluded from the
present study. No patients received chemotherapy or radiotherapy
prior to surgery. The clinical stage and histologic grade of the
tumor was defined according to the 7th edition of the Tumor Node
Metastasis (TNM) classification of the International Union Against
Cancer (21). Follow-up began on the
date of surgery and ended in July 2014. The median follow-up was
35.5 months (range, 6–70 months). Regular history and physical
examinations were performed in all patients every 3 months during
the first 2 years after surgery and then every 6 months
thereafter.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cancerous specimens
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). A total of 5 µg of each RNA sample was reverse
transcribed into complementary DNA using the Prime-Script™ one step
RT-PCR kit (Takara Bio, Inc., Otsu, Japan). SATB1 expression level
was determined by RT-qPCR using the following primer sequences:
forward, 5′-GTGGAAGCCTTGGGAATCC-3′ and reverse,
5′-CTGACAGCTCTTCTTCTAGTT-3′. β-actin was used as an internal
control, using the following primer sequences: forward,
5′-CTGGCACCACACCTTCTACAATG-3′ and reverse,
5′-CCTCGTAGATGGGCACAGTGTG-3′. The RT-qPCR reaction was performed
with an initial 95°C denaturation step for 10 min, followed by 40
cycles of 95°C for 30 sec 60°C for 30 sec and 72°C for 30 sec at
for 40 cycles using the ABI7500 system (Applied Biosystems; Thermo
Fisher Scientific, Inc.) and the SYBRGreen PCR Master Mix (Takara
Bio, Inc.). SATB1 mRNA levels were normalized to β-actin by the
2−ΔΔCq method (22).
Statistical analysis
All statistical analyses were performed using SPSS
software, version 17.0 (SPSS, Inc., Chicago, IL, USA). Spearman's
rank correlation coefficient test was used to analyze the
correlation between SATB1 expression in different groups of
patients. Comparison of continuous data was performed with an
independent t-test between two groups, whereas the correlation
among categorical variables was analyzed using a χ2
test. Disease-free survival (DFS) and overall survival (OS)
analyses were performed with the Kaplan-Meier estimator and
log-rank tests. Multiple factor analyses were performed using Cox
regression models to evaluate prognostic factor for patients with
ESCC. P<0.05 was considered to indicate a statistically
significant difference.
Results
Patient clinicopathological
characteristics
A total of 102 patients with ESCC were included in
the current study (78 male, 24 female). Their median age was 60.5
years old (range, 39–81 years old). A total of 35 patients (34.3%)
had lymph node metastases and none had distant nodal metastases
(Table I).
 | Table I.Association between SATB1 expression
and clinicopathologic parameters in 102 patients with ESCC. |
Table I.
Association between SATB1 expression
and clinicopathologic parameters in 102 patients with ESCC.
| Factor | SATB1 low expression
(n=53) N (%) | SATB1 high expression
(n=49) N (%) | P-value |
|---|
| Age (mean ± standard
deviation)a | 61.5±7.8 | 59.5±10.8 | 0.310b |
| Age group |
|
| 0.666c |
| ≤61
yrs | 25 (47.2) | 21 (42.0) |
|
| >61
yrs | 28 (52.8) | 28 (56.0) |
|
| Gender |
|
| 0.475c |
| Male | 39 (73.6) | 39 (79.6) |
|
|
Female | 14 (26.4) | 10 (20.4) |
|
| Smoking/drinking
alcohol |
|
| 0.701c |
|
Never | 17 (32.1) | 14 (28.6) |
|
| Ever and
current | 36 (67.9) | 35 (71.4) |
|
| Family history of
ESCC |
|
| 0.963c |
| No | 37 (69.8) | 34 (69.4) |
|
| Yes | 16 (30.2) | 15 (30.6) |
|
| Barium meal
examination |
|
| 0.760c |
| Mushroom
type | 10 (18.9) | 6 (12.0) |
|
| Medullary
type | 16 (30.2) | 16 (32.0) |
|
|
Ulcer | 25 (47.2) | 24 (46.0) |
|
| Mass | 0 | 1 (2.0) |
|
|
Coarctation | 2 (3.8) | 2 (4.0) |
|
| Tumor location |
|
| 0.908c |
|
Upper | 2 (3.8) | 1 (2.0) |
|
|
Middle | 38 (71.7) | 35 (71.4) |
|
| Low | 13 (24.5) | 13 (26.5) |
|
| Histological
type |
|
| 0.552c |
|
High | 22 (41.5) | 23 (46.9) |
|
|
Middle | 28 (50.9) | 24 (49) |
|
|
Low | 3 (5.7) | 2 (4.1) |
|
| pT status |
|
| 0.074c |
|
pT1 | 1 (1.9) | 5 (10.2) |
|
|
pT2-4 | 52 (98.1) | 44 (89.8) |
|
| Lymph node
metastasis |
|
| 0.361c |
|
Yes | 16 (30.2) | 19 (38.8) |
|
| No | 37 (69.8) | 30 (61.2) |
|
| pTNM stage |
|
| 0.003c |
| I | 15 (28.3) | 16 (32.7) |
|
| II | 25 (47.2) | 10 (20.4) |
|
|
III | 13 (24.5) | 16 (32.7) |
|
| IV | 0 (0) | 7 (14.3) |
|
| pTNM stage
group |
|
| 0.023c |
|
I–II | 40 (75.5) | 26 (53.1) |
|
|
III–IV | 13 (24.5) | 23 (46.9) |
|
| Relapse |
|
| 0.029c |
| No | 33 (62.3) | 19 (38.8) |
|
|
Yes | 20 (37.7) | 30 (61.2) |
|
Association between SATB1 expression
and clinicopathological characteristics
According to the average expression (the median
value=1.39) of SATB1 mRNA, the 102 patients with ESCC were split
into two groups. A high expression group with SATB1 mRNA expression
above 1.39 and a low expression group with SATB1 mRNA expression
below 1.39. A total of 49 (48.0%) specimens exhibited high
expression and 53 (54.0%) specimens exhibited low expression of
SATB1 mRNA. No significant associations were identified between the
expression level of SATB1 and patient age, gender, tumor
differentiation grade, adjuvant radio/chemotherapy or the use of
alcohol and cigarettes (P>0.05). However, the expression level
was correlated with clinical TNM stage (P<0.05) (Table I).
High expression of SATB1 mRNA is
associated with poor prognosis for patients with ESCC
To reveal whether pathological TNM (pTNM) stage and
SATB1 mRNA expression affected postoperative outcome, univariate
analyses were performed. The results indicated that SATB1
expression and pTNM stage were associated with postoperative
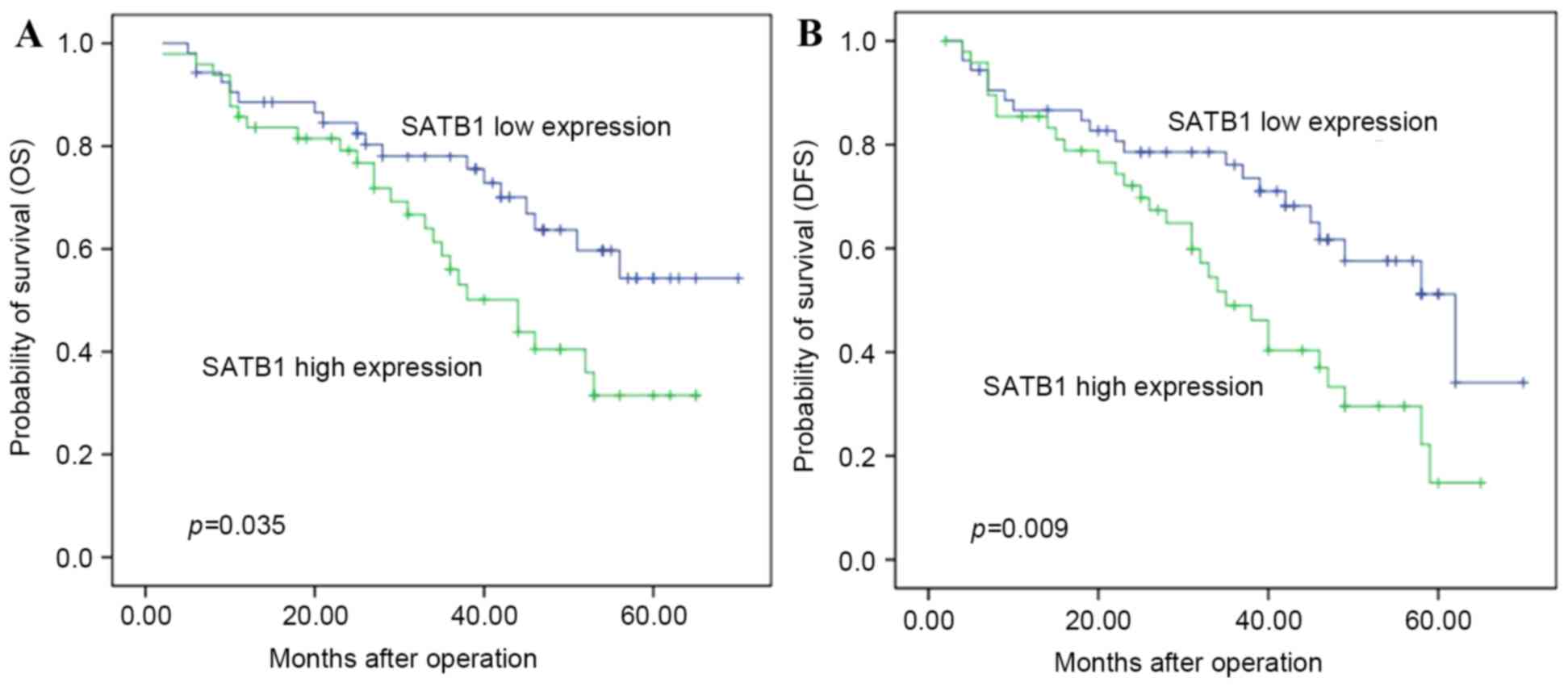
outcome (Table II). Patients with
high SATB1 mRNA expression had a shorter OS (median, 34.9 months)
compared with those with low SATB1 mRNA expression (median, 40.3
months) (P=0.035; Table III and
Fig. 1A). Multivariate analysis
suggested that SATB1 expression was an independent factor
associated with OS (RR=2.02; 95% CI: 1.10–3.68, P<0.05).
 | Table II.Univariate analyses of prognostic
factors in ESCC with respect to OS. |
Table II.
Univariate analyses of prognostic
factors in ESCC with respect to OS.
| Factor | RR | 95% CI | P-value |
|---|
| pT status
(T1/T2-4) | 1.358 | 0.472–3.908 | 0.571 |
| pN status
(N0/1-3) | 0.517 | 0.203–1.317 | 0.167 |
| pTNM stage
(I–II/III–IV) | 7.25 | 1.62–32.458 | 0.021a |
| Adjuvant
therapy | 1.134 | 0.668–1.924 | 0.642 |
| SATB1 expression:
low/high | 1.928 | 1.128–3.297 | 0.016a |
 | Table III.Cumulative proportion of OS at
different time points. |
Table III.
Cumulative proportion of OS at
different time points.
|
|
| Cumulative
proportion of OS |
|---|
|
|
|
|
|---|
| Group | No. of patients
that succumbed | 12 months (%) | 36 months (%) | 60 months (%) |
|---|
| SATB1 low
expression (n=53) | 18 | 88.6 | 63.7 | 54.3 |
| SATB1 high
expression (n=49) | 26 | 83.6 | 56.0 | 31.5 |
SATB1 mRNA expression is associated
with the relapse of ESCC
The duration of DFS of patients with ESCC with high
SATB1 mRNA expression (31.53±16.34 months) was significantly
shorter compared with that of patients with low SATB1 mRNA
expression (37.70±16.28 months, P=0.029) (Table IV and Fig.
1B).
 | Table IV.The cumulative proportion of DFS at
different time points. |
Table IV.
The cumulative proportion of DFS at
different time points.
|
|
| Cumulative
proportion of DFS |
|---|
|
|
|
|
|---|
| Group | N of relapse | 12 months (%) | 36 months (%) | 60 months (%) |
|---|
| SATB1 low
expression (n=53) | 20 | 86.6 | 76.1 | 51.2 |
| SATB1 high
expression (n=49) | 30 | 85.4 | 49.0 | 14.8 |
Discussion
Previous studies have indicated that SATB1 is
associated with prognosis in resected upper gastrointestinal tract
adenocarcinoma and other tumor forms (20,23,24). Data
from the present study revealed that patients with higher SATB1
mRNA expression had a higher rate of relapse. SATB1 expression was
detected in ESCC tumor tissues by measuring mRNA expression; high
SATB1 mRNA expression was associated with poor DFS and OS, as
determined by univariate analysis. Multivariate analysis revealed
that high SATB1 expression may be an independent factor for poor
prognosis in patients with ESCC following surgery. These results
were consistent with another previous report, in which SATB1
expression groups were divided using a semi-quantitative scoring
system for staining intensity and the percentage of positive
malignant cells (25). These results
verified the importance of the SATB1 gene in ESCC. The current
study defined the expression state according to the median value of
all ESCC tissues, which may exclude subjective bias. However, it is
challenging to obtain a ‘clean’ tumor sample without inflammatory
and stromal cells. Cong et al (25) reported that SATB1 expression measured
by immunohistochemistry could classify SATB1 expression in tumor
cells from inflammatory cells and stromal cells, whereas the
results obtained using semi-quantitative systems were not precise.
Furthermore, SATB1 expression was identified in inflammatory cells
but not in stromal cells (26). SATB1
integrates global epigenetic and transcriptional programs that
determine cellular phenotypes, differentiation, and the activity of
leukocyte subsets (27). In future
studies, more patients at respective pTNM stages will be recruited
to define the median level of SATB1 mRNA expression at different
stages, explore the expression of SATB1 expression between tumor
tissues and inflammatory cells, and investigate the role of SATB1
in the development and prognosis of ESCC.
The incidence of ESCC was different in different
regions of the world and the variation has changed over time
(2). This suggests that multiple risk
factors (genetic and environmental) contribute to the development
of ESCC (28–30). Morita et al (30) reported that cigarette smoking and
alcohol consumption exhibit synergistic effects on the development
of ESCC. Kasagi et al (31)
reviewed the clinicopathological characteristics of ESCC in
patients <50 years old and revealed that the incidence of ESCC
was associated with heavy exposure to smoking and/or drinking. Wu
et al (32) recruited 718
patients with ESCC in Taiwan and demonstrated that the habitual
drinking of alcohol was the strongest predictor for ESCC survival,
followed by areca chewing and smoking. In the current study,
smoking and drinking was associated with the prognosis of ESCC, but
had not with SATB1 mRNA expression, suggesting that other
mechanisms participate in ESCC development.
In conclusion, the present study identified that
high expression of SATB1 in ESCC was associated with a clinically
unfavorable prognosis independent of the patient's disease stage,
therefore SATB1 expression may be an independent factor of poor
prognosis in ESCC. Further studies are required to elucidate the
molecular mechanisms underlying the roles of SATB1 in the
progression of ESCC.
Acknowledgements
The present study was financially supported by
grants from the National Natural Science Foundation of China (grant
no. 81472808). The authors would like to thank Dr Nan Li (Sichuan
University, Chengdu, China) for language editing and
proofreading.
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang L, Parkin DM, Ferlay J, Li L and Chen
Y: Estimates of cancer incidence in China for 2000 and projections
for 2005. Cancer Epidemiol Biomarkers Prev. 14:243–250.
2005.PubMed/NCBI
|
|
3
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dickinson LA, Joh T, Kohwi Y and
Kohwi-Shigematsu T: A tissue-specific MAR/SAR DNA-binding protein
with unusual binding site recognition. Cell. 70:631–645. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Alvarez JD, Yasui DH, Niida H, Joh T, Loh
DY and Kohwi-Shigematsu T: The MAR-binding protein SATB1
orchestrates temporal and spatial expression of multiple genes
during T-cell development. Genes Dev. 14:521–535. 2000.PubMed/NCBI
|
|
6
|
Fessing MY, Mardaryev AN, Gdula MR, Sharov
AA, Sharova TY, Rapisarda V, Gordon KB, Smorodchenko AD,
Poterlowicz K, Ferone G, et al: p63 regulates Satb1 to control
tissue-specific chromatin remodeling during development of the
epidermis. J Cell Biol. 194:825–839. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kohwi-Shigematsu T, Poterlowicz K,
Ordinario E, Han HJ, Botchkarev VA and Kohwi Y: Genome organizing
function of SATB1 in tumor progression. Semin Cancer Biol.
23:72–79. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cai S, Han HJ and Kohwi-Shigematsu T:
Tissue-specific nuclear architecture and gene expression regulated
by SATB1. Nat Genet. 34:42–51. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao XD, Ji WY, Zhang W, He LX, Yang J,
Liang HJ and Wang LL: Overexpression of SATB1 in laryngeal squamous
cell carcinoma. ORL J Otorhinolaryngol Relat Spec. 72:1–5. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mokhtar NM, Ramzi NH, Yin-Ling W, Rose IM,
Hatta Mohd Dali AZ and Jamal R: Laser capture microdissection with
genome-wide expression profiling displayed gene expression
signatures in endometrioid endometrial cancer. Cancer Invest.
30:156–164. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tu W, Luo M, Wang Z, Yan W, Xia Y, Deng H,
He J, Han P and Tian D: Upregulation of SATB1 promotes tumor growth
and metastasis in liver cancer. Liver Int. 32:1064–1078. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Meng WJ, Yan H, Zhou B, Zhang W, Kong XH,
Wang R, Zhan L, Li Y, Zhou ZG and Sun XF: Correlation of SATB1
overexpression with the progression of human rectal cancer. Int J
Colorectal Dis. 27:143–150. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen H, Takahara M, Oba J, Xie L, Chiba T,
Takeuchi S, Tu Y, Nakahara T, Uchi H, Moroi Y and Furue M:
Clinicopathologic and prognostic significance of SATB1 in cutaneous
malignant melanoma. J Dermatol Sci. 64:39–44. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cheng C, Lu X, Wang G, Zheng L, Shu X, Zhu
S, Liu K, Wu K and Tong Q: Expression of SATB1 and heparanase in
gastric cancer and its relationship to clinicopathologic features.
APMIS. 118:855–863. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu X, Cheng C, Zhu S, Yang Y, Zheng L,
Wang G, Shu X, Wu K, Liu K and Tong Q: SATB1 is an independent
prognostic marker for gastric cancer in a Chinese population. Oncol
Rep. 24:981–987. 2010.PubMed/NCBI
|
|
16
|
Mao L, Yang C, Wang J, Li W, Wen R, Chen J
and Zheng J: SATB1 is overexpressed in metastatic prostate cancer
and promotes prostate cancer cell growth and invasion. J Transl
Med. 11:1112013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou LY, Liu F, Tong J, Chen QQ and Zhang
FW: Expression of special AT-rich sequence-binding protein mRNA and
its clinicopathological significance in non-small cell lung cancer.
Nan Fang Yi Ke Da Xue Xue Bao. 29:534–537. 2009.(In Chinese).
PubMed/NCBI
|
|
18
|
Selinger CI, Cooper WA, Al-Sohaily S,
Mladenova DN, Pangon L, Kennedy CW, McCaughan BC, Stirzaker C and
Kohonen-Corish MR: Loss of special AT-rich binding protein 1
expression is a marker of poor survival in lung cancer. J Thorac
Oncol. 6:1179–1189. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang S, Gao X, Ma Y, Jiang J, Dai Z, Yin
X, Min W, Hui W and Wang B: Expression and significance of SATB1 in
the development of breast cancer. Genet Mol Res. 14:3309–3317.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hedner C, Gaber A, Korkocic D, Nodin B,
Uhlén M, Kuteeva E, Johannesson H, Jirström K and Eberhard J: SATB1
is an independent prognostic factor in radically resected upper
gastrointestinal tract adenocarcinoma. Virchows Arch. 465:649–659.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rice TW, Blackstone EH and Rusch VW: 7th
Edition of the AJCC cancer staging manual: Esophagus and
esophagogastric junction. Ann Surg Oncol. 17:1721–1724. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wan F, Cheng C, Wang Z, Xiao X, Zeng H,
Xing S, Chen X, Wang J, Li S, Zhang Y, et al: SATB1 overexpression
regulates the development and progression in bladder cancer through
EMT. PLoS One. 10:e01175182015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Han HJ, Russo J, Kohwi Y and
Kohwi-Shigematsu T: SATB1 reprogrammes gene expression to promote
breast tumour growth and metastasis. Nature. 452:187–193. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cong QX, Zhang H, Sun SX, Li HF, Wang Y
and Jian S: Pilot study special AT-rich sequence-binding protein 1
investigating as a potential biomarker for esophageal squamous cell
carcinoma. Dis Esophagus. 29:621–626. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tesone AJ, Rutkowski MR, Brencicova E,
Svoronos N, Perales-Puchalt A, Stephen TL, Allegrezza MJ, Payne KK,
Nguyen JM, Wickramasinghe J, et al: Satb1 overexpression drives
tumor-promoting activities in cancer-associated dendritic cells.
Cell Rep. 14:1774–1786. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Borghesi L: Hematopoiesis in steady-state
versus stress: Self-renewal, lineage fate choice, and the
conversion of danger signals into cytokine signals in hematopoietic
stem cells. J Immunol. 193:2053–2058. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Song Y, Wang Y, Xu L, Ma J, Chen E, Zang
R, Jia W, Tao X and Hu L: A genetic variant in CHRNB3-CHRNA6
increases risk of esophageal squamous cell carcinoma in Chinese
populations. Carcinogenesis. 36:538–542. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen J, Kwong DL, Cao T, Hu Q, Zhang L,
Ming X, Chen J, Fu L and Guan X: Esophageal squamous cell carcinoma
(ESCC): Advance in genomics and molecular genetics. Dis Esophagus.
28:84–89. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Morita M, Kumashiro R, Kubo N, Nakashima
Y, Yoshida R, Yoshinaga K, Saeki H, Emi Y, Kakeji Y, Sakaguchi Y,
et al: Alcohol drinking, cigarette smoking, and the development of
squamous cell carcinoma of the esophagus: Epidemiology, clinical
findings, and prevention. Int J Clin Oncol. 15:126–134. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kasagi Y, Morita M, Otsu H, Kawano H, Ando
K, Hiyoshi Y, Ito S, Miyamoto Y, Saeki H, Oki E and Maehara Y:
Clinicopathological characteristics of esophageal squamous cell
carcinoma in patients younger than 50 years. Ann Surg Oncol.
22:311–315. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu IC, Wu CC, Lu CY, Hsu WH, Wu MC, Lee
JY, Chou SH, Lee JM, Chou YP, Wu DC and Wu MT: Substance use
(alcohol, areca nut and cigarette) is associated with poor
prognosis of esophageal squamous cell carcinoma. PLoS One.
8:e558342013. View Article : Google Scholar : PubMed/NCBI
|