Introduction
Liver cancer refers primarily to hepatocellular
carcinomas (HCC), which accounts for up to 90 percent cases
(1) and is the most frequent primary
malignancy of liver (2). Despite
progress having been achieved in the diagnosis and treatment of
hepatoma, it remains among the tumors with the poorest prognosis
(3). Previous studies have suggested
that alterations in genomics and epigenetics are involved with
tumor growth and metastasis, however, the specific mechanisms
remain to be elucidated (4). Genomic
and epigenetic changes, particularly the accumulation of mutations,
affect the normal growth and differentiation of hepatic cells, and
thus lead to hepatoma (5). Mutations
often break the expression balance of oncogenes and
oncogene-suppress genes (6).
DEPDC7 was first identified by Gawin et al in
1999 (7). However, the function of
DEPDC7 remains largely unknown. The protein consists of 511 amino
acids, with two potentially functional domains consisting of the
Dishevelled, EGL-10 and Pleckstrin (DEP) domain and the GTPase
activating protein (GAP) domain. The DEP domain is a globular
protein domain of ~90 amino acids that is present in numerous
signaling proteins involved in G-protein signaling pathways and Wnt
signaling pathways (8–10). Based on conserved domain analysis and
literature mining, it is hypothesized that the proteins containing
DEP domain maybe important in cell signal transduction and numerous
other biological processes (11). It
has previously been shown that DEPDC7 is highly and specifically
expressed in normal liver tissue and thus belongs to the class of
genes of liver-selective cell communication (LSCC) (12). It was identified that the DEPDC7 gene
(NM_001077242.1) is located at chromosome 11p13, where deletion
mutations were found in 31.6% of HCC cells (13). It was therefore considered that
chromosome 11p13 may contain genes, such as DEPDC7, that inhibit
tumor growth in the liver.
In the present study, the gene expression of DEPDC7
was examined in several hepatoma cell lines, and this was compared
with the expression in normal hepatic cells and liver cancer
tissues. Wild-type DEPDC7 was overexpressed in two hepatoma cell
lines, and cell proliferation, cell cycles progression and cell
migration and invasion of these cells were investigated.
Materials and methods
Cell culture
Human 293T cells, human hepatic cell L-02, human
hepatoma SMMC-7721, Huh-7, SK-Hep-1 and HepG2 cell lines were
cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml
penicillin and 100 mg/ml streptomycin. All cells were maintained at
37°C in a humidified atmosphere containing 5% CO2. Cells
were used in studies when they reach 75% confluence.
Plasmid construction
The expression vector pLV-EF1α-MCS-IRES-Puro and the
helper plasmids pMDLg, pRSV-REV and pVSV-G were kindly provided by
Professor Jiahuai Han (School of Life Sciences, Xiamen University,
Xiamen, China). The DEPDC7 was amplified using a human cDNA derived
from the L-02 liver cells as a template and the following primers:
BamHI-F-depdc7, 5′AGA GAA TTC GGA TCC ATG GCC ACC GTG
CAG3′; SmaI-R-DEPDC7, 5′TGG CTC GAG CCC GGGTCA GTC
TCA AAA TGC TCA 3′. The BamHI and SmaI sites are
underlined. The ATG translation initiation site and the TCA
termination site of DEPDC7 are shown in bold. Subsequently, the
amplified product was cloned into the BamHI and SmaI
sites of the vector pLV-EF1α-MCS-IRES-Puro using the Exonuclease
III-assisted ligase-free cloning method as previously described
(14). All clonal products were
identified by BamHI and SmaI and verified by DNA
sequencing (Sangon Biotech Co., Ltd., Shanghai, China). The empty
vector was used as a negative control for infection and subsequent
detection.
Virus packaging and infection
Both recombinant lentiviruses and negative control
were packaged in 293T cells in the presence of helper plasmids
(pMDLg, pRSV-REV and pVSV-G) using TurboFect Infection Reagent
(Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. The infected cells were cultured at 37°C for 48 h and the
virus was then collected for infection. Huh-7 and SK-Hep-1 were
seeded in a 12-well cell culture plate, grown overnight at 37°C to
achieve 30–40% confluence, and then infected with the virus in
fresh medium containing 10 µg/ml polybrene. After 12 h of
incubation, the culture medium was replaced with fresh medium and
the infected cells were harvested at 72 h after infection and then
used in subsequent experiments. All virus packaging and infection
experiments were conducted in triplicate.
RNA isolation and
reverse-transcription quantitative PCR (RT-qPCR)
Total RNA was isolated from the collected cells
using TRIzol reagent (TakaraBio, Inc., Otsu, Japan) and
reverse-transcribed into cDNA using PrimeScript™ RT reagent Kit
(TakaraBio, Inc.), according to the manufacturer's protocol. RNA
expression levels were normalized to an internal control, β-actin.
qPCR was conducted as follows: 95°C for 10 min; and 95°C for 15 sec
and 60°C for 1 min for 40 cycles. qPCR were performed on the Step
One™ Real-Time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.) using SYBER-Green qPCR Supermix (Roche
Diagnostics, Basel, Switzerland). The relative expression level of
DEPDC7 was normalized to that of β-actin by
2−ΔΔCq cycle threshold method (15). Following primers were used: DEPDC7
forward, 5′-ACCTTCCACTTCTTGACTCCTTAC-3′ and reverse,
5′-CGAGAGCCACTCATCTTCCTG-3′; β-Actin forward,
5′-CGTGCGTGACATTAAGGAGAAG-3′ and reverse,
5′-GGAAGGAAGGCTGGAAGAGTG-3′.
SDS-PAGE and Western blot analysis
detect DEPDC7
Culture plates were rinsed with ice-chilled PBS. The
cells were collected using a plastic cell scraper and lysed in
Lysis buffer (20 mM Tris/HCl pH 7.5; 120 mM NaCl; 1 mM EDTA; 1 mM
EGTA; 1% Triton X-100; 2.5 mM Sodium pyrophosphate; 1 mM
β-Glycerophosphate; 1 mM Na3VO4) in the
presence of protease inhibitor phenylmethylsulfonyl fluoride.
Protein were dissolved in 5X sample buffer (0.25 M Tris/HCl pH 6.8;
10% (w/v) SDS; 50% glycerol; 0.5% (w/v) bromophenol blue) and were
subsequently denatured at 100°C for 10 min. Subsequently, 30 µg
protein were loaded onto 10% Tris-Acrylamide gels and
electrotransferred onto PVDF membranes. The membranes were blocked
in 10% skimmed milk for 2 h at room temperature and incubated with
primary antibodies against β-actin at a dilution of 1:8,000
(catalog no. 3700; Cell Signaling Technology, Inc., Danvers, MA,
USA) and DEPDC7 at a dilution of 1:1,000 (catalog no. ab174659,
Abcam, Cambridge, UK) at 4°C overnight. Subsequent to washing, the
membranes were incubated in horse radish peroxidase
(HRP)-conjugated goat anti-mouse/anti-rabbit IgG secondary antibody
at a dilution of 1:10,000 (catalog nos. 7076-mouse and 7074-rabbit;
Cell Signaling Technology, Inc.) at room temperature for 2 h, with
three washes after incubations. Signals were detected using
enhanced chemiluminescence (WBKLS0100; EMD Millipore, Billerica,
MA, USA) and captured by the ImageQuant LAS 4000 mini Imaging
System (GE Healthcare, Chicago, IL, USA).
Immunohistochemical analysis and
assessment
Tissues
A total of 48 hepatoma tissues were used and they
were collected between April 2012 and July 2014. The present study
was approved by the Ethics Committee of Fuzhou Dongfang Hospital
(Fuzhou, China), and the experiments were conducted in accordance
with the Declaration of Helsinki and Good Clinical Practice.
Informed consent was obtained from all patients prior to the use of
tissues.
DEPDC7 immunostaining
DEPDC7 immunohistochemical staining of hepatoma
tissues was performed using EliVision™ plus Polymer HRP
IHC kit (Fuzhou Maixin Biotech Co., Ltd., Fuzhou, China). Briefly,
Formalin-fixed paraffin-embedded sections were deparaffinized in
xylene and dehydrated with ethanol. The tissues were heated in a
microwave at 700 W for 5 min in 10 mM citrate solution, for antigen
retrieval. Endogenous peroxidase activity was blocked by 3%
hydrogen peroxide in methanol for 10 min at room temperature. The
sections were then incubated overnight at 4°C with anti-human
DEPDC7 (dilution, 1:30; catalog no. HPA015800; Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) in PBS containing 1% bovine serum
albumin. The slides were rinsed and incubated at room temperature
with HRP-conjugate anti-mouse IgG (dilution, 1:3,000; catalog no.
W4021; Promega Corporation; Madison, WI, USA) for 30 min. The
immunoreactivity was visualized with diaminobenzidine (DAB Kit,
MAX-001; Fuzhou Maixin Biotech Co., Ltd.). The sections were
counter stained with hematoxylin at room temperature for 2 min,
dehydrated and evaluated under OLYMPUS BX51 light microscope
(Olympus, Tokyo, Japan). For quantification, the intensity of
DEPDC7 immunoreactivity was scored ’±’, ’+’, ’++’ or ’+++’, on the
basis of the brown-yellow staining area, where ’±’ represented no
distinct brown granules and ’+++’ represented the highest
strength.
Ki-67 immunostaining
Ki-67 immunostaining was performed using the
aforementioned procedure, using mouse monoclonal antibodies against
Ki-67 (Ready-to-Use; catalog no. MAB-0672, Fuzhou Maixin Biotech
Co., Ltd.). Staining was classified as positive if granular
brown-yellow-colored immunoreactivity was present in the nuclei.
For quantification, the number of positive cells of total cells was
counted.
MTT assay and colony formation assay
Cell proliferation was measured by MTT
(Sigma-Aldrich; Merck KGaA) and colony formation assay.
MTT assay
Both of infected and negative control (NC) hepatoma
cells were seeded onto 96-well plates at a density of
2×103 cells/well and incubated at 37°C in a 5%
CO2 atmosphere for 1–8 days, as previously described
(16). For every 24 h, cells were
incubated with 20 µl of MTT solution at a final concentration of
0.5 mg/ml MTT for 4 h. After removing supernatant, 150 µl of
dimethyl sulfoxide was added to solubilize the formazan salt. After
10 min, the optical density was measure at 570 nm using a
microplate reader (BioTekChina, Beijing, China).
Colony formation assay
In total, 200 cells were plated into each well of
6-well plate and cultured at 37°C for 2 weeks. The cell colonies
were stained with crystal violet for 15 min after fixation in
methanol at room temperature. Images of the colonies were captured
and the number of colonies in each well was counted, as previously
described (17).
Cell cycle assay
Cultured cells were harvested by trypsinization and
washed three times in PBS (pH 7.4), followed by fixation with
precooled 70% ethanol overnight at 4°C. Subsequent to
centrifugation (1,000 × g, 4°C, 5 min) and re-suspension in PBS,
the cells were incubated with 50 µg/ml propidium iodide
(Sigma-Aldrich; Merck KGaA) and 10 µg/ml RNaseA (Sigma-Aldrich;
Merck KGaA) for 30 min in the dark at 4°C. The cell cycle was
assessed by flow cytometry (FACSVerse; BD Biosciences, Franklin
Lakes, NJ, USA) with DRAQ5™ Fluorescent Probe
(Invitrogen; Thermo Fisher Scientific, Inc.) staining. The data
were analyzed using ModFit LT3.0 software (BD Biosciences).
Cell migration and invasion assay
Migration assays were performed using a 24-well
Transwell chamber system (Costar 3422; Corning Incorporated,
Corning, NY, USA). Cells were seeded in the upper chambers at
density of 1×104 cells/ml in 0.1 ml serum-free DMEM. The
lower chambers were filled with 0.8 ml DMEM with 10% fetal bovine
serum. Following incubation for 24 h at 37°C in a 5% CO2
atmosphere, the cells that migrated through the membrane were fixed
with methanol for 15 min, stained with crystal violet stain for 10
min at room temperature and counted using a microscope at ×200
magnification.
Invasion assays were performed using a 24-well
Transwell chamber coated with Matrigel (30 µl per filter; BD
Biosciences), according to the manufacturer's protocol. Cells were
seeded on the top of Matrigel-coated invasion chambers in
serum-free DMEM (2×104 cells/well) and DMEM containing
10% fetal bovine serum was added to the lower chambers. Subsequent
to incubation for 24 h at 37°C in a 5% CO2 atmosphere,
the cells that invaded through the membrane were fixed with
methanol for 15 min, stained with crystal violet stain for 10 min
at room temperature and counted using a microscope at ×200
magnification.
Statistical analysis
The band intensity was quantified using GeneTool
4.01 software (Syngene Inc., Frederick, MD, USA). Statistical
analysis was performed using the two-tailed Student's t-test, and
independent Student's t-test was used for comparisons of two
groups. The significances of the differences between the control
and each experimental group was evaluated using one-way analysis of
variance and the Dunnett's post hoc test. Data are expressed as the
mean ± standard deviation. P<0.05 was considered to indicate a
statistically significant difference.
Results
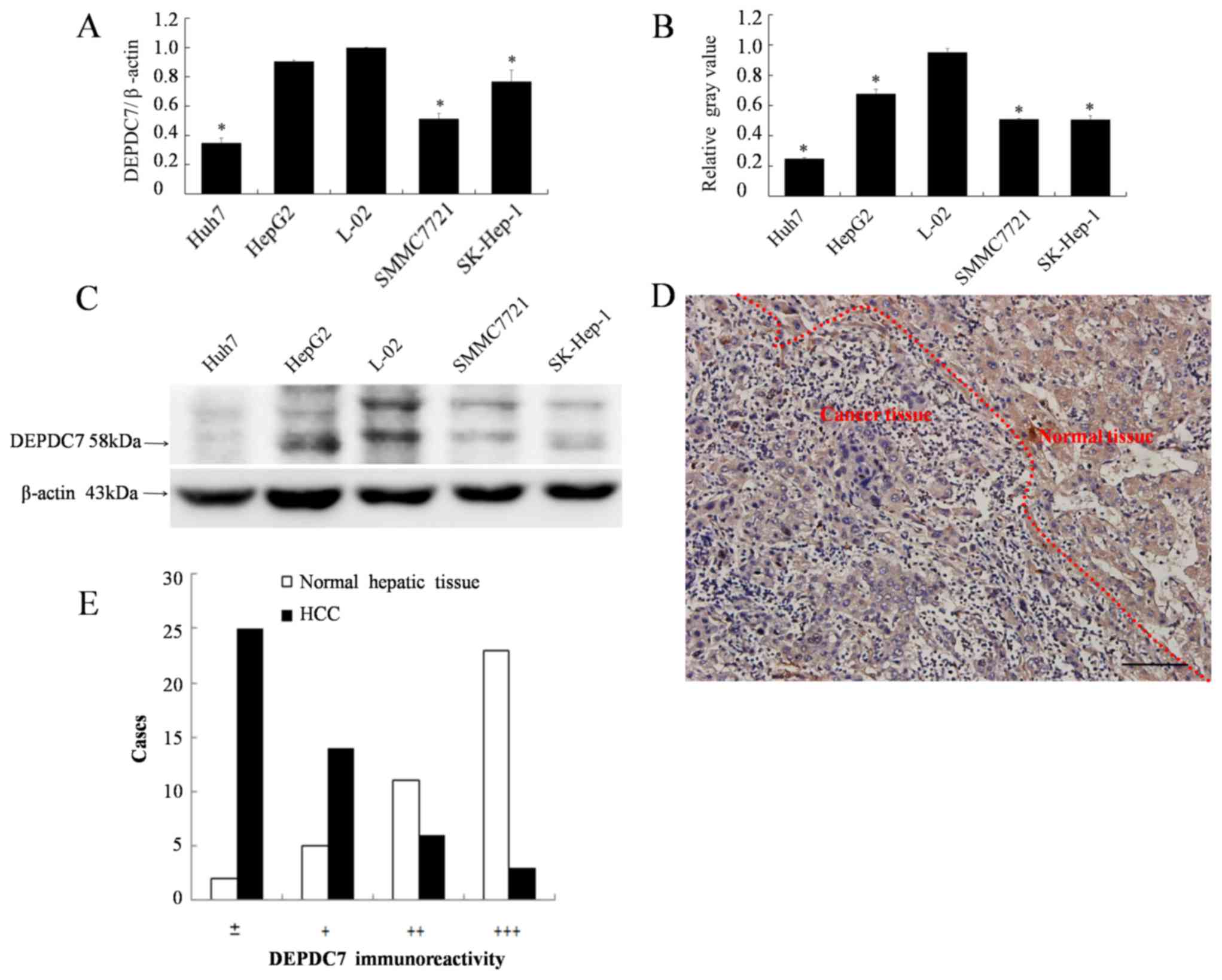
DEPDC7 is downregulated in hepatoma
cells
Since we have previously found that DEPDC7 is highly
and specifically expressed in normal liver tissue (12), it was hypothesized whether the
expression of DEPDC7 may be up or downregulated in hepatoma cells.
The expression levels of DEPDC7 in all four hepatoma cell lines
were significantly decreased compared with those in the normal
hepatic L-02 cell line (P<0.05; Fig.
1A-C). The expression of DEPDC7 in hepatoma tissues was also
examined. As demonstrated in Fig. 1D,
the granular brown-yellow-colored immunoreactivity was evidently
weaker in carcinoma cells compared with the surrounding normal
hepatic cells. This observation was confirmed in the 41 cases the
liver sections obtained hepatoma patients (Fig. 1E). Thus, DEPDC7 is downregulated in
the hepatoma cells. Since the Huh-7 and SK-Hep-1 cell lines had the
lowest DEPDC7 expression levels, they were used for further
functional examination.
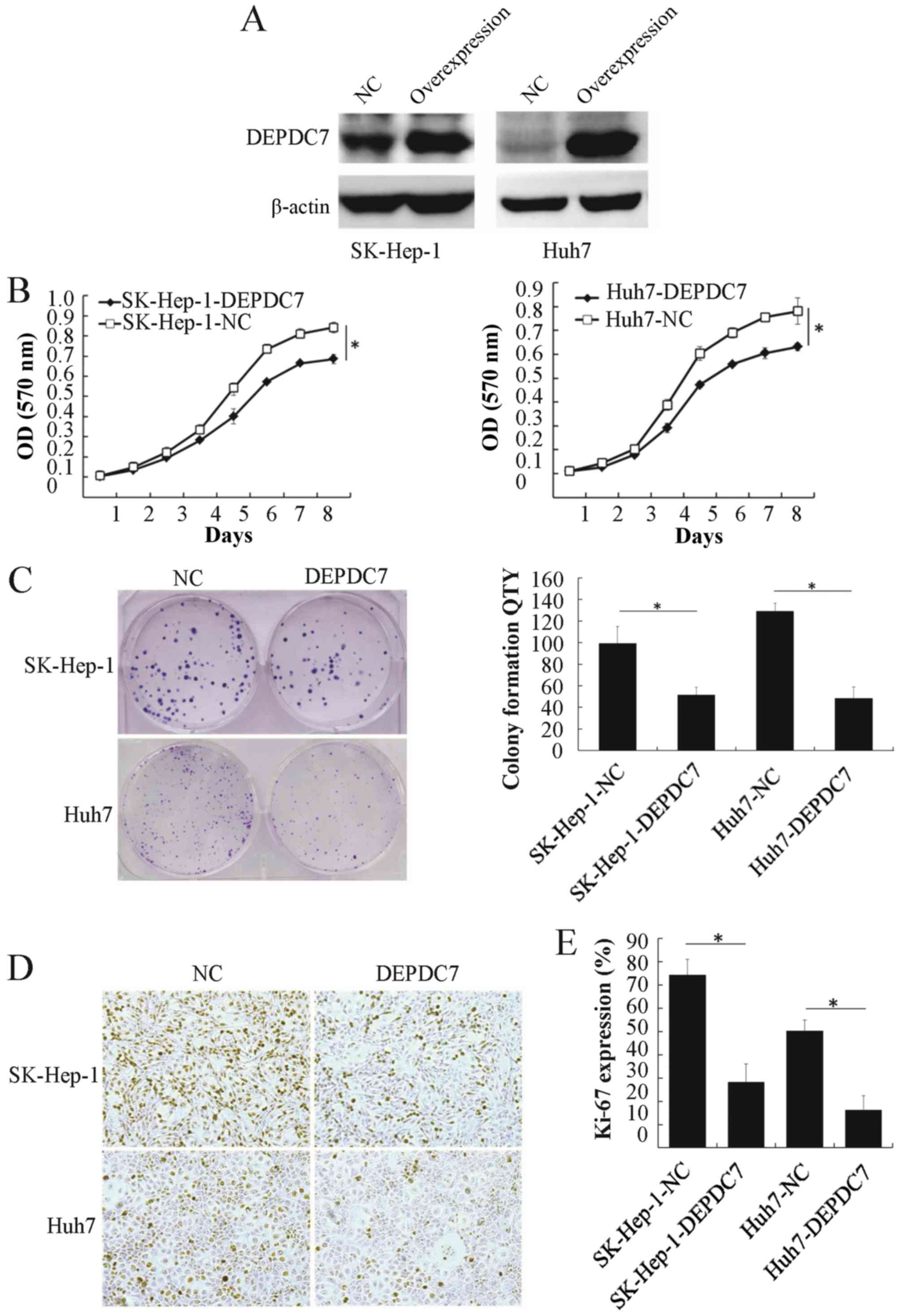
Overexpression of DEPDC7 inhibits the
proliferation of hepatoma cells
To gain insight into the potential function of
DEPDC7, wild-type DEPDC7 was cloned into the pLV-EF1α-MCS-IRES-Puro
vector (plv-DEPDC7) and infected into SK-Hep-1 and Huh-7 cell lines
(SK-Hep-1-DEPDC7 and Huh-7-DEPDC7). Cells infected with empty
vector acted as a negative control (SK-Hep-1-NC and Huh-7-NC). The
DEPDC7 was found to be significantly overexpressed in these two
cell lines compared with NC groups (Fig.
2A). The proliferation of these cells was then examined. After
96 h of cell seeding, the DEPDC7-transfected cells were
significantly decreased in number compared with the NC groups
(Fig. 2B and C), indicating that
overexpression of DEPDC7 leads to inhibition of cell proliferation
in SK-Hep-1 and Huh-7 cells. The colony formation assay showed that
the DEPDC7-overexpressing cells formed fewer colonies than the NC
groups (P<0.05; Fig. 2D and E),
confirming that growth is inhibited in these two hepatoma cell
lines when DEPDC7 is overexpressed. Since Ki-67 is a cell
proliferation marker (18), Ki-67
expression was examined by immunohistochemistry in SK-Hep-1-DEPDC7,
Huh-7-DEPDC7 cells and NC cells. The number of Ki-67-positive cells
was significantly decreased in the DEPDC7-overexpression groups
compared with the NC groups (Fig. 2D and
E). These data demonstrate that DEPDC7 inhibits the development
of hepatoma through accelerated cell proliferation.
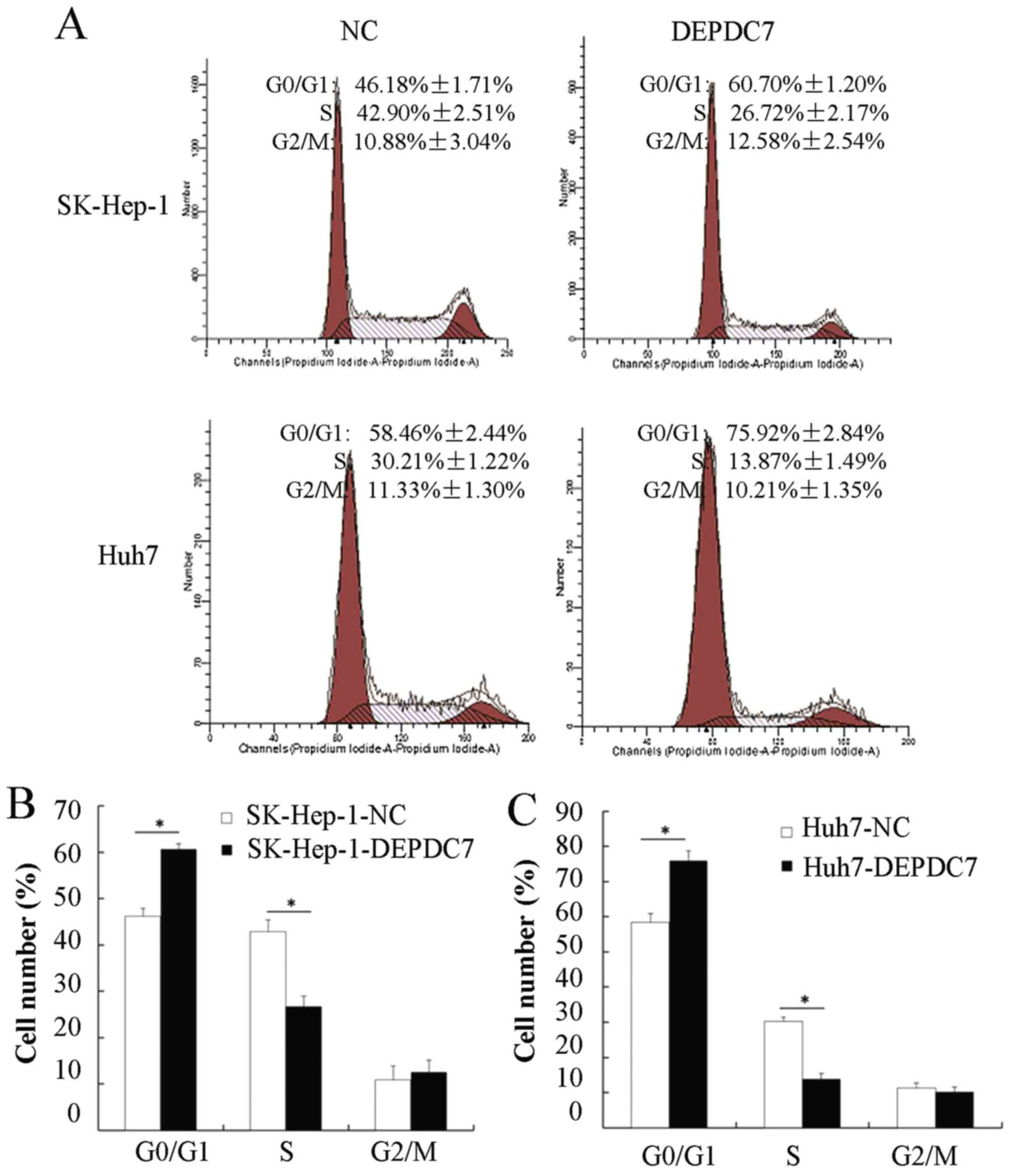
Overexpression of DEPDC7 blocks the
cell cycle transition from the G0/G1 to the S phase
To further investigate the mechanisms of DEPDC7
involved in regulating hepatoma cell proliferation, flow cytometry
was used to analyze cell cycles distribution of these cells. As
shown in Fig. 3A, the proportion of
cells in the G0/G1 phase in both SK-Hep-1-DEPDC7 and Huh7-DEPDC7
cells was increased compared with NC cells, whereas the proportion
of cells in the S phase was decreased in SK-Hep-1-DEPDC7 and
Huh7-DEPDC7 cells compared with NC cells. The results were
consistent in three independent experiments (Fig. 3B and C), indicating that the
overexpression of DEPDC7 induced cell cycle arrest in hepatoma
cells at the G1/S phase.
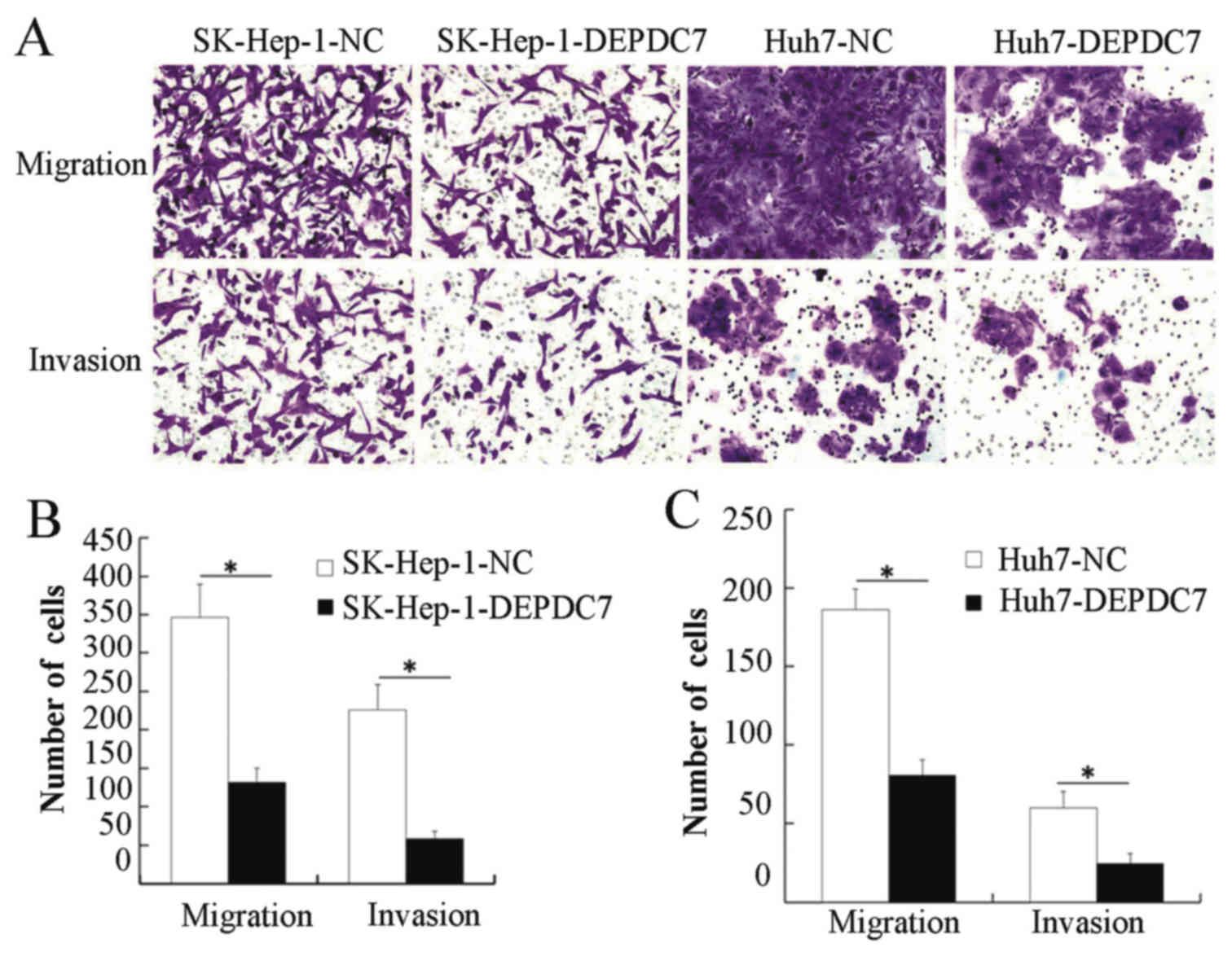
Overexpression of DEPDC7 inhibits the
migration and invasion of hepatoma cells
Migration and invasion assays were then performed,
to determine whether DEPDC7 has a functional significance on
hepatoma cells. SK-Hep-1-DEPDC7, Huh7-DEPDC7 cells and NC cells
were grown in Transwell plates, either with or without Matrigel.
When the cells that passed through the membrane or Matrigel-coated
membrane were stained and counted, SK-Hep-1-DEPDC7 and Huh7-DEPDC7
showed significantly decreased invasion and migration compared with
NC cells (Fig. 4), indicating that
overexpression of DEPDC7 led to inhibition of the cell motility and
cell invasion of hepatoma cells.
Discussion
The present authors have previously researched
liver-specific genes and looked for new targets in the treatment of
hepatoma. A previous study indicates that DEPDC7 is highly and
specifically expressed in normal liver tissue and thus belongs to
the class of genes of LSCC (12). It
was shown that expression of DEPDC7 is suppressed in all hepatoma
cell lines, both at mRNA and protein levels, suggesting the
regulation is transcriptional. Furthermore, DEPDC7 immunoreactivity
is frequently decreased in hepatoma tissues compared with normal
surrounding tissues. These results suggest that DEPDC7 may be
involved in the development of hepatoma cells. In a genome-wide
molecular profiles research using the Oncomine database, Wurmbach
et al (19) also found that
DEPDC7 expression is decreased in hepatoma compared with normal
liver tissues. It is notable that the expression levels of DEPDC7
vary between hepatoma cell lines and between hepatoma tissues.
Further study may investigate the association between DEPDC7 levels
and the malignancy of tumors.
Although DEPDC7 has been found for over a decade,
the function of it has remained largely unknown. In subsequent
functional studies, the role of DEPDC7 gene overexpressed in
hepatoma cells was explored, and the results of proliferation and
flow cytometry assays illustrated that DEPDC7 overexpression
markedly repressed proliferation in hepatoma cell lines by blocking
the cell cycle transition between G0/G1 and S phase, It was also
found that the proliferation index reflected by Ki-67 in the
DEPDC7-overexpressing hepatocellular carcinoma cells was
significantly decreased compared with that in the control
hepatocellular carcinoma cells. Additionally, it was further shown
that overexpression of DEPDC7 significantly inhibited hepatoma cell
migration and invasion. The present results are consistent with our
previous findings that the cell proliferation and colony formation
are promoted in DEPDC7-knockdown HepG2 cells (20), indicating that DEPDC7 is involved the
regulation of cell growth, cell cycle transition and cell mobility
and invasion. Although HepG2 was reported to be recognized as a
hepatoblastoma rather than a HCC cell line (21), this misidentification did not affect
our conclusion for the present study focusing on hepatoma.
In conclusion, the present study suggests that
DEPDC7 is a tumor suppress gene. The expression of DEPDC7 is
downregulated in hepatoma cells, leading to cell proliferation,
cell cycle progression, and migration and invasion. This finding
will encourage the identification of the molecular mechanism of
DEPDC7-associated inhibition of cell proliferation and motility,
and may also encourage the targeting of DEPDC7 as a treatment of
hepatoma.
Acknowledgements
The authors thank Dr Shuo Lin (Biozentrum,
University of Basel, Switzerland) for his critical reading and
helpful modification of the manuscript. This study was supported by
the Foundation for Young Talents of Fujian Province, China (grant
no. 2008F3045).
References
|
1
|
Tamai T, Hayato S, Hojo S, Suzuki T,
Okusaka T, Ikeda K and Kumada H: Dose finding of lenvatinib in
subjects with advanced hepatocellular carcinoma based on population
pharmacokinetic and exposure-response analyses. J Clin Pharmacol.
57:1138–1147. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang Q, Zhao S, Pang X and Chi B:
MicroRNA-381 suppresses cell growth and invasion by targeting the
liver receptor homolog-1 in hepatocellular carcinoma. Oncol Rep.
35:1831–1840. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Simoneau E, Hassanain M, Madkhali A,
Salman A, Nudo CG, Chaudhury P and Metrakos P:
(18)F-Fluorodeoxyglucose positron-emission tomography could have a
prognostic role in patients with advanced hepatocellular carcinoma.
Curr Oncol. 21:e551–e556. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Patel A and Sun W: Molecular targeted
therapy in hepatocellular carcinoma: From biology to clinical
practice and future. Curr Treat Options Oncol. 15:380–394. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chappell G, Kutanzi K, Uehara T, Tryndyak
V, Hong HH, Hoenerhoff M, Beland FA, Rusyn I and Pogribny IP:
Genetic and epigenetic changes in fibrosis-associated
hepatocarcinogenesis in mice. Int J Cancer. 134:2778–2788. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Martincorena I and Campbell PJ: Somatic
mutation in cancer and normal cells. Science. 349:1483–1489. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gawin B, Niederfuhr A, Schumacher N,
Hummerich H, Little PF and Gessler M: A 7.5 Mb sequence-ready PAC
contig and gene expression map of human chromosome 11p13-p14.1.
Genome Res. 9:1074–1086. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu W and He X: DEEP insights through the
DEP domain. Structure. 18:1223–1225. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Consonni SV, Maurice MM and Bos JL: DEP
domains: Structurally similar but functionally different. Nat Rev
Mol Cell Biol. 15:357–362. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tauriello DV, Jordens I, Kirchner K,
Slootstra JW, Kruitwagen T, Bouwman BA, Noutsou M, Rudiger SG,
Schwamborn K, Schambony A, et al: Wnt/beta-catenin signaling
requires interaction of the dishevelled DEP domain and C terminus
with a discontinuous motif in frizzled. Proc Natl Acad Sci USA.
109:pp. E812–E820. 2012, View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang XR, Yang H and Liao ZJ: Structure and
function of the DEP domain. Chemistry of Life. 35:264–271.
2015.
|
|
12
|
Liao ZJ, Ma WL, Liang S, Meng W, Shang T
and Zheng WL: Transcriptional regulation of genes involved in
liver-selective cell communication. Nan Fang Yi Ke Da Xue Xue Bao.
28:1582–1585. 2008.PubMed/NCBI
|
|
13
|
Kahng YS, Lee YS, Kim BK, Park WS, Lee JY
and Kang CS: Loss of heterozygosity of chromosome 8p and 11p in the
dysplastic nodule and hepatocellular carcinoma. J Gastroenterol
Hepatol. 18:430–436. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li L, Chen W, Liang Y, Ma H, Li W, Zhou Z,
Li J, Ding Y, Ren J, Lin J, et al: The Gbetagamma-Src signaling
pathway regulates TNF-induced necroptosis via control of necrosome
translocation. Cell Res. 24:417–432. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Meng QC, Wang HC, Song ZL, Shan ZZ, Yuan
Z, Zheng Q and Huang XY: Overexpression of NDC80 is correlated with
prognosis of pancreatic cancer and regulates cell proliferation. Am
J Cancer Res. 5:1730–1740. 2015.PubMed/NCBI
|
|
17
|
Dai J, Wu W, Zhou J, Gao K, Hu G, Lin C,
Zhang Y and Li X: Effect of antisense microRNA targeting survivin
on rectal cancer HRC-9698 cells and its mechanism. Int J Clin Exp
Pathol. 8:6057–6063. 2015.PubMed/NCBI
|
|
18
|
Su H, Ma C, Liu J, Li N, Gao M, Huang A,
Wang X, Huang W and Huang X: Downregulation of nuclear receptor FXR
is associated with multiple malignant clinicopathological
characteristics in human hepatocellular carcinoma. Am J Physiol
Gastrointest Liver Physiol. 303:G1245–G1253. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wurmbach E, Chen YB, Khitrov G, Zhang W,
Roayaie S, Schwartz M, Fiel I, Thung S, Mazzaferro V, Bruix J, et
al: Genome-wide molecular profiles of HCV-induced dysplasia and
hepatocellular carcinoma. Hepatology. 45:938–947. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liao ZJ, Wang XR, Yang H and Zeng YT:
Effects of shRNA-mediated knockdown of DEPDC7 on proliferation,
migration and invasion of human hepatoma cell HepG2. Chin J Cell
Biol. 37:977–983. 2015.
|
|
21
|
Lopez-Terrada D, Cheung SW, Finegold MJ
and Knowles BB: Hep G2 is a hepatoblastoma-derived cell line. Hum
Pathol. 40:1512–1515. 2009. View Article : Google Scholar : PubMed/NCBI
|