Introduction
Retinoblastoma (RB) is the most common primary type
of intraocular malignancy among children. It originates from the
primitive stem cells in the nuclear layer of the retina. Its
prevalence is between 1/15,000 and 1/18,000 cases/people, with 95%
of cases occurring before the age of 5 years (1). The main symptoms of RB are leukokoria
and strabismus. The RB transcriptional corepressor 1 (RB1) gene
located on chromosome 13 is associated with RB. The RB1 gene, which
produces the RB protein, serves an important role in regulating and
controlling the cell cycle (2).
Loss-of-function mutations in RB1 disrupt the cell cycle and have
been revealed to be an important initiating event prior to the
development of RB (3).
Besides the RB1 gene, other genes and proteins have
been observed to serve important roles in the pathogenesis of RB.
For example, 1% of all cases of RB have high levels of MYCN
proto-oncogene bHLH transcription factor amplification and no RB
mutations. Furthermore, a previous study demonstrated that proteins
associated with the redox signaling pathways are involved in RB
pathogenesis (4).
In order to further investigate the pathogenesis of
RB, the present study performed the quantitative proteomic strategy
of using isobaric tags for relative and absolute quantitation
(iTRAQ) coupled with two-dimensional liquid chromatography-tandem
mass spectrometry (MS) in order to identify associated proteins.
The main advantage of the iTRAQ technique for proteomic analysis is
that its multiplexing capability allows various protein samples to
be simultaneously quantified with a control-standard sample that is
processed in the same run (5). The
insights the present study have gained may be used for further
research of RB in pursuit of a novel therapy target.
Materials and methods
Subjects
Patients and control subjects were recruited from
the Department of Ophthalmology, Peking University People's
Hospital (Beijing, China). The present study was approved by the
Clinic Institutional Review Board of Peking University People's
Hospital and complied with the Declaration of Helsinki. Written
informed consent was obtained from all patients prior to enrollment
in the present study. A total of 10 patients (2 women and 8 men;
mean age, 3.8 years; range, 2–5 years) diagnosed with RB between
September 2014 and March 2015 were included and 10 patients with
cataracts (3 women and 7 men; mean age, 70.4 years; range, 65–79
years) were recruited as controls. The inclusion criteria were as
follows: Diagnosis of group D RB in accordance with the
International Classification of Retinoblastoma (6) with clear optical media in poor
responders to chemotherapy, laser or cryotherapy. Patients with RB
or control subjects with a history of any systemic or ocular
disorder or condition (including ocular surgery, trauma and
disease) were excluded from the current study.
Sample collection
Aqueous humor (AH) samples from patients with RB and
control subjects were collected. The whole procedure was performed
using a microscope viewing through a dilated pupil, followed by
intravitreal injections. As previously described (7), anterior chamber paracentesis was
performed through a clear cornea limbus track created with a 25G
MVR blade without perforating the Descemet's membrane. A 32G needle
mounted on a tuberculin syringe was then introduced through the
track tangentially into the anterior chamber periphery, parallel to
the iris. A volume of 0.1 ml aqueous fluid was aspirated,
registered and stored at −80°C until processing. For patients with
RB, three cycles of freeze and thaw (6 sec each) were applied at
the injection site at the time of removal of the needle.
Protein extraction and digestion
Total AH protein concentration was determined using
a Bradford protein assay kit (Bio-Rad Laboratories, Inc., Hercules,
CA, USA), according to the manufacturer's protocol and as described
in a previous study (8). Each sample
(200 µg) was reconstituted in SDS-PAGE sample buffer with 5%
β-mercaptoethanol, and electrophoresed on a (10–14.5%) SDS-PAGE
precast gel (Criterion; Bio-Rad Laboratories, Inc.). A total of
nine gel slices were excised from each lane. In-gel digestion was
performed as previously reported (8).
Briefly, the excised bands were de-stained using 40 mM ammonium
bicarbonate in 50% acetonitrile (ACN) solution (45°C for 20 min).
The de-stained gel bands were then subjected to reduction using 5
mM dithiothreitol (60°C for 45 min), followed by alkylation using
10 mM iodoacetamide (56°C for 60 min). The gel pieces were
dehydrated using 100% ACN, followed by digestion with trypsin
(modified sequencing grade; Promega Corporation, Madison, WI, USA)
at 37°C for 12–16 h. The peptides were extracted from the gel
fragments by treating the gel bands twice with 0.4% formic acid and
3% ACN solution once with 0.4% formic acid and 50% ACN solution and
finally with 100% ACN solution (all at room temperature for 15 min
each). The samples were labeled with iTRAQ® reagents by
adding the contents of the iTRAQ Reagent-8Plex Multiplex kit
(Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) to the sample solutions. The two samples were iTRAQ-labeled as
follows: R0 (RB) and C4 (Control).
Two-dimensional liquid
chromatography-electrospray ionization MS
Sample analysis was performed using a QTRAP 5500
system (Applied Biosystems; Thermo Fisher Scientific, Inc.) to
generate MS and tandem MS (MS/MS) data. Peptides were loaded onto a
Kinetex 100.0×2.1-mm C18 column (300 Å, 2.6 µm; Phenomenex,
Torrance, CA, USA) and then submitted to mobile-phase elution in
buffer A (0.1% formic acid in water) and buffer B (0.1 % formic
acid in acetonitrile), according to the manufacturer's protocol.
The peptides were eluted at a flow rate of 400 µl/min. The liquid
chromatography eluent was directed to an electrospray ionization
source for quantitative time-of-flight MS analysis. Electrospray
ionization was performed for information-dependent acquisition in
positive-ion mode with a spray voltage of 1.8 kV and a selected
mass range of 350–2,000 m/z. The QTRAP 5500 system
was operated in data-dependent acquisition mode. The three most
abundantly charged peptides above a 5-count threshold were selected
for MS/MS.
Database search
All MS/MS samples were analyzed using Mascot
(version 2.4.1; Matrix Science, London, UK). Mascot was set up to
search the Uniprot2014_human database (www.uniprot.org; accessed January 2014). The analysis
and search parameters were as follows: Trypsin as the digestion
enzyme with allowance for a maximum of one missed cleavage;
Carbamidomethyl (C) as a fixed modification; Oxidation (M),
Gln→Pyro-Glu (N-term Q) and iTRAQplex modification (K, Y and N
termini) as a variable modification; peptide mass tolerance of 15
ppm; and fragment mass tolerance of 20 mmu.
Expression changes of the identified peptides in
human AH were determined and compared with the controls using the
iTRAQ reporter ion intensities. Based on the relative
quantification and statistical analysis, a 1.2-fold change cut-off
was selected to categorize proteins as significantly altered.
Therefore, proteins with iTRAQ ratios >1.2 were considered to be
upregulated, whereas those with iTRAQ ratios <0.83 were
considered to be downregulated.
Bioinformatics analysis
In order to characterize the function of the
proteins identified in the quantitative proteomics analysis,
information from the DAVID Bioinformatics website (https://david.ncifcrf.gov/) was applied to the
functional enrichment and gene ontology (G) analyses, as previously
described (9). In the present study,
the GO categories with P<0.05 were considered to indicate a
statistically significant expression in patients with RB.
Results
Protein identification in AH
Proteins with corrected P<0.05 and a fold change
of >1.2 or <0.83 were considered to be significantly
differentially expressed. In total, 83 proteins were identified in
the AH of patients with RB by iTRAQ analysis (Table I). Of these proteins, 44 were
upregulated and 39 were downregulated. The proteins with an
increased fold change of >2.0 included the following: Vitamin
D-binding protein, angiotensinogen, carbonic anhydrase 1, Ig
κ-chain V–III region B6, Ig α-1 chain C region, α-1-antitrypsin,
prothrombin, anti-thrombin-III stimulated by retinoic acid 6
(STRA6), fibrinogen λ-chain, Ig λ-2 chain C region,
thyroxine-binding globulin, α-2-HS-glycoprotein and hemoglobin
subunit α. The five proteins with a decreased fold change of
>0.5 were β-crystallin S, prosaposin, glutathione peroxidase 3,
cathepsin D and retinol-binding protein (RBP) 3.
 | Table I.Proteins identified in the aqueous
humor of patients with retinoblastoma using iTRAQ analysis. |
Table I.
Proteins identified in the aqueous
humor of patients with retinoblastoma using iTRAQ analysis.
| Fold change | Protein | Description |
|---|
| 0.269 | CRYGS | β-crystallin S |
| 0.351 | PSAP | Prosaposin |
| 0.387 | GPX3 | Glutathione
peroxidase 3 |
| 0.389 | CTSD | Cathepsin D |
| 0.478 | RBP3 | Retinol-binding
protein 3 |
| 0.52 | LGALS3BP | Galectin-3-binding
protein |
| 0.522 | LDHA | L-lactate
dehydrogenase α-chain |
| 0.528 | CLSTN1 | Calsyntenin-1 |
| 0.571 | C4A | Complement
C4-A |
| 0.572 | Ig κ chain V–III
region SIE | Ig κ-chain V–III
region SIE |
| 0.593 | Ig κ chain V–I
region Roy | Ig κ-chain V–I
region Roy |
| 0.594 | LACRT | Extracellular
glycoprotein lacritin |
| 0.596 | FBLN | Fibulin-1 |
| 0.598 | LUM | Lumican |
| 0.614 | SERPING1 | Plasma protease C1
inhibitor |
| 0.621 | APLP | Amyloid-like
protein 2 |
| 0.625 | PTGDS | Prostaglandin-H2
D-isomerase |
| 0.631 | CLEC3B | Tetranectin |
| 0.66 | A2M |
α-2-macroglobulin |
| 0.681 | KRT9 | Keratin, type I
cytoskeletal 9 |
| 0.682 | SPON1 | Spondin-1 |
| 0.687 | ENPP | Ectonucleotide
pyrophosphatase/phosphodiesterase family member 2 |
| 0.727 | GSN | Gelsolin |
| 0.73 | LYZ | Lysozyme C |
| 0.739 | KRT14 | Keratin, type I
cytoskeletal 14 |
| 0.743 | CP | Ceruloplasmin |
| 0.75 | RBP | Retinol-binding
protein 4 |
| 0.758 | Ig λ chain V–I
region NEW | Ig λ-chain V–I
region NEW |
| 0.773 | SERPINF1 | Pigment
epithelium-derived factor |
| 0.777 | IGFBP7 | Insulin-like growth
factor-binding protein 7 |
| 0.787 | KRT10 | Keratin, type I
cytoskeletal 10 |
| 0.788 | TTR | Transthyretin |
| 0.803 | CST3 | Cystatin-C |
| 0.814 | Ig λ chain V–III
region SH | Ig λ-chain V–III
region SH |
| 0.82 | KRT1 | Keratin, type II
cytoskeletal 1 |
| 0.82 | C2 | Complement C2 |
| 0.82 | IGLL5 | Immunoglobulin
λ-like polypeptide 5 |
| 0.832 | HP | Haptoglobin |
| 0.833 | KRT2 | Keratin, type II
cytoskeletal 2 epidermal |
| 1.238 | Ig heavy chain
V–III region GAL | Ig heavy chain
V–III region GAL |
| 1.24 | AZGP1 |
Zinc-α-2-glycoprotein |
| 1.261 | ORM1 | α-1-acid
glycoprotein 1 |
| 1.261 | Ig heavy chain V–I
region EU | Ig heavy chain V–I
region EU |
| 1.274 | OPTC | Opticin OS=Homo
sapiens |
| 1.278 | AMBP | Protein AMBP |
| 1.288 | APOA1 | Apolipoprotein
A-I |
| 1.293 | LRG1 | Leucine-rich
α-2-glycoprotein |
| 1.306 | Ig heavy chain
V–III region BRO | Ig heavy chain
V–III region BRO |
| 1.321 | CHI3L1 | Chitinase-3-like
protein 1 |
| 1.354 | VTN | Vitronectin |
| 1.378 | HPX | Hemopexin |
| 1.378 | IGHG1 | Ig γ-1 chain C
region |
| 1.388 | LCN1P1 | Putative lipocalin
1-like protein 1 |
| 1.408 | APOA4 | Apolipoprotein
A-IV |
| 1.457 | ALB | Serum albumin |
| 1.468 | ORM2 | α-1-acid
glycoprotein 2 |
| 1.482 | A1BG |
α-1B-glycoprotein |
| 1.538 | HBD | Hemoglobin subunit
delta |
| 1.546 | FGA | Fibrinogen α
chain |
| 1.547 | Ig heavy chain
V–III region TIL | Ig heavy chain
V–III region TIL |
| 1.549 | HRG | Histidine-rich
glycoprotein |
| 1.55 | IGHM | Ig μ-chain C
region |
| 1.552 | SMTN | Smoothelin |
| 1.642 | EFEMP | EGF-containing
fibulin-like extracellular matrix protein 1 |
| 1.705 | HBB | Hemoglobin subunit
β |
| 1.746 | Ig heavy chain V–I
region HG3 | Ig heavy chain V–I
region HG3 |
| 1.765 | NOL6 | Nucleolar protein
6 |
| 1.768 | KNG1 | Kininogen-1 |
| 1.975 | SERPINA3 |
α-1-antichymotrypsin |
| 2.012 | GC | Vitamin D-binding
protein |
| 2.089 | AGT |
Angiotensinogen |
| 2.124 | CA1 | Carbonic anhydrase
1 |
| 2.191 | Ig κ chain V–III
region B6 | Ig κ-chain V–III
region B6 |
| 2.218 | IGHA1 | Ig α-1 chain C
region |
| 2.267 | SERPINA1 |
α-1-antitrypsin |
| 2.429 | F2 | Prothrombin |
| 2.437 | SERPINC1 |
Antithrombin-III |
| 2.479 | STRA6 | Stimulated by
retinoic acid gene 6 protein homolog |
| 2.573 | FGG | Fibrinogen γ
chain |
| 2.605 | IGHG2 | Ig γ-2 chain C
region |
| 2.973 | SERPINA7 | Thyroxine-binding
globulin |
| 2.98 | AHSG |
α-2-HS-glycoprotein |
| 3.146 | HBA1 | Hemoglobin subunit
α |
GO analysis
In order to identify the functions of proteins
identified using the iTRAQ technique, the present study performed
GO analysis with the assistance of DAVID Bioinformatics Resources.
The DAVID classification of proteins by biological process
demonstrated that the proteins were primarily involved in the
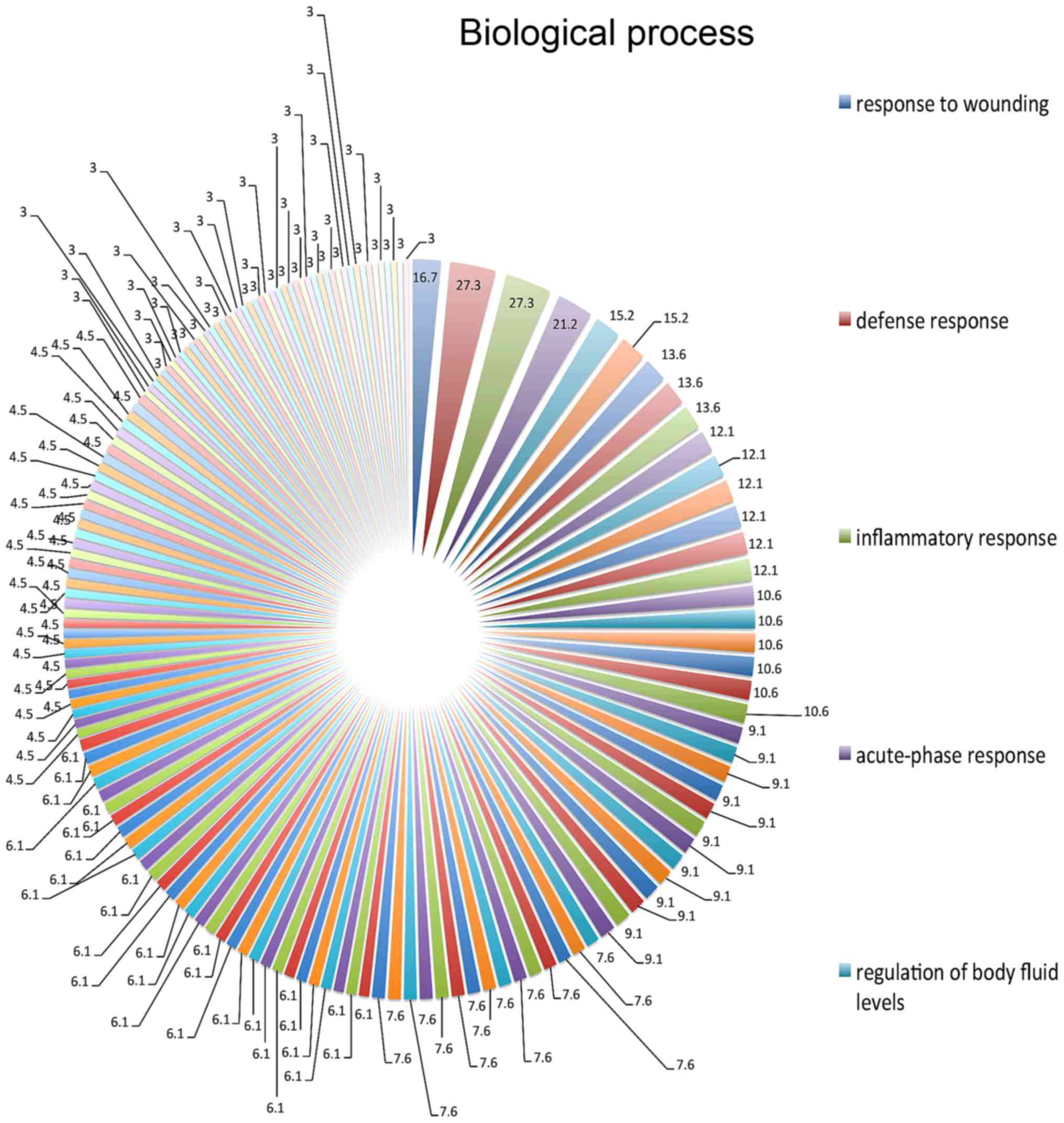
defensive (27.3%), inflammatory (27.3%) and acute-phase (27.3%)
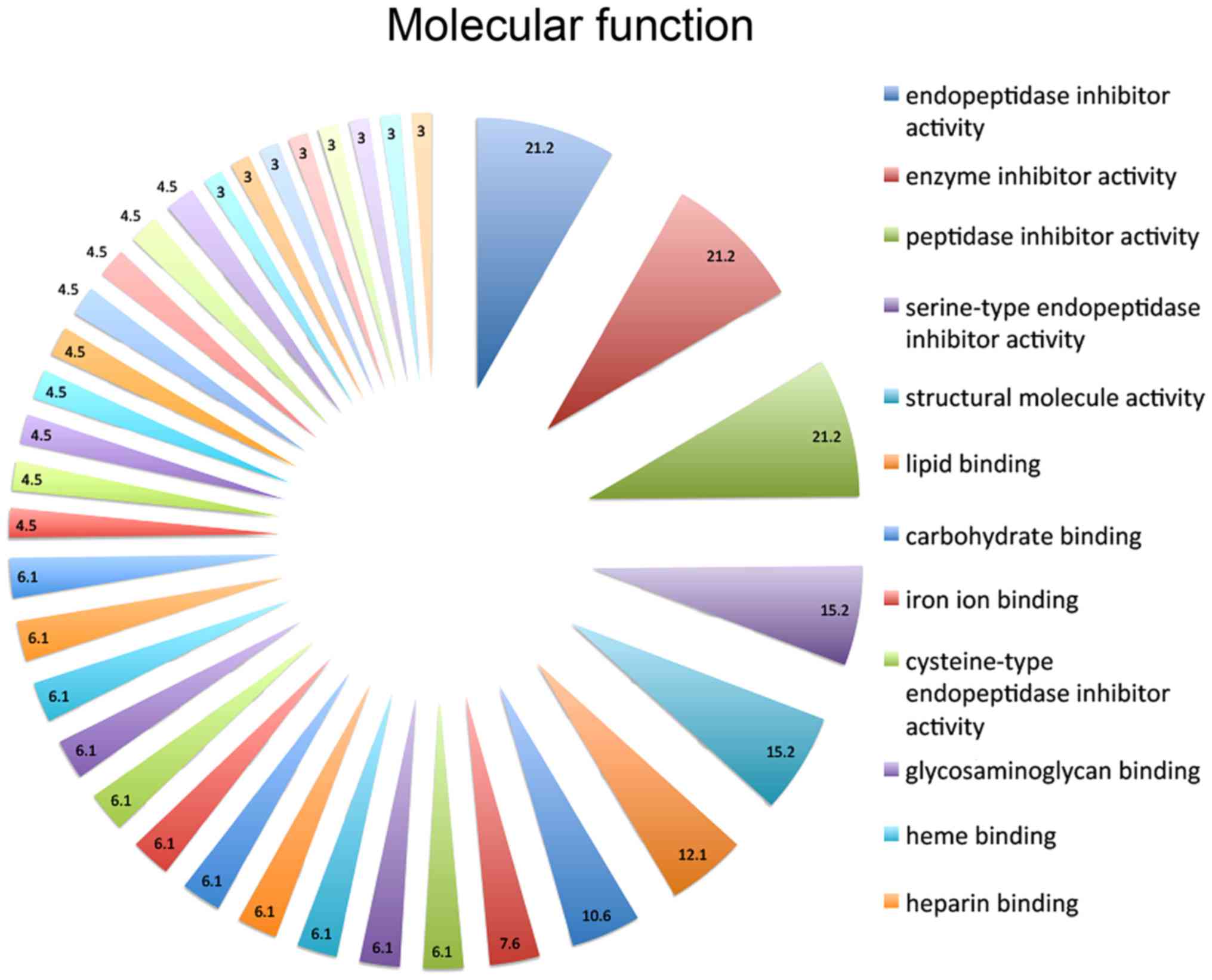
responses, and the response to wounding (16.7%) (Fig. 1). On the basis of molecular function
annotations, the proteins in the present study were implicated in
endopeptidase inhibitor activity (21.2%), peptidase inhibitor
(21.2%), enzyme inhibitor (21.2%), serine-type endopeptidase
inhibitor (15.2%), structural molecule (15.2%), lipid binding
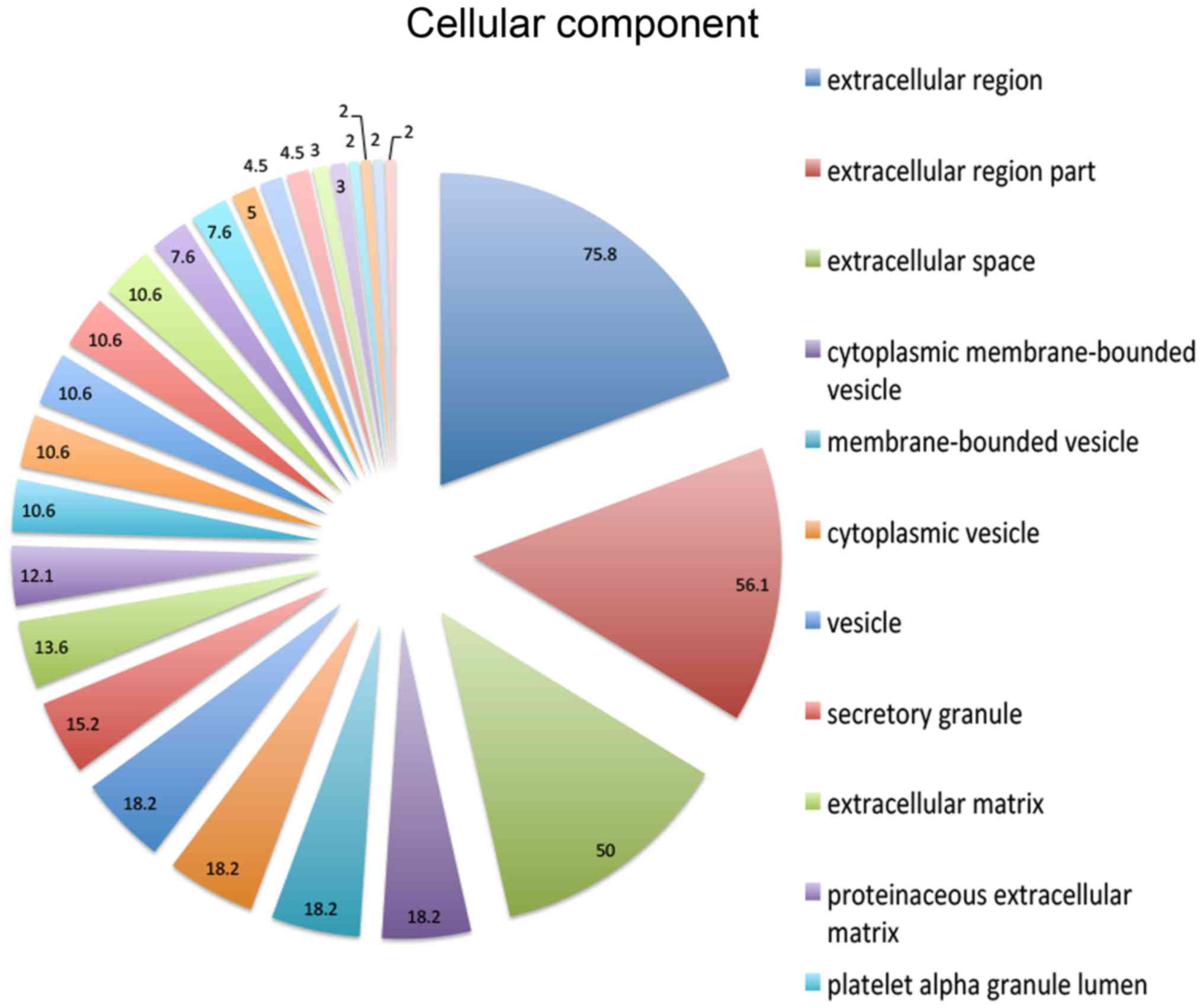
(12.1%) and carbohydrate binding (10.6%) activities (Fig. 2). In the cellular component ontology,
the present study revealed that the majority of enriched categories
were associated with extracellular construction, including
extracellular region (75.8%), extracellular region part (56.1%) and
extracellular space (50%) (Fig. 3).
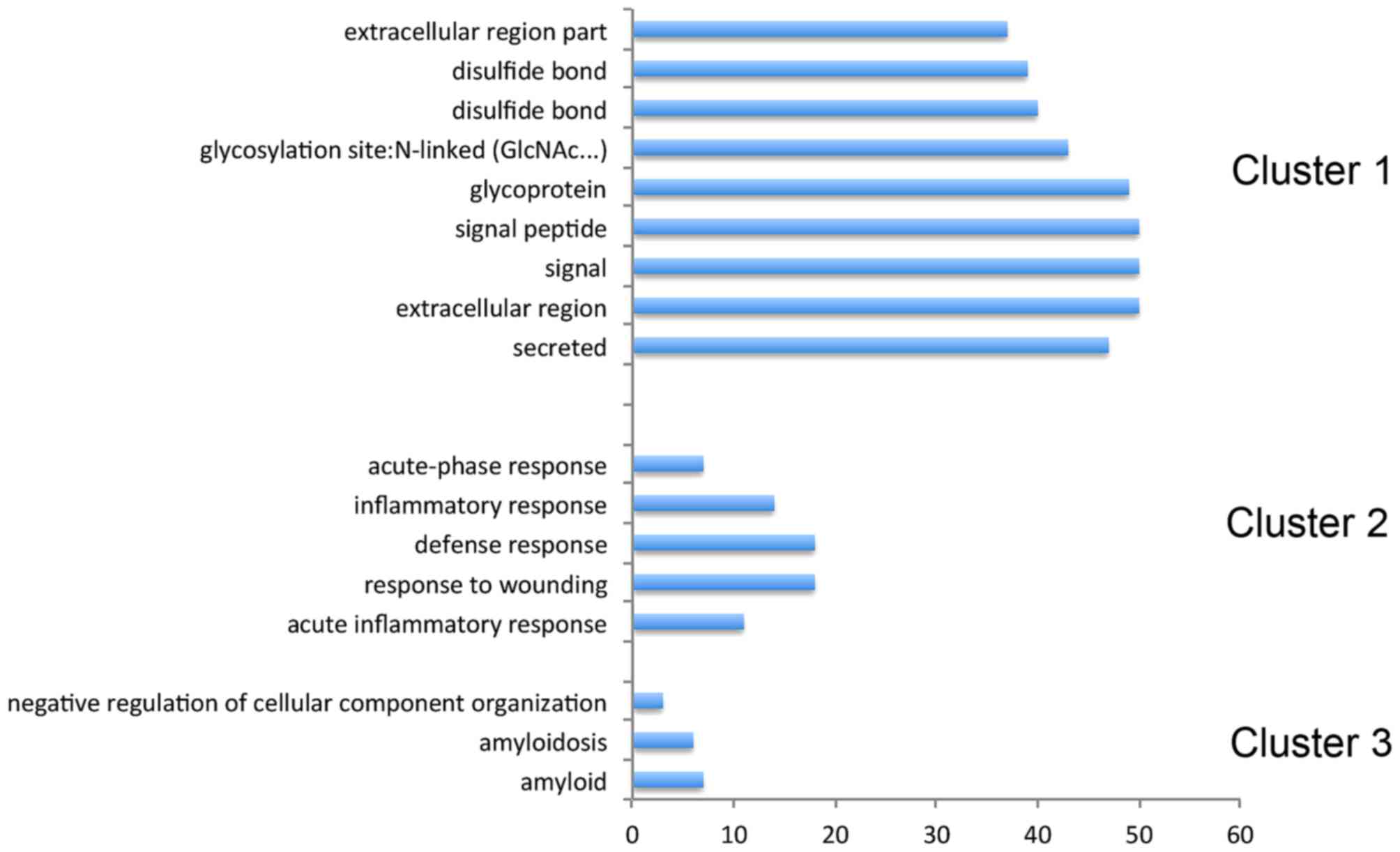
Functional annotation clustering demonstrated that they belong to
glycoprotein, amyloid, acute-inflammatory and defense responses
(Fig. 4).
Discussion
RB has become curable in the majority of cases in
children, so long as there is early diagnosis and accurate
prognosis. In order to investigate RB, the present study performed
iTARQ analysis to identify proteins that may serve a role in RB in
children. A total of 44 upregulated proteins and 39 downregulated
proteins were identified. Functional annotation clustering revealed
that they belong to glycoprotein, amyloid, acute-inflammatory and
defense responses. Among these proteins, the present study
speculated that pigment epithelium-derived factor precursor (PEDF)
serves a potential role in the treatment of RB and STRA6 may be
involved in RB development.
PEDF is a member of the serine protease inhibitor
(serpin) superfamily. As a nerotrophic factor, PEDF promotes the
differentiation of RB cells and other tumor cells of neuronal
origin (10). It has potent
anti-angiogenesis activity, and it has been revealed that PEDF may
delay tumor growth and decrease the expression level of vascular
endothelial growth factor (VEGF) (11). Of note, PEDF has been demonstrated to
be an inhibitor of tumor cell invasion, migration and metastasis
in vitro (12), and in
numerous in vivo models (13–15). It is
also known that the Fas cell surface death receptor
(Fas)/Fas-L/caspase-8 apoptotic signaling pathway is involved in
the apoptosis of PEDF-induced endothelial cells (16). Dawson et al (11) identified PEDF as a potent inhibitor of
angiogenesis in the eye. A number of studies have identified that
PEDF overexpression could prevent ocular neovascularization, and
delay photoreceptor and neural retinal cell death in vivo
(17–19). Yang et al (20) revealed that PEDF inhibited tumor
angiogenesis, as microvessel density was demonstrated to have
decreased in PEDF-treated tumor tissues. Further research
demonstrated that PEDF may downregulate VEGF expression in
vitro and in vivo by inhibition of hypoxia-inducible
factor-1α. The anti-angiogenic effect of PEDF makes it an excellent
candidate as a therapy target for RB. The present study revealed
that the expression level of PEDF was lower in RB compared with
control samples, which was consistent with the results in a
previous study (20). These results
suggest the potentially pivotal role that PEDF may serve in RB
treatment.
STRA6 is widely expressed during embryonic
development and in adult organ systems (21). STRA6 is the receptor of RBP and
transports retinol from extracellular RBP into cells (22). It also serves as a cytokine and
transduces a signaling cascade via Janus kinase 2, and the
transcription factors, signal transducer and activator of
transcription (STAT)3 and STAT5, leading to induction of STAT
target gene expression that promote oncogenic transformation
(23). Overexpression of STRA6 has
been observed in numerous types of human cancer, including Wilm
kidney tumors, melanomas, and colorectal, ovarian and endometrium
cancer (24). In vitro, STRA6
may facilitate cell proliferation, migration and invasion in cells.
However, it was recently reported that STRA6 contributes to
p53-induced apoptosis in response to DNA damage (25). The present study demonstrated that
STRA6 was upregulated in patients with RB, and further research is
required in order to identify the function of STRA6 in the
development of RB.
The GO analysis of the present study revealed that
the identified proteins were involved in glycoprotein, amyloid,
acute-inflammatory and defense responses. Glycosylation serves an
important role in posttranslational modifications, and modulates
the physical, chemical and biological properties of a protein
(26,27). Glycan structures are important for
numerous processes, including protein-protein interactions, protein
trafficking and folding, immune recognition, cell adhesion, and
migration and inter-cellular signaling (28). A number of studies have identified
that aberrant glycans may serves an essential role in cancer
biology by mediating tumor cell adhesion, motility and invasiveness
(29–31). The first group identified in the
functional clustering analysis involved glycoproteins.
α-1B-glycoprotein (A1BG) is a plasma glycoprotein that has sequence
similarity to the variable regions of certain immunoglobulin
supergene family member proteins. Recent proteomics studies have
revealed that A1BG may act as a biomarker for numerous types of
tumors, including bladder (32) and
pancreatic (33) cancer. Leucine-rich
α-2-glycoprotein (LRG1) is a secreted glycoprotein of the
leucine-rich repeat family. It was reported that LRG1 is associated
with cancer metastasis and poor prognosis, resulting from its
effects on promoting cell invasion, angiogenesis, and migration
(33). Chitinase 3-like 1 (CHI3L1) is
a member of the glycosyl hydrolase 18 family. CHI3L1 is considered
to serve a role in the process of inflammation and tissue
remodeling (34). A previous study
demonstrated the association of high serum CHI3L1 expression level
with poor patient prognosis and short survival time in human solid
tumors, including lung cancer and liver cancer (35).
Additionally, the present study indicated that
proteins involved in acute phase responses are associated with RB.
The acute phase response is a rapid inflammatory response that
provides protection against microorganisms using non-specific
defense mechanisms (36,37). It was observed that inflammation may
serve a dominant role in the pathogenesis of various types of
cancer (38,39). Fibrinogen γ, serpin family A member 1
(SerpinA1) and orosomucoid 2 (ORM2) are all acute-phase associated
proteins. Fibrinogen is involved in numerous processes, including
blood clotting, fibrinolysis, the inflammatory response and wound
healing. Previously, the role of fibrinogen and fibrinogen
degradation products in carcinogenesis of certain tumor types has
been suggested (40,41). Proteomic analysis has also
demonstrated that fibrinogen γ is overexpressed in patients with
pancreatic cancer (42). SerpinA1 is
produced in the liver and secreted into serum. It has been reported
to exhibit an invasive and metastatic capacity in lung cancer,
gastric cancer, and colorectal cancer (43,44).
Previous studies have revealed that SerpinA1 may serve as a
biomarker for gastric cancer as it induces the invasion and
migration of gastric cancer cells, and its expression is associated
with the progression of gastric cancer (45). ORM2 is an important acute phase plasma
protein due to its increased abundance during acute inflammation.
ORM2 may function in the modulation of the immune system in the
acute phase. Upregulation of ORM2 has been reported in patients
with colorectal cancer (46).
In conclusion, to the best of our knowledge, the
present study is the first to identify proteins associated with RB
using iTRAQ technology. The results of the present study may aid in
providing an improved understanding of RB and contribute to
developing a novel therapy target in the future. Further studies
are required to explore the function of proteins in RB
development.
Acknowledgements
The present study was supported by Peking University
People's Hospital Research and Development Funds (grant no.
RDC-2014-24).
References
|
1
|
Bishop JO and Madson EC: Retinoblastoma.
Review of the current status. Surv Ophthalmol. 19:342–366.
1975.PubMed/NCBI
|
|
2
|
Knudson AG: Cancer genetics. Am J Med
Genet. 111:96–102. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Corson TW and Gallie BL: One hit, two
hits, three hits, more? Genomic changes in the development of
retinoblastoma. Genes Chromosomes Cancer. 46:617–634. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vandhana S, Lakshmi TS, Indra D, Deepa PR
and Krishnakumar S: Microarray analysis and biochemical
correlations of oxidative stress responsive genes in
retinoblastoma. Curr Eye Res. 37:830–841. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tannu NS and Hemby SE: Methods for
proteomics in neuroscience. Prog Brain Res. 158:41–82. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shields CL, Mashayekhi A, Au AK, Czyz C,
Leahey A, Meadows AT and Shields JA: The International
Classification of Retinoblastoma predicts chemoreduction success.
Ophthalmology. 113:2276–2280. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Munier FL, Soliman S, Moulin AP, Gaillard
MC, Balmer A and Beck-Popovic M: Profiling safety of intravitreal
injections for retinoblastoma using an anti-reflux procedure and
sterilisation of the needle track. Br J Ophthalmol. 96:1084–1087.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Duan X, Lu Q, Xue P, Zhang H, Dong Z, Yang
F and Wang N: Proteomic analysis of aqueous humor from patients
with myopia. Mol Vis. 14:370–377. 2008.PubMed/NCBI
|
|
9
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Steele FR, Chader GJ, Johnson LV and
Tombran-Tink J: Pigment epithelium-derived factor: Neurotrophic
activity and identification as a member of the serine protease
inhibitor gene family. Proc Natl Acad Sci USA. 90:pp. 1526–1530.
1993, View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dawson DW, Volpert OV, Gillis P, Crawford
SE, Xu H, Benedict W and Bouck NP: Pigment epithelium-derived
factor: A potent inhibitor of angiogenesis. Science. 285:245–248.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Subramanian P, Deshpande M,
Locatelli-Hoops S, Moghaddam-Taaheri S, Gutierrez D, Fitzgerald DP,
Guerrier S, Rapp M, Notario V and Becerra SP: Identification of
pigment epithelium-derived factor protein forms with distinct
activities on tumor cell lines. J Biomed Biotechnol.
2012:4259072012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guan M, Jiang H, Xu C, Xu R, Chen Z and Lu
Y: Adenovirus-mediated PEDF expression inhibits prostate cancer
cell growth and results in augmented expression of PAI-2. Cancer
Biol Ther. 6:419–425. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guan M, Pang CP, Yam HF, Cheung KF, Liu WW
and Lu Y: Inhibition of glioma invasion by overexpression of
pigment epithelium-derived factor. Cancer Gene Ther. 11:325–332.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Orgaz JL, Ladhani O, Hoek KS,
Fernández-Barral A, Mihic D, Aguilera O, Seftor EA, Bernad A,
Rodríguez-Peralto JL, Hendrix MJ, et al: ‘Loss of pigment
epithelium-derived factor enables migration, invasion and
metastatic spread of human melanoma’. Oncogene. 28:4147–4161. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li L, Yao YC, Fang SH, Ma CQ, Cen Y, Xu
ZM, Dai ZY, Li C, Li S, Zhang T, et al: Pigment epithelial-derived
factor (PEDF)-triggered lung cancer cell apoptosis relies on p53
protein-driven Fas ligand (Fas-L) up-regulation and Fas protein
cell surface translocation. J Biol Chem. 289:30785–30799. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bouck N: PEDF: Anti-angiogenic guardian of
ocular function. Trends Mol Med. 8:330–334. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mori K, Duh E, Gehlbach P, Ando A,
Takahashi K, Pearlman J, Mori K, Yang HS, Zack DJ, Ettyreddy D, et
al: Pigment epithelium-derived factor inhibits retinal and
choroidal neovascularization. J Cell Physiol. 188:253–263. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Amaral J and Becerra SP: Effects of human
recombinant PEDF protein and PEDF-derived peptide 34-mer on
choroidal neovascularization. Invest Ophthalmol Vis Sci.
51:1318–1326. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang H, Cheng R, Liu G, Zhong Q, Li C, Cai
W, Yang Z, Ma J, Yang X and Gao G: PEDF inhibits growth of
retinoblastoma by anti-angiogenic activity. Cancer Sci.
100:2419–2425. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chazaud C, Bouillet P, Oulad-Abdelghani M
and Dollé P: Restricted expression of a novel retinoic acid
responsive gene during limb bud dorsoventral patterning and
endochondral ossification. Dev Genet. 19:66–73. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kawaguchi R, Yu J, Honda J, Hu J,
Whitelegge J, Ping P, Wiita P, Bok D and Sun H: A membrane receptor
for retinol binding protein mediates cellular uptake of vitamin A.
Science. 315:820–825. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Berry DC, Levi L and Noy N:
Holo-retinol-binding protein and its receptor STRA6 drive oncogenic
transformation. Cancer Res. 74:6341–6351. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Szeto W, Jiang W, Tice DA, Rubinfeld B,
Hollingshead PG, Fong SE, Dugger DL, Pham T, Yansura DG, Wong TA,
et al: Overexpression of the retinoic acid-responsive gene Stra6 in
human cancers and its synergistic induction by Wnt-1 and retinoic
acid. Cancer Res. 61:4197–4205. 2001.PubMed/NCBI
|
|
25
|
Carrera S, Cuadrado-Castano S, Samuel J,
Jones GD, Villar E, Lee SW and Macip S: Stra6, a retinoic
acid-responsive gene, participates in p53-induced apoptosis after
DNA damage. Cell Death Differ. 20:910–919. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dube DH and Bertozzi CR: Glycans in cancer
and inflammation-potential for therapeutics and diagnostics. Nat
Rev Drug Discov. 4:477–488. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bertozzi CR and Kiessling LL: Chemical
glycobiology. Science. 291:2357–2364. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen S, LaRoche T, Hamelinck D, Bergsma D,
Brenner D, Simeone D, Brand RE and Haab BB: Multiplexed analysis of
glycan variation on native proteins captured by antibody
microarrays. Nat Methods. 4:437–444. 2007.PubMed/NCBI
|
|
29
|
Taniguchi N and Kizuka Y: Glycans and
cancer: Role of N-glycans in cancer biomarker, progression and
metastasis, and therapeutics. Adv Cancer Res. 126:11–51. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pinho SS and Reis CA: Glycosylation in
cancer: Mechanisms and clinical implications. Nat Rev Cancer.
15:540–555. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Compagno D, Gentilini LD, Jaworski FM,
Pérez IG, Contrufo G and Laderach DJ: Glycans and galectins in
prostate cancer biology, angiogenesis and metastasis. Glycobiology.
24:899–906. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kreunin P, Zhao J, Rosser C, Urquidi V,
Lubman DM and Goodison S: Bladder cancer associated glycoprotein
signatures revealed by urinary proteomic profiling. J Proteome Res.
6:2631–2639. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tian M, Cui YZ, Song GH, Zong MJ, Zhou XY,
Chen Y and Han JX: Proteomic analysis identifies MMP-9, DJ-1 and
A1BG as overexpressed proteins in pancreatic juice from pancreatic
ductal adenocarcinoma patients. BMC cancer. 8:2412008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Libreros S and Iragavarapu-Charyulu V:
YKL-40/CHI3L1 drives inflammation on the road of tumor progression.
J Leukoc Biol. 98:931–936. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang XW, Cai CL, Xu JM, Jin H and Xu ZY:
Increased expression of chitinase 3-like 1 is a prognosis marker
for non-small cell lung cancer correlated with tumor angiogenesis.
Tumour Biol. 36:901–907. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kushner I: Regulation of the acute phase
response by cytokines. Perspect Biol Med. 36:611–622. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Davalieva K, Kiprijanovska S, Komina S,
Petrusevska G, Zografska NC and Polenakovic M: Proteomics analysis
of urine reveals acute phase response proteins as candidate
diagnostic biomarkers for prostate cancer. Proteome Sci. 13:22015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hoenerhoff MJ: Inflammation and cancer:
Partners in crime. Vet J. 206:1–2. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Raposo TP, Beirão BC, Pang LY, Queiroga FL
and Argyle DJ: Inflammation and cancer: Till death tears them
apart. Vet J. 205:161–174. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gerner C, Steinkellner W, Holzmann K, Gsur
A, Grimm R, Ensinger C, Obrist P and Sauermann G: Elevated plasma
levels of crosslinked fibrinogen gamma-chain dimer indicate
cancer-related fibrin deposition and fibrinolysis. Thromb Haemost.
85:494–501. 2001.PubMed/NCBI
|
|
41
|
Palumbo JS, Talmage KE, Massari JV, La
Jeunesse CM, Flick MJ, Kombrinck KW, Jirousková M and Degen JL:
Platelets and fibrin(ogen) increase metastatic potential by
impeding natural killer cell-mediated elimination of tumor cells.
Blood. 105:178–185. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bloomston M, Zhou JX, Rosemurgy AS,
Frankel W, Muro-Cacho CA and Yeatman TJ: Fibrinogen gamma
overexpression in pancreatic cancer identified by large-scale
proteomic analysis of serum samples. Cancer Res. 66:2592–2599.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang J, Xiong X, Wang X, Guo B, He K and
Huang C: Identification of peptide regions of SERPINA1 and ENOSF1
and their protein expression as potential serum biomarkers for
gastric cancer. Tumour Biol. 36:5109–5118. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kwon CH, Park HJ, Choi JH, Lee JR, Kim HK,
Jo HJ, Kim HS, Oh N, Song GA and Park DY: Snail and serpinA1
promote tumor progression and predict prognosis in colorectal
cancer. Oncotarget. 6:20312–20326. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kwon CH, Park HJ, Lee JR, Kim HK, Jeon TY,
Jo HJ, Kim DH, Kim GH and Park DY: Serpin peptidase inhibitor clade
A member 1 is a biomarker of poor prognosis in gastric cancer. Br J
Cancer. 111:1993–2002. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang X, Xiao Z, Liu X, Du L, Wang L, Wang
S, Zheng N, Zheng G, Li W, Zhang X, et al: The potential role of
ORM2 in the development of colorectal cancer. PLoS One.
7:e318682012. View Article : Google Scholar : PubMed/NCBI
|