Introduction
Colorectal cancer (CRC) is the third most frequently
occurring cancer, and remains the second most common cause of
cancer-associated mortality worldwide (1,2). Even with
the use of standard or enhanced treatments, a relatively high
proportion of patients develop drug resistance, consequently
suffering recurrence and relapse (3).
In recent years, several potential therapeutic targets have been
used in clinical practice; however, only a small subset of patients
with CRC are able to benefit from undergoing treatment with drugs
against such targets (4). This
situation highlights the importance of identifying novel
therapeutic targets that may be exploited for CRC treatment.
Although the identification of novel targets is
attractive for cancer treatment, the processes by which this
identification is achieved are often tedious and have a high
failure rate. Recently, a ‘repurposing’ strategy has been proposed,
in which established non-cancer drug targets are evaluated for
anticancer activity (5). This
strategy provides an opportunity to rapidly develop and clinically
trial novel therapeutic strategies against cancer. For example,
using the gene expression data from the Oncomine database, Rhodes
et al (6) identified that an
anti-hypertensive drug target, angiotensin II receptor type I
(AGTR1), was overexpressed in a subset of breast cancer tissues,
which demonstrated sensitivity to the AGTR1 antagonist
losartan.
In the present study, using a similar strategy, the
different expression profiles of approved or candidate drug-target
genes between CRC and normal colorectal tissues were evaluated.
Phosphoserine aminotransferase 1 (PSAT1), a serine
biosynthesis-related target gene, was identified to be specifically
and reproducibly overexpressed in CRC tumors, and its expression
was associated with resistance to irinotecan, 5-fluorouracil and
leucovorin (FOLFIRI) chemotherapy. Subsequently, it was
experimentally validated that PSAT1 is significantly associated
with a lack of response to chemotherapy and poor clinical outcomes
in an independent cohort of CRC samples.
Materials and methods
In silico comparison of mRNA
expression profiles of food and drug administration (FDA)-approved
and literature-defined drug-target genes
Initially, mining of published microarray studies on
CRC mRNA expression was performed using the Oncomine tool
(www.oncomine.org) as described previously
(7). A total of 6 CRC mRNA expression
datasets deposited in the Oncomine database were included in the
systematic comparative analysis, according to the following
filtering criteria available within the database: i) CRC
pathological types; ii) clinical specimens; and iii) primary CRC
and normal colorectal samples included. The 6 datasets that met
these criteria were analyzed further. Information on these 6
datasets is summarized in Table I
(8–13). In addition to the common clinical and
pathological characteristics (including tumor size, lymph node
metastasis and distant metastasis) included in each dataset,
chemotherapy response information was available in the Graudens
colon dataset, and survival data were available in The Cancer
Genome Atlas (TCGA) CRC dataset.
 | Table I.Colorectal cancer mRNA expression
datasets used in the present study. |
Table I.
Colorectal cancer mRNA expression
datasets used in the present study.
| Oncomine name | GEO ID | Platform | Cases, n | (Refs.) |
|---|
| Gaedcke
colorectal | GSE20842 | Agilent Human Genome
44K | 130 | (8) |
| Graudens colon | GSE3964 | 11K_VJF-ARRAY | 60 | (9) |
| Hong colorectal | GSE9348 | Human Genome U133
Plus 2.0 Array | 82 | (10) |
| Kaiser colon | GSE5206 | Human Genome U133
Plus 2.0 Array | 105 | (11) |
| Ki colon | GSE6988 | Human 17K
cDNA-GeneTrack | 123 | (12) |
| TCGA colorectal | – | Agilent 244K Custom
Gene Array Illumina RNA-Seq | 237 | (13) |
Subsequently, an in silico analysis was
performed to compare the differentially expressed drug-target genes
(2-fold variation; P<0.0001) between CRC and normal colorectal
tissues across the 6 independent CRC datasets, implementing the
‘Concepts: Drugbank Targets-FDA approved-Literature-defined’ and
‘Analysis Type: Cancer vs. Normal Analysis’ functions available in
the Oncomine database (7). Rank is
used to compare the over- or underexpressed genes using the
Oncomine tools as described previously (7). To determine the expression pattern of
PSAT1, CRC datasets were further filtered by ‘Gene: PSAT1’. The
expression of PSAT1 in each histological type was also examined,
and the associations between PSAT1 expression levels and the
clinical and pathological characteristics of the patients in each
dataset were analyzed.
CRC specimens for immunohistochemistry
(IHC)
Formalin-fixed and paraffin-embedded specimens from
88 patients with CRC who underwent surgical resection at Huzhou
Maternity and Child Care Hospital (Huzhou, China) between January
2007 and October 2013 were included in the present study, following
the acquisition of informed consent. No patient received
neo-adjuvant chemotherapy or radiotherapy. In 30 cases,
chemotherapy with the FOLFIRI regimen was administered due to the
occurrence of metastasis. Drug response was determined according to
criteria described previously (14).
Briefly, metastatic lesions were evaluated using computed
tomography scanning, and cases with a ≥50% decrease in lesion size
were classified as responders, whereas patients with a decrease of
<50% or with an increase in lesion size were classified as
non-responders. In all cases, complete data regarding
clinicopathological characteristics and follow-up were available,
as well as an adequate volume of tumor specimen for IHC. The
disease-specific survival time was defined as the interval between
surgical resection and mortality due to cancer. All procedures used
in the present study were approved by the research ethics committee
of Huzhou Maternity & Child Care Hospital.
IHC
Tissue sections (4-µm thick) were subjected to
immunohistochemical staining with an EnVision
avidin-biotin-peroxidase complex kit (Dako; Agilent Technologies,
Inc., Santa Clara, CA, USA). Briefly, following deparaffinization
and rehydration, 10 mM citrate buffer at pH 6.0 was used for
antigen retrieval at 100°C for 10 min. Hydrogen peroxide (3%) was
used to block the activity of endogenous peroxidases. The slides
were then stained with primary antibody (rabbit anti-PSAT1
antibody; cat. no. sc-133929; Santa Cruz Biotechnology, Inc., Santa
Cruz, CA, USA; dilution, 1:100) at 4°C overnight, followed by
secondary staining by the chromogenic substrate
3,3′-diaminobenzidine, followed by counterstaining with hematoxylin
using the EnVision kit according to the manufacturer's protocol.
For the negative control, anti-PSAT1 antibody was replaced by
non-specific rabbit IgG (at 1:100 dilution; cat. no. SC2027; Santa
Cruz Biotechnology, Inc.). The staining was evaluated by two
independent pathologists under a light microscope (magnification,
×40) by calculating H-scores, obtained by multiplying the
percentage and staining intensity scores in >10 fields, as
described previously (15). Cases
with a score of more or less than the median value were defined as
having high or low expression, respectively.
Statistical analysis
The associations between PSAT1 expression levels and
clinicopathological features were evaluated using a χ2
test. Kaplan-Meier estimator curves and log-rank test were used for
the survival analysis. Multivariate survival analyses were
performed using the Cox's proportional hazards model. P<0.05 was
considered to indicate a statistically significant difference. All
statistical analyses were performed using MedCalc software (version
15.22; MedCalc Software bvba, Ostend, Belgium).
Results
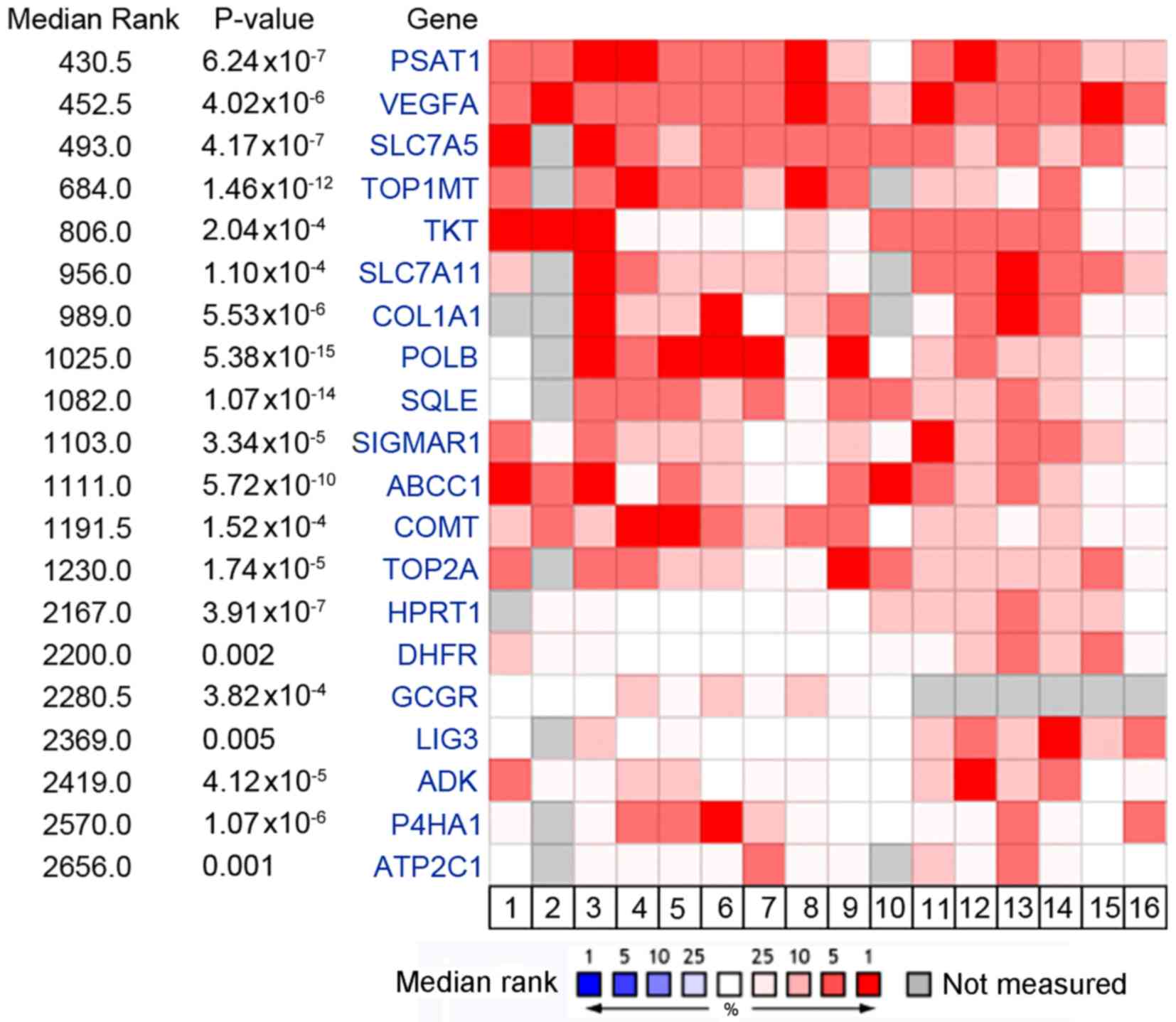
PSAT1 is identified as the most
upregulated drug target gene using in silico analysis
An in silico analysis of the 6 published
microarray datasets was performed to determine which drug-target
genes were reproducibly upregulated in CRC tumors vs. normal
colorectal tissues. Using mRNA gene expression datasets from the
Oncomine database, a panel of drug targets that were significantly
overexpressed in CRC tumors compared with normal colorectal tissues
was identified (Fig. 1). Among this
target list, several genes have previously been validated as CRC
therapeutic targets, such as vascular endothelial growth factor A
(16). Notably, PSAT1, a key serine
biosynthesis enzyme, ranked top of the upregulated targets (median
rank, 430.5; P=6.24×10−7) across all CRC cohorts
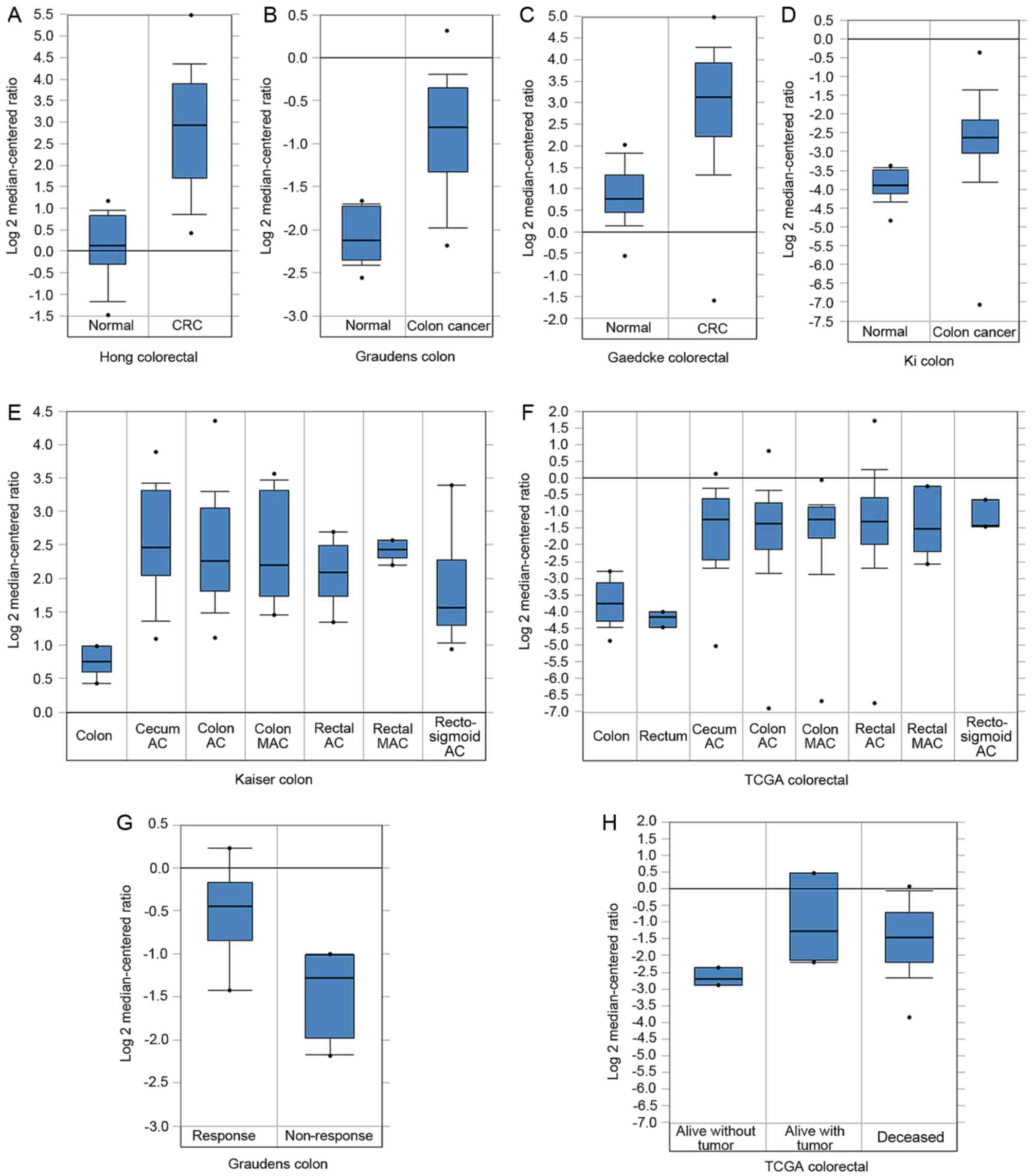
(Fig. 2A-F). Furthermore, PSAT1 was
not only overexpressed in the whole set of CRC tissues, but also
significantly upregulated in each histological subtype in the
Kaiser colon and TCGA CRC datasets compared with the normal
colorectal tissues (Fig. 2E and
F).
Clinical and prognostic value of PSAT1
across CRC datasets
The clinical and prognostic significance of PSAT1
expression in the 6 Oncomine CRC datasets was evaluated. No
association between PSAT1 and clinical traits was observed, except
that patients with CRC who did not exhibit a response to FOLFIRI
chemotherapy had an increased PSAT1 expression level compared with
those who did exhibit a response (Graudens colon dataset); and
patients who were alive without tumor at 3 years exhibited
relatively decreased PSAT1 expression compared with those who were
alive with tumor and deceased at 3 years (TCGA CRC dataset)
(Fig. 2G and H).
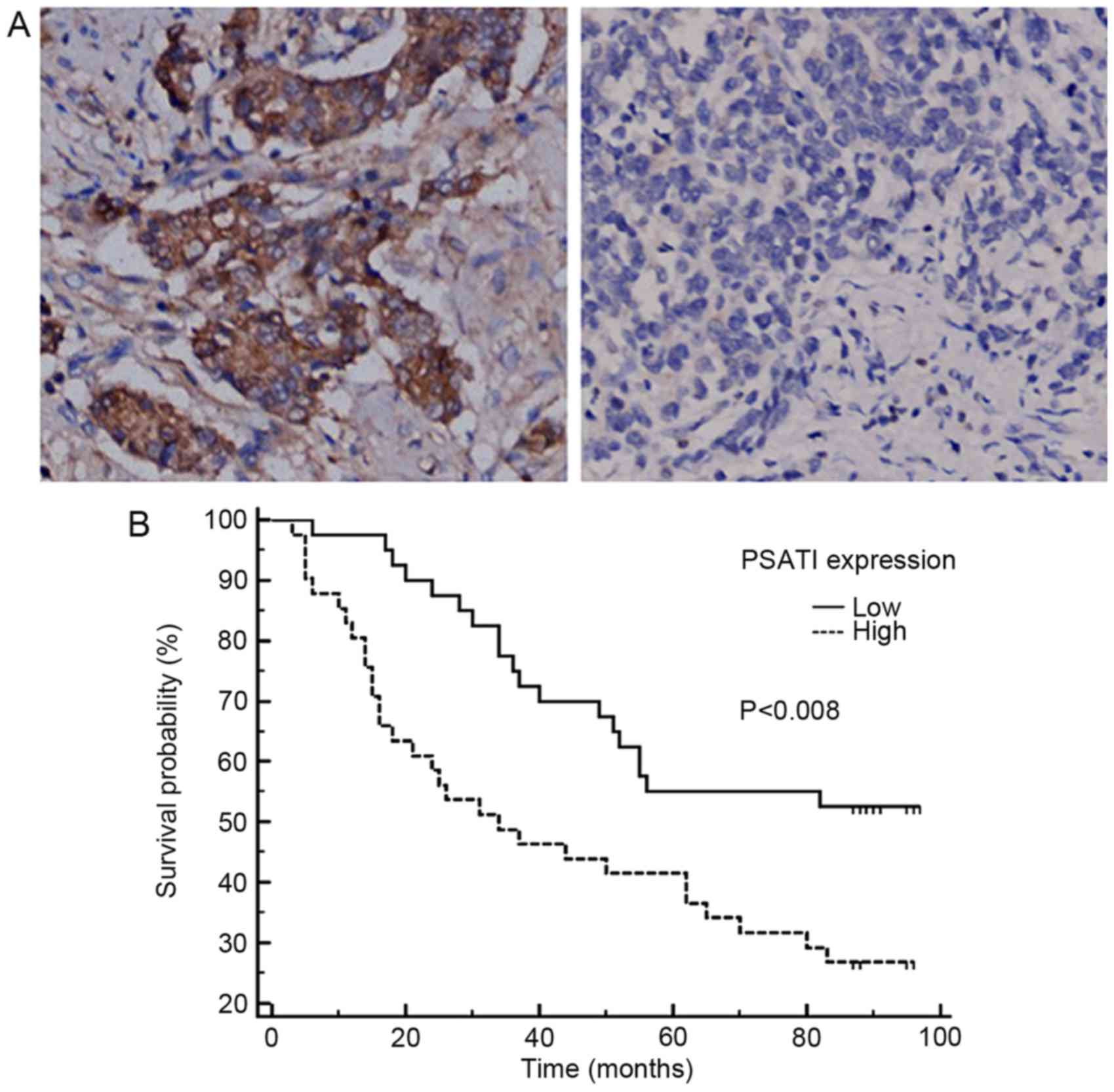
PSAT1 is a biomarker and prognostic
factor for CRC
Immunohistochemical validation of the expression
pattern and clinical significance of PSAT1 was performed in an
independent CRC cohort. The IHC results identified that PSAT1
staining was positive in the cytoplasm and membrane of cancer cells
(Fig. 3A). As expected, PSAT1
staining was observed in CRC tissues with various degrees of
intensity and density, whereas no staining was detected in
tumor-adjacent normal colorectal tissues, which is consistent with
the results of the in silico analysis. Heterogeneous PSAT1
expression among CRC tumors was observed. On the basis of the
median H-score, the 88 cases were divided into high- and
low-expression subgroups. PSAT1 expression was identified to be
associated with FOLFIRI treatment efficacy; as presented in
Table II, PSAT1 overexpression was
more frequently observed in FOLFIRI non-responders (84.2%) than in
FOLFIRI responders (36.4%) (P=0.0228). No significant association
with other clinical characteristics was identified.
 | Table II.Comparison of clinicopathological
characteristics between high and low PSAT1 expression subgroups
evaluated by immunohistochemistry. |
Table II.
Comparison of clinicopathological
characteristics between high and low PSAT1 expression subgroups
evaluated by immunohistochemistry.
|
|
| PSAT1 expression |
|
|---|
|
|
|
|
|
|---|
| Characteristics | Total patients,
n | Low, n | High, n | P-value |
|---|
| Age, years |
|
|
| 0.6043 |
|
<60 | 19 | 11 | 8 |
|
| ≥60 | 69 | 33 | 36 |
|
| Sex |
|
|
| 0.5220 |
| Male | 46 | 21 | 25 |
|
|
Female | 42 | 23 | 19 |
|
| Tumor grade |
|
|
| 0.1016 |
| 1 | 10 | 4 | 6 |
|
| 2 | 46 | 28 | 18 |
|
| 3 | 32 | 12 | 20 |
|
| Tumor size |
|
|
| 0.4817 |
|
T1-T2 | 9 | 3 | 6 |
|
|
T3-T4 | 79 | 41 | 38 |
|
| Lymph node
metastasis |
|
|
| 0.8267 |
|
Negative | 54 | 27 | 27 |
|
|
Positive | 34 | 17 | 17 |
|
| Distant
metastasis |
|
|
| 0.4744 |
|
Negative | 86 | 43 | 43 |
|
|
Positive | 2 | 1 | 1 |
|
| Stage |
|
|
| 0.8267 |
|
1–2 | 54 | 27 | 27 |
|
|
2–4 | 34 | 17 | 17 |
|
| FOLFIRI
treatment |
|
|
| 0.0228 |
|
Response | 11 | 7 | 4 |
|
| No
response | 19 | 3 | 16 |
|
Furthermore, survival data analysis revealed that a
high expression level of PSAT1 was associated with poor
disease-specific survival [hazard ratio (HR), 2.1802; 95%
confidence interval (CI), 1.2289–3.8680; P=0.008, log-rank test;
Fig. 3B; Table III], and was identified as an
independent prognostic factor on multivariate analysis (HR, 2.5950;
95% CI, 1.4317–4.7035; P=0.0018; Table
III).
 | Table III.Univariate and multivariate analysis
for overall survival according to PSAT1 expression evaluated
immunohistochemistry. |
Table III.
Univariate and multivariate analysis
for overall survival according to PSAT1 expression evaluated
immunohistochemistry.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Prognostic
factor | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Tumor grade | 1.4765
(0.9477–2.3003) | 0.0865 | 1.3006
(0.8007–2.1126) | 0.2907 |
| Tumor size | 1.2658
(0.5037–3.1807) | 0.6179 | 1.6396
(0.5110–5.2613) | 0.4083 |
| Lymph node
metastasis | 2.3543
(1.3400–4.1364) | 0.0030 | 1.9354
(1.0478–3.5751) | 0.0359 |
| Distant
metastasis | 9.8512
(2.2493–43.1446) | 0.0025 | 22.8051
(3.4643–150.1243) | 0.0012 |
| PSAT1
expression | 2.1802
(1.2289–3.8680) | 0.0080 | 2.5950
(1.4317–4.7035) | 0.0018 |
Discussion
In the present study, the expression profiles of a
list of approved or candidate drug-target genes were compared in
silico between cancerous and normal tissues across independent
CRC cohorts. The PSAT1 gene and protein were identified and
validated to be reproducibly overexpressed in CRC tumors, and PSAT1
expression levels were identified to be associated with response to
FOLFIRI treatment. The analysis also suggested that PAST1 may serve
as a potential independent prognostic factor for CRC. To the best
of our knowledge, the present study is the first to report the
clinicopathological and prognostic significance of PSAT1 in
CRC.
PSAT1 encodes a key enzyme involved in serine
synthesis, a process that serves a pivotal role in proliferating
cancer cells and is associated with clinical aggressiveness
(17). Mutation or decreased levels
of PSAT1 may be associated with schizophrenia or phosphoserine
aminotransferase deficiency. Marked PSAT1 expression has been
identified in various cancer types, including breast cancer, and
non-small cell lung cancer (18,19).
Martens et al (20) reported
that DNA methylation of PSAT1 was inversely associated with its
mRNA expression and with poor clinical outcome. Furthermore, PSAT1
may be a predictor of tamoxifen therapy response in breast cancer
(21). In a previous study of a small
population of patients with CRC, using microarray and
semi-quantitative PCR methods, Vié et al (22) observed that PSAT1 mRNA was increased
in colon tumors compared with normal colorectal tissues. The
present study verified and extended these earlier results in
clinical setting and a larger cohort of patients, and further
confirmed that PSAT1-positive CRC cases may represent a subgroup of
more aggressive disease, with resistance to the currently used
regimens and with poorer clinical outcomes.
Experimental studies have indicated that inhibiting
PSAT1 could aid in the treatment of cancer (16,22). Vié
et al (22) also suggested
that PSAT1 may be implicated in cancer progression and
chemoresistance in human colon cells, reporting that a
PSAT1-positive cell line was more resistant to oxaliplatin
treatment compared with a non-transfected cell line lacking PSAT1
expression. Additionally, Yang et al (18) found that PSAT1 could sustain the
proliferation of non-small cell lung cancer cells by inhibiting
cyclin D1 degradation and subsequently activating the
retinoblastoma-E2F transcription factor signaling pathway. On the
basis of the present and previous results, we hypothesize that
PSAT1 may be a prognostic biomarker as well as a promising
therapeutic target, and that a PSAT1-positive CRC subgroup may
benefit from therapies directed against this target. Investigation
of PSAT1 inhibition in animal models and development of
small-molecule inhibitors of PSAT1 merit attention in future
research.
In summary, the present study identified and
validated the overexpression of PSAT1 in CRC tumors compared with
normal colorectal tissues, and demonstrated that its overexpression
is associated with response to chemotherapy and poor clinical
outcome. Therefore, it is possible that PSAT1 may be used as a
prognostic marker and a potential therapeutic target for patients
with CRC.
Acknowledgements
The present study was supported by the National
Science Foundation of China (grant no. 81471605), a Shanghai
Shenkang Grant (grant no. CHDC22014014) and Changhai Hospital
(grant no. CH125530300).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zheng R, Zeng H, Zhang S, Chen T and Chen
W: National estimates of cancer prevalence in China, 2011. Cancer
Lett. 370:33–38. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kanwar SS, Poolla A and Majumdar AP:
Regulation of colon cancer recurrence and development of
therapeutic strategies. World J Gastrointest Pathophysiol. 15:1–9.
2012. View Article : Google Scholar
|
|
4
|
Meyerhardt JA and Mayer RJ: Systemic
therapy for colorectal cancer. N Engl J Med. 352:476–487. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gupta SC, Sung B, Prasad S, Webb LJ and
Aggarwal BB: Cancer drug discovery by repurposing: Teaching new
tricks to old dogs. Trends Pharmacol Sci. 34:508–517. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rhodes DR, Ateeq B, Cao Q, Tomlins SA,
Mehra R, Laxman B, Kalyana-Sundaram S, Lonigro RJ, Helgeson BE,
Bhojani MS, et al: AGTR1 overexpression defines a subset of breast
cancer and confers sensitivity to losartan, an AGTR1 antagonist.
Proc Natl Acad Sci USA. 106:pp. 10284–10289. 2009, View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno
V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ,
Kincead-Beal C, Kulkarni P, et al: Oncomine 3.0: Genes, pathways,
and networks in a collection of 18,000 cancer gene expression
profiles. Neoplasia. 9:166–180. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gaedcke J, Grade M, Jung K, Camps J, Jo P,
Emons G, Gehoff A, Sax U, Schirmer M, Becker H, et al: Mutated KRAS
results in overexpression of DUSP4, a MAP-kinase phosphatase, and
SMYD3, a histone methyltransferase, in rectal carcinomas. Genes
Chromosomes Cancer. 49:1024–1034. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Graudens E, Boulanger V, Mollard C,
Mariage-Samson R, Barlet X, Grémy G, Couillault C, Lajémi M,
Piatier-Tonneau D, Zaborski P, et al: Deciphering cellular states
of innate tumor drug responses. Genome Bio. 7:R192006. View Article : Google Scholar
|
|
10
|
Hong Y, Downey T, Eu KW, Koh PK and Cheah
PY: A ‘metastasis-prone’ signature for early-stage mismatch-repair
proficient sporadic colorectal cancer patients and its implications
for possible therapeutics. Clin Exp Metastasis. 27:83–90. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kaiser S, Park YK, Franklin JL, Halberg
RB, Yu M, Jessen WJ, Freudenberg J, Chen X, Haigis K, Jegga AG, et
al: Transcriptional recapitulation and subversion of embryonic
colon development by mouse colon tumor models and human colon
cancer. Genome Bio. 8:R1312007. View Article : Google Scholar
|
|
12
|
Ki DH, Jeung HC, Park CH, Kang SH, Lee GY,
Lee WS, Kim NK, Chung HC and Rha SY: Whole genome analysis for
liver metastasis gene signatures in colorectal cancer. Int J
cancer. 121:2005–2012. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cancer Genome Atlas Network: Comprehensive
molecular characterization of human colon and rectal cancer.
Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Therasse P, Arbuck SG, Eisenhauer EA,
Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van
Oosterom AT, Christian MC and Gwyther SG: New guidelines to
evaluate the response to treatment in solid tumors. J Natl Cancer
Inst. 92:205–216. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang HJ, Neven P, Drijkoningen M,
Paridaens R, Wildiers H, Van Limbergen E, Berteloot P, Amant F,
Vergote I and Christiaens MR: Association between tumour
characteristics and HER-2/neu by immunohistochemistry in 1362 women
with primary operable breast cancer. J Clin Path. 58:611–616. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Khan K, Cunningham D and Chau I: Targeting
angiogenic pathways in colorectal cancer: Complexities, challenges
and future directions. Curr Drug Targets. 18:56–71. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Antonov A, Agostini M, Morello M, Minieri
M, Melino G and Amelio I: Bioinformatics analysis of the serine and
glycine pathway in cancer cells. Oncotarget. 5:11004–11013. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang Y, Wu J, Cai J, He Z, Yuan J, Zhu X,
Li Y, Li M and Guan H: PSAT1 regulates cyclin D1 degradation and
sustains proliferation of non-small cell lung cancer cells. Int J
Cancer. 136:E39–E50. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim YH, Jung WH and Koo JS: Expression of
metabolism-related proteins in invasive lobular carcinoma:
Comparison to invasive ductal carcinoma. Tumor Bio. 35:10381–10393.
2014. View Article : Google Scholar
|
|
20
|
Martens JW, Nimmrich I, Koenig T, Look MP,
Harbeck N, Model F, Kluth A, Bolt-de Vries J, Sieuwerts AM,
Portengen H, et al: Association of DNA methylation of phosphoserine
aminotransferase with response to endocrine therapy in patients
with recurrent breast cancer. Cancer Res. 65:4101–4117. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
De Marchi T, Timmermans MA, Sieuwerts AM,
Smid M, Look MP, Grebenchtchikov N, Sweep FCGJ, Smits JG, Magdolen
V, van Deurzen CHM, et al: Phosphoserine aminotransferase 1 is
associated to poor outcome on tamoxifen therapy in recurrent breast
cancer. Sci Rep. 7:20992017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vié N, Copois V, Bascoul-Mollevi C, Denis
V, Bec N, Robert B, Fraslon C, Conseiller E, Molina F, Larroque C,
et al: Overexpression of phosphoserine aminotransferase PSAT1
stimulates cell growth and increases chemoresistance of colon
cancer cells. Mol Cancer. 7:142008. View Article : Google Scholar : PubMed/NCBI
|