Introduction
Hepatocellular carcinoma (HCC) is the sixth most
common type of cancer worldwide, ranking fifth and eighth in males
and females, respectively, and exhibits one of the highest
mortality rates (1). Hepatitis B
virus (HBV) and hepatitis C virus (HCV) are primary causes of HCC.
Chronic HBV infection is a dominant risk factor in the majority of
areas of Asia and Sub-Saharan Africa that have a high incidence of
HCC (2). The majority of patients
with HCC whoexperience HBV infectionexhibit cirrhosis, secondary to
the chronic necroinflammation (3).
HBV, an oncogenic virus, promotes HCC via indirect
(necroinflammation and regeneration injury) and direct (integration
of its DNA in the host genome) pathways (4). The aberrant expression of genes and
regulatory RNA moleculesare key nodes for the occurrence and
development of HCC.
Circular RNAs (circRNA/ciR), initially observed in
RNA viruses in the 1970s, have been identified as unique non-coding
RNA molecules (5). CircRNAsare a type
of endogenous RNA with a stable structure and tissue-specific
expression (6) and are widely present
in the cytoplasm of eukaryotic organisms, in the circular form
(7). CircRNA forms a covalently
closed continuous loop by means of unique non-canonical
‘head-to-tail’ splice without a free 3′ or 5′ end (8–10).
CircRNAs derive from non-linear reverse splicing or gene
rearrangement and circRNAs dominate the total spliced transcripts
(11). High-throughput sequencing has
enabled >25,000 types of circRNAs to be discovered in human
fibroblasts (12). In addition,
circRNAs may be formed in exons and introns, and cirRNAs with
either origin may function in the regulation of gene expression
(13).
A previous study demonstrated that circRNA served a
role in the level of miRNA-mediated regulation of gene expression
by sequestering the miRNAs. Furthermore, circRNAs are able to
regulate gene expression by acting as competing endogenous RNAs and
also termed miRNA ‘sponges’ (14).
CircRNAs contain multiple, tandem miRNA binding sites. CircRNAs
adsorb and sequester miRNAs to terminate the suppression of their
targets, and to modulate the expression levels of other associated
RNA molecules which share the same miRNA response elements (MREs)
(14–16). The interaction between circRNAs and
disease-associated miRNAs indicates that circRNAs are important for
disease regulation (17).
CircRNAs serve crucial roles in the development of
diseases, including nervous system disorders and atherosclerosis
(18,19). In addition, circRNAs have been
demonstrated to be involved in the neoplastic process (20); however, the molecular mechanisms
underlying the association of circRNAs with cancer remain unclear
(21).
To the best of our knowledge, a large-scale
microarray screening of HCC and the focus of circRNAs as biomarkers
of HCC has not been previously reported. The present study screened
dysregulated circRNAs expression in HCC tissues using a microarray
and annotated them for circRNA/microRNA (miRNA/miR)
interactions.
Patients and methods
Patients and clinical specimens
The total three paired HCC tissues and adjacent
non-tumorous tissues (NTs; Table I)
were collected from HCC surgical specimens between June 2012 and
December 2013 at Beijing YouAn Hospital, Capital Medical University
(Beijing, China). All tissue specimens were immediately preserved
in RNA-fixer reagent (BioTeke Corporation, Beijing, China)
following removal from the body and were stored at −80°C until use.
The corresponding adjacent NTs were taken 5 cm from the edge of the
cancer and contained no obvious tumor cells, as evaluated by an
experienced pathologist.
 | Table I.Clinical parameters of the three
patients with hepatitis B-associated HCC. |
Table I.
Clinical parameters of the three
patients with hepatitis B-associated HCC.
| Patient | Sex | Age, years | HBV-DNA, IU/ml | Cirrhosis |
Differentiation | HCC stage, TNM |
|---|
| 1 | M | 63 |
4.01×106 | Yes | Middle | T1N0M0 |
| 2 | M | 44 |
9.85×102 | Yes | Poor | T3aN0M0 |
| 3 | F | 45 |
1.98×103 | Yes | Middle | T1N0M0 |
All three HCC patients were diagnosed with HBV
infection. Tumors were staged according to the
tumor-node-metastasis (TNM) staging system (22). The three patients were diagnosed with
T1N0M0, T1N0M0, and T3aN0M0, respectively (Table I). No radiotherapy, chemotherapy or
targeted therapy was administered prior to surgery.
The present study protocol conforms to the ethical
guidelines of the 1975 Declaration of Helsinki and was approved by
the Ethics Committee of Beijing YouAn Hospital, Capital Medical
University (Beijing, China). Written informed consent was obtained
from all participants.
Total RNA extraction, labeling,
hybridization, and array scanning
Total tissue RNA was extracted from the HCC tissues
and paired adjacent NTs using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), following the
manufacturer's protocol. CircRNAs were treated with RNase R
(Epicentre; Illumina, Inc., San Diego, CA, USA) to remove linear
RNAs, according to the manufacturer's protocol. Each sample was
amplified and transcribed into fluorescent complementary RNA
utilizing a random priming method (Arraystar Super RNA Labeling
kit; Arraystar, Inc., Rockville, MD, USA). The labeled circRNAs
were hybridized onto the Arraystar Human circRNA Array (5,396 human
circRNA probes; cat. no. 6×7K; Arraystar, Inc.).
The labeled circRNAs were purified using an RNeasy
Mini kit (Qiagen, Inc., Valencia, CA, USA). The concentration and
specific activity of the labeled circRNAs (pmol Cy3/µg circRNA)
were measured using NanoDrop ND-1000 spectrophotometer (NanoDrop;
Thermo Fisher Scientific, Inc., Wilmington, DE, USA). A total of 1
µg of each labeled circRNA was dispensed into the gasket slide and
assembled to the circRNA expression microarray slide. The slides
were incubated for 17 h at 65°C in an Agilent Hybridization Oven
(Agilent Technologies, Inc., Santa Clara, CA, USA). The hybridized
arrays were washed, fixed and scanned using the Axon GenePix 4000B
microarray scanner (Molecular Devices, Sunnyvale, CA, USA).
Detection of expression profiling data
and differentially expressed data
Scanned images were imported into GenePix Pro
version 6.0 software (Axon; Molecular Devices) for grid alignment
and raw data extraction. Quantile normalization of raw data and
subsequent data processing were performed using the R software
package (version 3.1.2; Lucent Technologies, Inc.; Nokia, Espoo,
Finland). Low-intensity filtering was performed, and the circRNAs
with ≥2 of the 6 samples having ‘flags expressed’ (≥2 times
background standard deviation) were retained for further analysis.
The analysis outputs were filtered and the differentially expressed
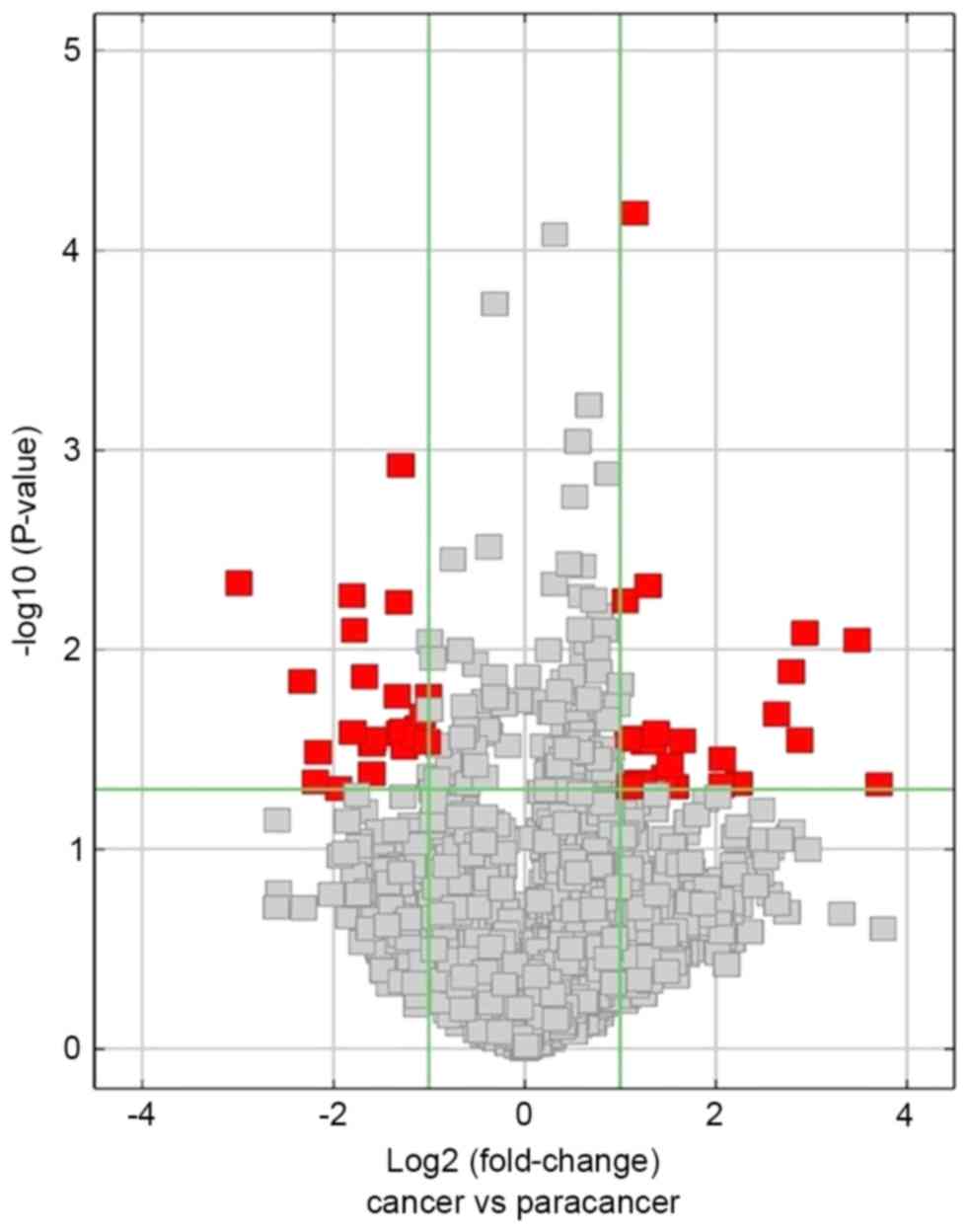
circRNAs were ranked according to fold-change and P-value.
Differentially expressed circRNAs were filtered and illustrated as
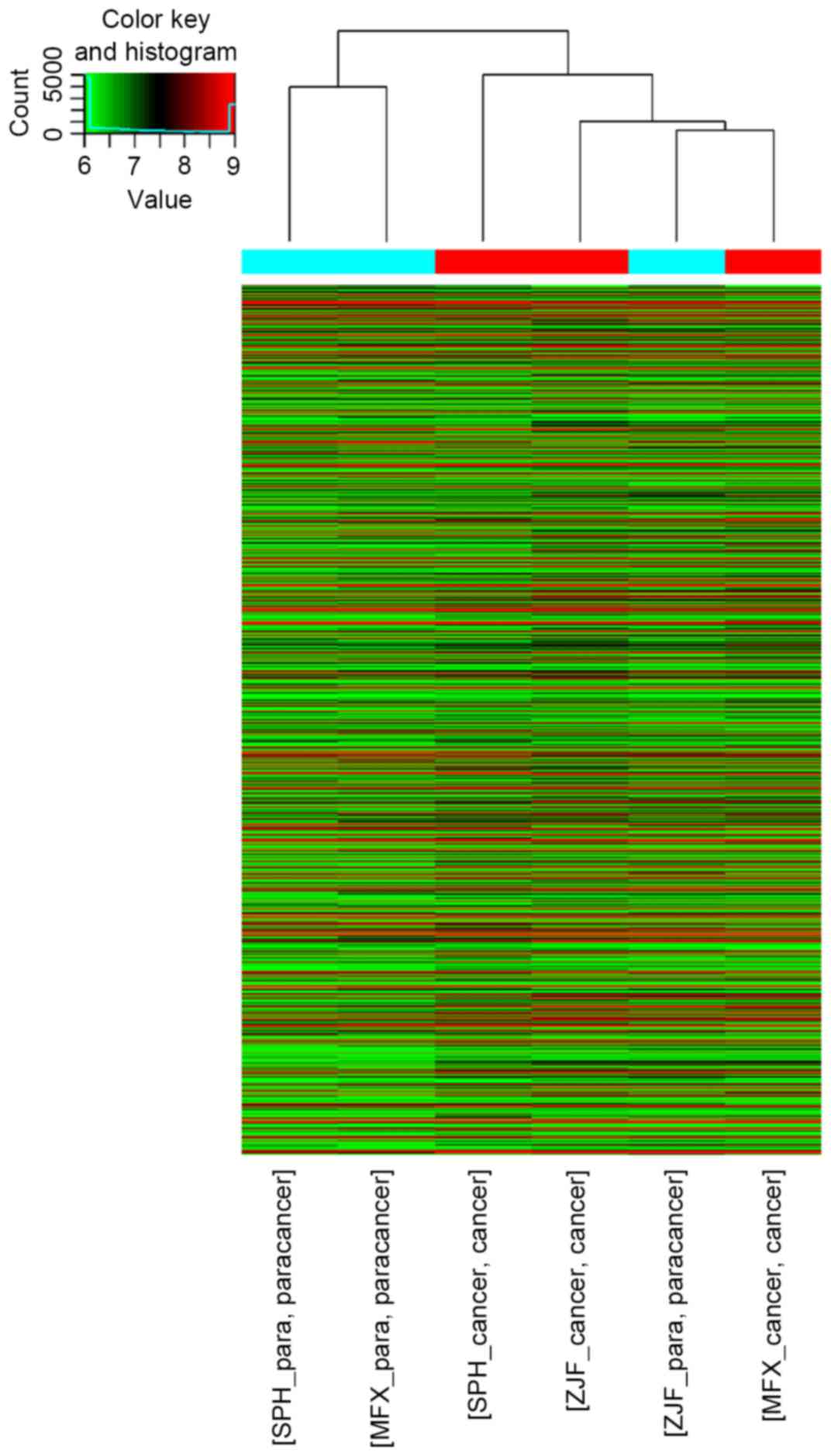
a volcano plot. Hierarchical clustering was performed to reveal the
distinguishable circRNAs expression pattern among samples.
Annotation for circRNA/miRNA
interaction
The circRNA/miRNA interaction was predicted using
Target Scan (www.targetscan.org/vert_71) and Miranda (www.microrna.org/microrna/home.do). All
differentially expressed circRNAs were annotated in detail using
the circRNA/miRNA interaction information.
Statistical analysis
All data were analyzed using SPSS (version 21.0; IBM
Corp., Armonk, NY, USA) and all results were presented as the mean
± standard deviation. Differences between two groups were estimated
using the Student's t-test, and fold-change ≥2.0 and P≤0.05 were
considered to indicate a statistically significant difference.
Results
circRNA expression profiles
A total of 5,396circRNAs were scanned and the array
image of each sample was demonstrated. Quantile normalization of
raw data and subsequent data processing were performed using the R
software package. The data demonstrated that 222,567,556 circRNAs
were upregulated (fold-change ≥2) and 125,439,219 circRNAs were
downregulated (fold-change ≥2; Fig.
1).
Differentially expressed circRNAs
The differentially expressed circRNAs with
statistical significance between the two groups (HCC tissues group
vs. NT group) were identified through using volcano plot filtering.
A total of 24 upregulatedcircRNAs and 23 downregulated circRNAs
were identified to be significant in HCC tissues compared with NTs
(fold-change ≥2; P≤0.05; Fig. 2;
Tables II and III). The top five upregulated circRNAs
were hsa_circRNA_104351, hsa_circRNA_102814, hsa_circRNA_103489,
hsa_circRNA_102109 and hsa_circRNA_100381. Furthermore, the top
five downregulated circRNAs were hsa_circRNA_100327,
hsa_circRNA_101764, hsa_circRNA_101092, hsa_circRNA_001225 and
hsa_circRNA_102904.
 | Table II.Significantly upregulated circRNAs in
HCC vs. NTs. |
Table II.
Significantly upregulated circRNAs in
HCC vs. NTs.
| circRNA | Aliasa | Gene symbol | FCb |
P-valuec |
circRNA_typed | Chromosome | Strand |
|---|
|
hsa_circRNA_103436 |
hsa_circ_0006884 | TIMMDC1 | 2.3799788 | 0.02920409 | Exonic | chr3 | + |
|
hsa_circRNA_103920 |
hsa_circ_0001513 | LNPEP | 2.1170474 | 0.02782662 | Exonic | chr5 | + |
|
hsa_circRNA_101842 |
hsa_circ_0007669 | SLC7A6 | 2.9740602 | 0.048252979 | Exonic | chr16 | + |
|
hsa_circRNA_104497 |
hsa_circ_0008419 | EXOC4 | 2.108166 | 0.028206035 | Exonic | chr7 | + |
|
hsa_circRNA_103792 |
hsa_circ_0003037 | TRIO | 2.7598056 | 0.043619449 | Exonic | chr5 | + |
|
hsa_circRNA_102814 |
hsa_circ_0003789 | TSN | 11.121925 | 0.008936648 | Exonic | chr2 | + |
|
hsa_circRNA_101169 |
hsa_circ_0028630 | RFC5 | 2.0770889 | 0.005712231 | Exonic | chr12 | + |
|
hsa_circRNA_102583 |
hsa_circ_0003449 | MEIS3 | 2.221169 | 0.00006516883 | Exonic | chr19 | − |
|
hsa_circRNA_102541 |
hsa_circ_0050834 | RYR1 | 2.2713685 | 0.045762338 | Exonic | chr19 | + |
|
hsa_circRNA_103588 |
hsa_circ_0068925 | FAM193A | 4.725986 | 0.046621193 | Exonic | chr4 | + |
|
hsa_circRNA_101828 |
hsa_circ_0039787 | C16orf70 | 2.4441344 | 0.004790339 | Exonic | chr16 | + |
|
hsa_circRNA_104543 |
hsa_circ_0083172 | DNAJB6 | 2.5453667 | 0.028933433 | Exonic | chr7 | + |
|
hsa_circRNA_000596 |
hsa_circ_0000661 | ADAMTS17 | 3.1358992 | 0.028697139 | Antisense | chr15 | + |
|
hsa_circRNA_102189 |
hsa_circ_0006942 | ATP5H | 6.2062939 | 0.020961504 | Exonic | chr17 | − |
|
hsa_circRNA_102230 |
hsa_circ_0046215 | HGS | 4.1868338 | 0.03517735 | Exonic | chr17 | + |
|
hsa_circRNA_100021 |
hsa_circ_0009456 | NPHP4 | 2.5935132 | 0.026262605 | Exonic | chr1 | − |
|
hsa_circRNA_100381 |
hsa_circ_0009109 | DCAF6 | 6.9136901 | 0.012898864 | Exonic | chr1 | + |
|
hsa_circRNA_102109 |
hsa_circ_0003650 | KPNB1 | 7.3568855 | 0.028283407 | Exonic | chr17 | + |
|
hsa_circRNA_100770 |
hsa_circ_0021506 | ANO5 | 2.3549632 | 0.04635107 | Exonic | chr11 | + |
|
hsa_circRNA_103616 |
hsa_circ_0069380 | TBC1D19 | 4.1619716 | 0.048093616 | Exonic | chr4 | + |
|
hsa_circRNA_104351 |
hsa_circ_0008537 | GLI3 | 13.0357676 | 0.04722754 | Exonic | chr7 | − |
|
hsa_circRNA_103489 |
hsa_circ_0067717 | RNF13 | 7.5996366 | 0.008247639 | Exonic | chr3 | + |
|
hsa_circRNA_102630 |
hsa_circ_0003287 | NBAS | 2.866929 | 0.038026903 | Exonic | chr2 | − |
|
hsa_circRNA_101539 |
hsa_circ_0005402 | ANXA2 | 2.141776 | 0.048215125 | Exonic | chr15 | − |
 | Table III.Significantly downregulated circRNAs
in HCC vs. NTs. |
Table III.
Significantly downregulated circRNAs
in HCC vs. NTs.
| circRNA | Aliasa | Gene symbol | FCb |
P-valuec |
circRNA_typed | Chromosome | Strand |
|---|
|
hsa_circRNA_101405 |
hsa_circ_0003670 | NEK9 | 2.4741971 | 0.026208247 | Exonic | chr14 | − |
|
hsa_circRNA_101764 |
hsa_circ_0038645 | PRKCB | 5.0118456 | 0.014366467 | Exonic | chr16 | + |
|
hsa_circRNA_104342 |
hsa_circ_0003162 | BBS9 | 2.5248442 | 0.017053943 | Exonic | chr7 | + |
|
hsa_circRNA_103627 |
hsa_circ_0069559 | APBB2 | 2.485444 | 0.005793933 | Exonic | chr4 | − |
|
hsa_circRNA_104044 |
hsa_circ_0075447 | GMDS | 2.3901855 | 0.030706961 | Exonic | chr6 | − |
|
hsa_circRNA_102446 |
hsa_circ_0049356 | CARM1 | 3.1862585 | 0.013653006 | Exonic | chr19 | + |
|
hsa_circRNA_104865 |
hsa_circ_0004928 | LPAR1 | 3.0272178 | 0.029853247 | Exonic | chr9 | − |
|
hsa_circRNA_001225 |
hsa_circ_0000305 | CELF1 | 4.4818374 | 0.032503965 | Intronic | chr11 | − |
|
hsa_circRNA_100327 |
hsa_circ_0014022 | TARS2 | 7.9507428 | 0.004638981 | Exonic | chr1 | + |
|
hsa_circRNA_100147 |
hsa_circ_0004240 | EIF3I | 2.1647679 | 0.024721895 | Exonic | chr1 | + |
|
hsa_circRNA_000963 |
hsa_circ_0001644 | BCLAF1 | 2.4269392 | 0.025372306 | Intronic | chr6 | − |
|
hsa_circRNA_103137 |
hsa_circ_0061817 | C2CD2 | 3.0063191 | 0.0279151 | Exonic | chr21 | − |
|
hsa_circRNA_100904 |
hsa_circ_0007767 | ALG8 | 2.0097943 | 0.016978441 | Exonic | chr11 | − |
|
hsa_circRNA_104349 |
hsa_circ_0079929 | CDK13 | 3.4763303 | 0.026028214 | Exonic | chr7 | + |
|
hsa_circRNA_102904 |
hsa_circ_0008459 | KANSL1L | 3.8502422 | 0.049696658 | Exonic | chr2 | − |
|
hsa_circRNA_100883 |
hsa_circ_0005918 | FCHSD2 | 3.028108 | 0.042033201 | Exonic | chr11 | − |
|
hsa_circRNA_101092 |
hsa_circ_0008594 | SRGAP1 | 4.566634 | 0.045816812 | Exonic | chr12 | + |
|
hsa_circRNA_100705 |
hsa_circ_0008898 | OAT | 2.4566529 | 0.001192501 | Exonic | chr10 | − |
|
hsa_circRNA_103361 |
hsa_circ_0001296 | SMARCC1 | 3.50005 | 0.005363723 | Exonic | chr3 | − |
|
hsa_circRNA_100395 |
hsa_circ_0015278 | KLHL20 | 2.0964102 | 0.021512811 | Exonic | chr1 | + |
|
hsa_circRNA_100748 |
hsa_circ_0020926 | STIM1 | 3.4384778 | 0.007965023 | Exonic | chr11 | + |
|
hsa_circRNA_102600 |
hsa_circ_0000958 | PPP1R12C | 2.2008434 | 0.027502872 | Exonic | chr19 | − |
|
hsa_circRNA_102261 |
hsa_circ_0046534 | TBCD | 2.0251354 | 0.028926837 | Exonic | chr17 | + |
Annotation for circRNA/miRNA
interactions
The circRNA/miRNA interaction was predicted using
the miRNA target prediction software. All differentially expressed
circRNAs (fold-change ≥2; P≤0.05) were annotated in detail using
the circRNA/miRNA interaction information (Tables IV and V). The most upregulated circRNA,
hsa_circRNA_104351, adjusts its MREs: hsa-miR-490-5p,
hsa-miR-876-5p, hsa-miR-619-3p, hsa-miR-619-3p, hsa-miR-331-3p and
hsa-miR-411-3p. Similarly, the most downregulated circRNA,
hsa_circRNA_100327, targets the following MREs: Hsa-miR-637,
hsa-miR-326, hsa-miR-330-5p, hsa-miR-646 and hsa-miR-24-3p.
 | Table IV.Upregulated circRNAs annotated with
circRNA/miRNA interaction information. |
Table IV.
Upregulated circRNAs annotated with
circRNA/miRNA interaction information.
| circRNA | MRE1 | MRE2 | MRE3 | MRE4 | MRE5 |
|---|
|
hsa_circRNA_103436 | hsa-miR-363-3p | hsa-miR-32-5p | hsa-miR-501-3p | hsa-miR-600 | hsa-miR-502-3p |
|
hsa_circRNA_103920 |
hsa-miR-1298-3p |
hsa-miR-29b-1-5p | hsa-miR-452-3p | hsa-miR-34c-5p | hsa-miR-9-5p |
|
hsa_circRNA_101842 | hsa-miR-489-3p | hsa-miR-892b |
hsa-miR-449c-5p | hsa-miR-665 | hsa-miR-138-5p |
|
hsa_circRNA_104497 | hsa-miR-139-5p | hsa-miR-141-5p | hsa-miR-597-3p | hsa-miR-632 | hsa-miR-648 |
|
hsa_circRNA_103792 | hsa-miR-215-3p | hsa-miR-627-5p | hsa-miR-532-3p |
hsa-miR-181b-5p | hsa-miR-630 |
|
hsa_circRNA_102814 |
hsa-miR-516a-5p | hsa-miR-224-5p | hsa-miR-501-5p | hsa-miR-429 |
hsa-miR-500a-5p |
|
hsa_circRNA_101169 | hsa-miR-591 | hsa-miR-1264 |
hsa-miR-517b-3p |
hsa-miR-517a-3p | hsa-miR-22-5p |
|
hsa_circRNA_102583 | hsa-miR-762 | hsa-miR-23b-3p | hsa-miR-765 | hsa-miR-675-5p | hsa-miR-423-5p |
|
hsa_circRNA_102541 | hsa-miR-486-3p | hsa-miR-328-5p |
hsa-miR-125a-3p | hsa-miR-296-5p | hsa-miR-873-5p |
|
hsa_circRNA_103588 | hsa-miR-127-3p | hsa-miR-452-5p | hsa-miR-22-5p |
hsa-miR-219a-2-3p | hsa-miR-509-5p |
|
hsa_circRNA_101828 | hsa-miR-619-3p | hsa-miR-877-5p |
hsa-miR-520f-3p | hsa-miR-452-5p | hsa-miR-490-3p |
|
hsa_circRNA_104543 |
hsa-miR-196b-3p |
hsa-miR-1298-3p | hsa-miR-345-3p | hsa-miR-1-3p |
hsa-miR-1224-3p |
|
hsa_circRNA_000596 | hsa-miR-647 |
|
|
|
|
|
hsa_circRNA_102189 | hsa-miR-588 | hsa-miR-659-3p | hsa-miR-490-3p | hsa-miR-152-5p | hsa-miR-330-3p |
|
hsa_circRNA_102230 | hsa-miR-588 | hsa-miR-10b-3p | hsa-miR-657 | hsa-miR-150-5p | hsa-miR-636 |
|
hsa_circRNA_100021 | hsa-miR-593-5p | hsa-miR-93-3p | hsa-miR-30b-3p | hsa-miR-766-3p | hsa-miR-484 |
|
hsa_circRNA_100381 | hsa-miR-525-5p | hsa-miR-544a | hsa-miR-345-3p | hsa-miR-577 | hsa-miR-587 |
|
hsa_circRNA_102109 |
hsa-miR-1301-3p | hsa-miR-20b-3p | hsa-miR-505-5p | hsa-miR-616-3p | hsa-miR-761 |
|
hsa_circRNA_100770 |
hsa-miR-19b-2-5p |
hsa-miR-19b-1-5p | hsa-miR-767-3p | hsa-miR-506-5p |
hsa-miR-550a-3p |
|
hsa_circRNA_103616 | hsa-miR-629-3p | hsa-miR-761 | hsa-miR-603 | hsa-miR-150-5p | hsa-miR-186-5p |
|
hsa_circRNA_104351 | hsa-miR-490-5p | hsa-miR-876-5p | hsa-miR-619-3p | hsa-miR-331-3p | hsa-miR-411-3p |
|
hsa_circRNA_103489 | hsa-miR-654-3p | hsa-miR-511-5p | hsa-miR-632 | hsa-miR-643 | hsa-miR-889-5p |
|
hsa_circRNA_102630 |
hsa-miR-19b-2-5p | hsa-miR-105-5p |
hsa-miR-29b-1-5p | hsa-miR-15b-3p | hsa-miR-29a-5p |
|
hsa_circRNA_101539 | hsa-miR-224-5p | hsa-miR-182-5p |
hsa-miR-208a-5p | hsa-miR-9-5p | hsa-miR-33b-5p |
 | Table V.Downregulated circRNAs annotated with
circRNA-miRNA interaction information. |
Table V.
Downregulated circRNAs annotated with
circRNA-miRNA interaction information.
| circRNA | MRE1 | MRE2 | MRE3 | MRE4 | MRE5 |
|---|
|
hsa_circRNA_101405 | hsa-miR-646 | hsa-miR-452-5p | hsa-miR-504-5p | hsa-miR-661 | hsa-miR-383-3p |
|
hsa_circRNA_101764 |
hsa-miR-181b-5p |
hsa-miR-181c-5p |
hsa-miR-181d-5p |
hsa-miR-181a-5p | hsa-miR-329-5p |
|
hsa_circRNA_104342 | hsa-miR-411-5p |
hsa-miR-520c-5p | hsa-miR-526a |
hsa-miR-518d-5p |
hsa-miR-518f-5p |
|
hsa_circRNA_103627 | hsa-miR-215-3p | hsa-miR-105-3p | hsa-miR-877-3p | hsa-miR-605-5p | hsa-miR-615-5p |
|
hsa_circRNA_104044 | hsa-miR-188-3p | hsa-miR-335-3p | hsa-miR-580-5p | hsa-miR-659-5p | hsa-miR-139-5p |
|
hsa_circRNA_102446 | hsa-miR-377-5p | hsa-miR-658 | hsa-miR-889-5p | hsa-miR-23b-5p | hsa-let-7i-5p |
|
hsa_circRNA_104865 |
hsa-miR-135a-3p | hsa-miR-7-5p | hsa-miR-588 | hsa-miR-383-5p | hsa-miR-620 |
|
hsa_circRNA_001225 | hsa-miR-30b-3p | hsa-miR-887-5p | hsa-miR-26b-3p | hsa-miR-485-5p | hsa-miR-363-5p |
|
hsa_circRNA_100327 | hsa-miR-637 | hsa-miR-326 | hsa-miR-330-5p | hsa-miR-646 | hsa-miR-24-3p |
|
hsa_circRNA_100147 |
hsa-miR-219a-1-3p | hsa-miR-486-3p | hsa-miR-493-3p |
hsa-miR-92a-2-5p | hsa-miR-495-3p |
|
hsa_circRNA_000963 | hsa-miR-324-3p | hsa-miR-214-5p | hsa-miR-423-3p |
|
|
|
hsa_circRNA_103137 | hsa-miR-497-5p |
hsa-miR-487b-3p | hsa-miR-25-3p | hsa-miR-92a-3p | hsa-miR-92b-3p |
|
hsa_circRNA_100904 | hsa-miR-627-3p | hsa-miR-539-5p | hsa-let-7i-5p | hsa-miR-190b | hsa-miR-98-5p |
|
hsa_circRNA_104349 | hsa-miR-212-5p |
hsa-miR-26a-1-3p |
hsa-miR-26a-2-3p | hsa-miR-639 |
hsa-miR-1271-3p |
|
hsa_circRNA_102904 |
hsa-miR-519c-3p |
hsa-miR-148a-3p |
hsa-miR-519a-3p | hsa-miR-567 | hsa-miR-205-3p |
|
hsa_circRNA_100883 | hsa-miR-217 |
hsa-miR-376a-2-5p | hsa-miR-92b-3p | hsa-miR-511-5p | hsa-miR-92a-3p |
|
hsa_circRNA_101092 | hsa-miR-631 | hsa-miR-612 | hsa-miR-221-5p | hsa-miR-889-3p |
hsa-miR-1298-3p |
|
hsa_circRNA_100705 |
hsa-miR-365b-3p |
hsa-miR-365a-3p | hsa-miR-670-3p | hsa-miR-34b-5p | hsa-miR-101-3p |
|
hsa_circRNA_103361 |
hsa-miR-449c-3p | hsa-miR-582-5p | hsa-miR-509-3p | hsa-miR-510-5p | hsa-miR-369-5p |
|
hsa_circRNA_100395 | hsa-miR-141-3p | hsa-miR-588 | hsa-miR-660-3p | hsa-miR-136-5p |
hsa-miR-200a-3p |
|
hsa_circRNA_100748 | hsa-miR-136-3p | hsa-miR-598-3p | hsa-miR-556-3p | hsa-miR-335-3p |
hsa-miR-499a-3p |
|
hsa_circRNA_102600 | hsa-miR-214-3p | hsa-miR-324-3p | hsa-miR-770-5p | hsa-miR-484 |
hsa-miR-513a-5p |
|
hsa_circRNA_102261 | hsa-miR-510-5p |
hsa-miR-1271-3p | hsa-miR-604 | hsa-miR-339-5p |
hsa-miR-146b-3p |
Discussion
Previously, circRNAs have been identified to serve a
role in a number of types of disease, including cancer. The
majority of circRNAs exhibit distinct tissue/developmental-stage
and diseases-specific expression in the process of organismal
differentiation, development and diseases (6). CircRNAs regulate cancer development via
a number of mechanisms, including miRNA sponges, modulating the Wnt
signaling pathway and epithelial-mesenchymal transition (23). The abnormal expression levels of
circRNAs have been observed in a number of types of cancer
(24), including: glioma (25–27), renal
cell carcinoma (28), bladder
carcinoma (29,30), laryngeal cancer (31), lung cancer (32,33),
breast cancer (34,35), esophageal cancer (36,37),
gastric cancer (38–40), colorectal cancer (41–46),
pancreatic ductal adenocarcinoma (47), cutaneous squamous cell carcinoma
(48), basal cell carcinoma (49) and ovarian cancer (50).
Abnormal circRNAs have been identified to be
involved in the occurrence and development of HCC (51). In a previous study, the RNA-seq data
from 50 paired HCC tissues and NTs were analyzed to identify the
function of circRNAs in HCC (51).
Protein-coding genes (PCGs) associated with the 2091 circRNAs were
identified to be enriched predominantly on
liver/cardiovascular-related diseases, and participated in a number
of metabolic processes (51). A total
of 45 circRNAs and 23 PCGs exhibited significant expression
alterations between HCC and normal tissues (51).
Representative circRNA, antisense to cerebellar
degeneration-related protein 1 (Cdr1as, also termed ciRS-7), has
been identified to act as an oncogene through targeting miR-7 in
HCC (52). Cdr1as expression was
upregulated and miR-7 expression was downregulated in HCC tissues.
Knockdown of Cdr1as downregulated the expression of miR-7, and
inhibited the expression of Cyclin E1 (CCNE1) and
phosphatidylinositol 3-kinase catalytic subunit delta (PIK3CD),
resulting in the suppression of proliferation and invasion of HCC
cells through targeting miR-7 (52).
Increased Cdr1as expression was significantly associated with
hepatic microvascular invasion (MVI), alpha-fetoprotein (AFP)
level, younger age and deterioration of HCC. The expression of
Cdr1as in HCC tissues with concurrent MVI was inversely associated
with miR-7 and positively associated with two miR-7-targeted genes
(PIK3CD and p70S6K) (53).
The expression of circZKSCAN1 (zinc finger with KRAB
and SCAN domains 1) and ZKSCAN1mRNA was significantly decreased in
the HCC samples compared with matched adjacent non-tumorous
tissues. The circZKSCAN1 levels varied in patients with tumor
numbers, cirrhosis, vascular invasion and the tumor grade.
ZKSCAN1mRNA primarily regulated cellular metabolism, whereas
circZKSCAN1 mediated a number of cancer-related signaling pathways,
suggesting a non-redundant role for ZKSCAN1mRNA and circZKSCAN1
(54).
CircRNAs are abundant, evolutionally conserved and
relatively stable in the cytoplasm and therefore may be valuable
for cancer diagnosis (12,14). A previous study demonstrated that a
total of 174 and 353 circRNAs were upregulated and downregulated in
HCC tissues, respectively, according to microarray analysis
(55). hsa_circ_0004018 may be
involved in cancer-related pathways via interactions with miRNAs
(55). hsa_circ_0004018 is
downregulated in HCC tissues and HCC lines, and a decreased
hsa_circ_0004018 level is associated with serum AFP level, tumor
diameters, differentiation, Barcelona Clinic Liver Cancer stage
(56) and TNM stage. An additional
study identified that hsa_circ_0001649 expression was significantly
downregulated in HCC tissues, using the reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
(57). hsa_circ_0001649 expression
was associated with tumor size and the occurrence of tumor embolus
in HCC; therefore, hsa_circ_0001649 may function in the
tumorigenesis and metastasis of HCC and serve as a novel potential
biomarker (57). In addition, GO
analysis revealed that hsa_circ_0005075 may participate in cell
adhesion during HCC development. Upregulated hsa_circ_0005075
exhibited an association with HCC tumor size and revealed
diagnostic potential (58).
The present study compared the circRNA expression
profiles between HCC tissue and adjacent NTs using microarray
analysis with 5,396 circRNA probes. On the basis of the microarray
data, 222,567,556 upregulated circRNAs and 125,439,219
downregulated circRNAs were identified in HCC tissues compared with
adjacent NTs. Further analysis identified 24 upregulated and 23
downregulated significantly circRNAs (fold-change ≥2; P≤0.05) in
HCC tissues compared with NTs. The results of the present study
demonstrated that the circRNA expression profiles of HCC tissues
differ from that of NTs.
Computer and database analysis annotated the MREs
associated with the abnormally expressed circRNAs. The upregulated
circRNAs may suppress miRNA expression. On the contrary,
downregulated circRNAs may increase miRNA expression. The circRNAs,
as a sponge for miRNAs, may be associated with the occurrence and
progression of HCC, and may provide a novel approach to identify
the underlying molecular basis of HCC. Furthermore, the identified
differentially expressed circRNAs may be used as biomarkers for
HCC.
In the present study, the abnormal expression levels
of three paired HCC tissues were analyzed. Additional studies, with
larger cohorts and using RT-qPCR, are required to validate the
results from the present study. The data from the present study
indicated that abnormal expression of certain circRNAs in HCC
tissues and abnormal circRNAs may be novel biomarkers for the
diagnosis of HCC.
Acknowledgements
The present study was supported by the Capital
Science and Technology Development Fund (grant no. 2014-1-2181),
the Beijing Municipal Administration of Hospitals Clinical Medicine
Development of Special Funding (grant no. ZYLX201610), the Liver
and AIDS Fund of Beijing YouAn Hospital (grant no. BJYAH-2011-032)
and National Sci-Tech Support Plan (grant no. 2015BAI02B00).
References
|
1
|
Zhu RX, Seto WK, Lai CL and Yuen MF:
Epidemiology of hepatocellular carcinoma in the Asia-Pacific
region. Gut Liver. 10:332–339. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
de Martel C, Maucort-Boulch D, Plummer M
and Franceschi S: World-wide relative contribution of Hepatitis B
and C viruses in hepatocellular carcinoma. Hepatology.
62:1190–1200. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
El-Serag HB: Epidemiology of viral
hepatitis and hepatocellular carcinoma. Gastroenterology.
142:1264–1273.e1. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Michielsen P and Ho E: Viral hepatitis B
and hepatocellular carcinoma. Acta Gastroenterol Belg. 74:4–8.
2011.PubMed/NCBI
|
|
5
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:pp. 3852–3856. 1976;
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hsu MT and Coca-Prados M: Electron
microscopic evidence for the circular form of RNA in the cytoplasm
of eukaryotic cells. Nature. 280:339–340. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hentze MW and Preiss T: Circular RNAs:
Splicing's enigma variations. EMBO J. 32:923–925. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wilusz JE and Sharp PA: Molecular biology.
A circuitous route to noncoding RNA. Science. 340:440–441. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu Q, Wang Y, Cao M, Pantaleo V, Burgyan
J, Li WX and Ding SW: Homology-independent discovery of replicating
pathogenic circular RNAs by deep sequencing and a new computational
algorithm. Proc Natl Acad Sci USA. 109:pp. 3938–3943. 2012;
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wilusz JE and Sharp PA: Molecular biology.
A circuitous route to noncoding RNA. Science. 340:440–441. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ghosal S, Das S, Sen R, Basak P and
Chakrabarti J: Circ2Traits: A comprehensive database for circular
RNA potentially associated with disease and traits. Front Genet.
4:2832013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z
and Sharpless NE: Expression of linear and novel circular forms of
an INK4/ARF-associated non-coding RNA correlates with
atherosclerosis risk. PLoS Genet. 6:e10012332010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen YT, Rettig WJ, Yenamandra AK, Kozak
CA, Chaganti RS, Posner JB and Old LJ: Cerebellar
degeneration-related antigen: A highly conserved neuroectodermal
marker mapped to chromosomes X in human and mouse. Proc Natl Acad
Sci USA. 87:pp. 3077–3081. 1990; View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hansen TB, Kjems J and Damgaard CK:
Circular RNA and miR-7 in cancer. Cancer Res. 73:5609–5612. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li J, Yang J, Zhou P, Le Y, Zhou C, Wang
S, Xu D, Lin HK and Gong Z: Circular RNAs in cancer: Novel insights
into origins, properties, functions and implications. Am J Cancer
Res. 5:472–480. 2015.PubMed/NCBI
|
|
22
|
Okuda K, Ohtsuki T, Obata H, Tomimatsu M,
Okazaki N, Hasegawa H, Nakajima Y and Ohnishi K: Natural history of
hepatocellular carcinoma and prognosis in relation to treatment.
Study of 850 patients. Cancer. 56:918–928. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He J, Xie Q, Xu H, Li J and Li Y: Circular
RNAs and cancer. Cancer Lett. 396:138–144. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rong D, Tang W, Li Z, Zhou J, Shi J, Wang
H and Cao H: Novel insights into circular RNAs in clinical
application of carcinomas. Onco Targets Ther. 10:2183–2188. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Song X, Zhang N, Han P, Moon BS, Lai RK,
Wang K and Lu W: Circular RNA profile in gliomas revealed by
identification tool UROBORUS. Nucleic Acids Res. 44:e872016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang P, Qiu Z, Jiang Y, Dong L, Yang W, Gu
C, Li G and Zhu Y: Silencing of cZNF292 circular RNA suppresses
human glioma tube formation via the Wnt/β-catenin signaling
pathway. Oncotarget. 7:63449–63455. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Barbagallo D, Condorelli A, Ragusa M,
Salito L, Sammito M, Banelli B, Caltabiano R, Barbagallo G, Zappalà
A, Battaglia R, et al: Dysregulated miR-671-5p/CDR1-AS/CDR1/VSNL1
axis is involved in glioblastoma multiforme. Oncotarget.
7:4746–4759. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang K, Sun Y, Tao W, Fei X and Chang C:
Androgen receptor (AR) promotes clear cell renal cell carcinoma
(ccRCC) migration and invasion via altering the
circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett.
394:1–12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhong Z, Lv M and Chen J: Screening
differential circular RNA expression profiles reveals the
regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in
bladder carcinoma. Sci Rep. 6:309192016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang M, Zhong Z, Lv M, Shu J, Tian Q and
Chen J: Comprehensive analysis of differentially expressed profiles
of lncRNAs and circRNAs with associated co-expression and competing
endogenous RNAs networks in bladder carcinoma. Oncotarget.
7:47186–47200. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xuan LJ, Qu LM, Zhou H, Wang P, Yu H, Wu
T, Wang X, Li Q, Tian L, Liu M and Sun Y.: Circular RNA: A novel
biomarker for progressive laryngeal cancer. Am J Transl Res.
8:932–939. 2016.PubMed/NCBI
|
|
32
|
Yao JT, Zhao SH, Liu QP, Lv MQ, Zhou DX,
Liao ZJ and Nan KJ: Over-expression of CircRNA-100876 in non-small
cell lung cancer and its prognostic value. Pathol Res Pract.
213:453–456. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wan L, Zhang L, Fan K, Cheng ZX, Sun QC
and Wang JJ: Circular RNA-ITCH suppresses lung cancer proliferation
via inhibiting the Wnt/β;-catenin pathway. Biomed Res Int.
2016:15794902016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lü L, Sun J, Shi P, Kong W, Xu K, He B,
Zhang S and Wang J: Identification of circular RNAs as a promising
new class of diagnostic biomarkers for human breast cancer.
Oncotarget. 8:44096–44107. 2017.PubMed/NCBI
|
|
35
|
Nair AA, Niu N, Tang X, Thompson KJ, Wang
L, Kocher JP, Subramanian S and Kalari KR: Circular RNAs and their
associations with breast cancer subtypes. Oncotarget.
7:80967–80979. 2016.PubMed/NCBI
|
|
36
|
Su H, Lin F, Deng X, Shen L, Fang Y, Fei
Z, Zhao L, Zhang X, Pan H, Xie D, et al: Profiling and
bioinformatics analyses reveal differential circular RNA expression
in radioresistant esophageal cancer cells. J Transl Med.
14:2252016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li F, Zhang L, Li W, Deng J, Zheng J, An
M, Lu J and Zhou Y: Circular RNA ITCH has inhibitory effect on ESCC
by suppressing the Wnt/β-catenin pathway. Oncotarget. 6:6001–6013.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li P, Chen S, Chen H, Mo X, Li T, Shao Y,
Xiao B and Guo J: Using circular RNA as a novel type of biomarker
in the screening of gastric cancer. Clin Chim Acta. 444:132–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li P, Chen H, Chen S, Mo X, Li T, Xiao B,
Yu R and Guo J: Circular RNA 0000096 affects cell growth and
migration in gastric cancer. Br J Cancer. 116:626–633. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen S, Li T, Zhao Q, Xiao B and Guo J:
Using circular RNA hsa_circ_0000190 as a new biomarker in the
diagnosis of gastric cancer. Clin Chim Acta. 466:167–171. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tang W, Ji M, He G, Yang L, Niu Z, Jian M,
Wei Y, Ren L and Xu J: Silencing CDR1as inhibits colorectal cancer
progression through regulating microRNA-7. Onco Targets Ther.
10:2045–2056. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Huang G, Zhu H, Shi Y, Wu W, Cai H and
Chen X: cir-ITCH plays an inhibitory role in colorectal cancer by
regulating the Wnt/β-catenin pathway. PLoS One. 10:e01312252015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang X, Zhang Y, Huang L, Zhang J, Pan F,
Li B, Yan Y, Jia B, Liu H, Li S and Zheng W: Decreased expression
of hsa_circ_001988 in colorectal cancer and its clinical
significances. Int J Clin Exp Pathol. 8:16020–16025. 2016.
|
|
44
|
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y,
Yang S, Zeng Z, Liao W, Ding Y and Liang L: Emerging roles of
circRNA_001569 targeting miR-145 in the proliferation and invasion
of colorectal cancer. Oncotarget. 7:26680–26691. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhu M, Xu Y, Chen Y and Yan F: Circular
BANP, an upregulated circular RNA that modulates cell proliferation
in colorectal cancer. Biomed Pharmacother. 88:138–144. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Dou Y, Cha DJ, Franklin JL, Higginbotham
JN, Jeppesen DK, Weaver AM, Prasad N, Levy S, Coffey RJ, Patton JG
and Zhang B: Circular RNAs are down-regulated in KRAS mutant colon
cancer cells and can be transferred to exosomes. Sci Rep.
6:379822016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Qu S, Song W, Yang X, Wang J, Zhang R,
Zhang Z, Zhang H and Li H: Microarray expression profile of
circular RNAs in human pancreatic ductal adenocarcinoma. Genom
Data. 5:385–387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sand M, Bechara FG, Gambichler T, Sand D,
Bromba M, Hahn SA, Stockfleth E and Hessam S: Circular RNA
expression in cutaneous squamous cell carcinoma. J Dermatol Sci.
83:210–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sand M, Bechara FG, Sand D, Gambichler T,
Hahn SA, Bromba M, Stockfleth E and Hessam S: Circular RNA
expression in basal cell carcinoma. Epigenomics. 8:619–632. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ahmed I, Karedath T, Andrews SS, Al-Azwani
IK, Mohamoud YA, Querleu D, Rafii A and Malek JA: Altered
expression pattern of circular RNAs in primary and metastatic sites
of epithelial ovarian carcinoma. Oncotarget. 7:36366–36381. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li Y, Dong Y, Huang Z, Kuang Q, Wu Y, Li Y
and Li M: Computational identifying and characterizing circular
RNAs and their associated genes in hepatocellular carcinoma. PLoS
One. 12:e01744362017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yu L, Gong X, Sun L, Zhou Q, Lu B and Zhu
L: The circular RNA Cdr1as act as an oncogene in hepatocellular
carcinoma through targeting miR-7 expression. PLoS One.
11:e01583472016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Xu L, Zhang M, Zheng X, Yi P, Lan C and Xu
M: The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of
hepatic microvascular invasion in hepatocellular carcinoma. J
Cancer Res Clin Oncol. 143:17–27. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yao Z, Luo J, Hu K, Lin J, Huang H, Wang
Q, Zhang P, Xiong Z, He C, Huang Z, et al: ZKSCAN1 gene and its
related circular RNA (circZKSCAN1) both inhibit hepatocellular
carcinoma cell growth, migration, and invasion but through
different signaling pathways. Mol Oncol. 11:422–437. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Fu L, Yao T, Chen Q, Mo X, Hu Y and Guo J:
Screening differential circular RNA expression profiles reveals
hsa_circ_0004018 is associated with hepatocellular carcinoma.
Oncotarget. 8:58405–58416. 2017.PubMed/NCBI
|
|
56
|
Bruix J and Sherman M: American
Association for the Study of Liver Diseases: Management of
hepatocellular carcinoma: An update. Hepatology. 53:1020–1022.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z,
Yang J, Fan J, Liu L and Qin W: Hsa_circ_0001649: A circular RNA
and potential novel biomarker for hepatocellular carcinoma. Cancer
Biomark. 16:161–169. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Shang X, Li G, Liu H, Li T, Liu J, Zhao Q
and Wang C: Comprehensive circular RNA profiling reveals that
hsa_circ_0005075, a new circular RNA biomarker, is involved in
hepatocellular carcinoma development. Medicine. 95:e38112016.
View Article : Google Scholar : PubMed/NCBI
|