Introduction
Colorectal cancer is one of the most prevalent types
of malignant tumor and is a frequent cause of cancer-associated
mortality worldwide (1,2). The pathogenesis of colorectal cancer is
known to occur through progression between adenomas and carcinomas
(3). Loss of cell polarity and
disruption of intracellular adhesion are frequently observed during
this process and serve important functions in cancer progression
(4,5).
The histological types of colorectal adenocarcinoma depend on the
degree of glandular differentiation and cellular polarity (6). Tight junctions (TJs), which are the most
apical type of cellular junction, maintain cell-cell adhesion,
perform important roles in selective barriers and establish
cellular polarity in epithelial cells (7–9). TJs have
been associated with the regulation of cell proliferation and
differentiation (10). TJs are
typically lost in cancer, and this loss is involved in the invasive
and metastatic phenotypes of tumor cells (11–14).
Membrane proteins, including claudin (15) and occludin (16), have been associated with TJs and are
involved in the regulation of cell proliferation (17,18).
Claudin and occludin are tetraspanin proteins that extend their
extracellular loops across adjacent cells (19). The C-terminal domain of claudins has a
PDZ-binding motif that interacts with PDZ-domain proteins,
including zona occludins (ZO)-1, −2 and −3 (20). The claudin family is composed of 27
members that form homotypic and/or heterotypic interactions with
each other (21,22). Among the numerous TJ proteins,
claudins are key functional proteins, and are expressed in various
types of tissues and cells (23,24).
Additionally, claudins have been shown to have a significant effect
on the biological behavior of tumor progression (22,25).
Previous studies reported abnormal expression of
several claudins. The expression of claudin-1 was observed to be
upregulated in colorectal cancer (19,26),
melanoma (27) and nasopharyngeal
carcinoma cells (28). The expression
of claudin-2 was also upregulated in lung adenocarcinoma cells
(29), whereas claudin-7 was
demonstrated to be downregulated in invasive ductal carcinomas of
the breast (30). The expression of
claudins-1, −3 and −4 was previously reported to be upregulated in
human colorectal cancer (31).
Furthermore, several studies have reported that the translocation
of claudins is associated with tumor cell proliferation and
survival (32,33). The expression of claudin-1 is
upregulated in human colon cancers, particularly in the metastatic
region, and is frequently mislocalized from the cell membrane to
the cytoplasm and nucleus (19). The
predominant cytoplasmic and nuclear localization of claudin-1 was
revealed to exhibit anti-apoptotic activity in nasopharyngeal
carcinoma cells (28). In addition,
the nuclear distribution of claudin-2 has been shown to contribute
to enhancing the proliferation of lung adenocarcinoma cells
(34). However, the mechanisms
underlying the different functions of claudins in tumorigenesis
have not been clearly defined to date. Based on the aforementioned
findings, it was hypothesized that the localization of claudins has
specific significance in the process of cellular transformation in
colorectal adenocarcinomas.
Therefore, the present study examined the cellular
localization of claudins and determined whether claudins are
associated with clinicopathological parameters of colorectal
adenocarcinomas. Immunocytochemical and immunohistochemical
staining were conducted using primary antibodies against claudin-3
and claudin-7. Cellular expression of claudin-3 and claudin-7 was
investigated in the human colorectal adenocarcinoma cell lines
Caco-2 and SW620. The clinicopathological associations (age, sex,
histological type, lymphatic invasion, venous invasion, lymph node
metastasis and depth of tumor invasion) with the expression of
claudin-3 and claudin-7 were then examined in colorectal
adenocarcinoma tissues.
Materials and methods
Cell culture
SW620 cells were obtained from Dainippon Sumimoto
Pharma Co., Ltd. (Osaka, Japan), while Caco-2 cells were obtained
from the American Type Culture Collection (Manassas, VA, USA).
These cells were cultured in Dulbecco's modified Eagle's medium
(Wako Pure Chemical Industries, Ltd., Osaka, Japan) supplemented
with 10% heat-inactive fetal bovine serum (Biological Industries
Israel Beit-Haemek, Kibbutz Beit-Haemek, Israel) and 1%
penicillin-streptomycin at 37°C in a humidified atmosphere of 5%
CO2/air.
RNA analysis
Total RNA was purified using an RNeasy Plus Mini kit
(Qiagen China Co., Ltd., Shanghai, China). In the reverse
transcription polymerase chain reaction (RT-PCR) analysis of
claudin-3, claudin-7 and GAPDH expression, RT was performed with 1
µg of RNA using a SuperScript VILO cDNA Synthesis kit (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), and the reaction
product was subsequently amplified by PCR using primers as follows:
Claudin 3 forward, 5′-CAACACCATTATCCGGGACT-3′ and reverse,
5′-CTTGGTGGCCGTGTACTTCT-3′; claudin 7 forward,
5′-AATTTTCATCGTGGCAGGTC-3′ and reverse, 5′-AGGACAGGAACAGGAGAGCA-3′;
and GAPDH forward, 5′-CAACGACCACTTTGTCAAGC-3′ and reverse,
5′-TCTTCAAGGGGTCTACATGG-3′. The PCR reactions were conducted under
the following conditions: 95°C for 10 min followed by 35 cycles of
95°C for 30 sec, 63°C for 30 sec, 71°C for 1 min. The PCR products
were separated on a 2% agarose gel and stained with ethidium
bromide. Quantitative (q)PCR was performed with a Power SYBR-Green
RNA-to-CT 1-Step kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.) using the Applied Biosystems StepOnePlus™ Real-Time PCR
system (Thermo Fisher Scientific, Inc.). The qPCR reactions were
conducted under the following conditions: 48°C for 30 min, 95°C for
10 min followed by 40 cycles of 95°C for 15 sec, 60°C for 1 min and
melting curve at 95°C for 15 sec, 60°C for 15 sec and 95°C for 15
sec. The aforementioned primers were used, and the expression
levels of the gene of interest were normalized to those of the
endogenous control GAPDH messenger RNA (mRNA) (35). This protocol was repeated 3 times. The
relative quantity of each gene was determined using the StepOnePlus
Software (version 2.3; Applied Biosystems; Thermo Fisher
Scientific, Inc.).
Protein extraction and western
blotting
Caco-2 (3×105) and SW620
(3×105) cells were seeded on 40-mm dishes and incubated
at 37°C until confluent. Cells were washed with ice-cold PBS (Wako
Pure Chemical Industries, Ltd.) twice, harvested with ice-cold
lysis buffer (1% Triton X-100, 150 mM NaCl, 10 mM MgCl2,
1 mM EDTA and 50 mM Tris-HCl, pH 7.5), and incubated on ice for 20
min. The lysates were centrifuged at 20,600 × g for 20 min at 4°C.
The supernatants were collected and stored at −80°C for subsequent
analysis. Western blotting was performed subsequent to protein
separation on a 15% polyacrylamide gel with 20 µl of lysis buffer
per lane. Protein samples separated by SDS-PAGE were transferred to
a polyvinylidene fluoride (PVDF) membrane. The PVDF membrane was
blocked at room temperature for 1 h in TBS-Tween-20 containing 2%
skimmed milk powder, and then incubated with a primary antibody
against rabbit polyclonal claudin-3 (cat. no. ab15102; 1:300;
Abcam, Cambridge, UK), rabbit polyclonal claudin-7 (cat. no.
ab27487; 1:1,000; Abcam) or rabbit monoclonal β-actin (cat. no.
4970S; 1:1,000; Cell Signaling Technology Inc., Danvers, MA, USA).
Upon washing, the membranes were incubated with a horseradish
peroxidase-conjugated anti-rabbit antibody (cat. no. 7074S;
1:5,000; Cell Signaling Technology, Inc., Osaka, Japan) at room
temperature for 1 h. Immunoreactivity was detected with an enhanced
chemiluminescence kit Chemi-Lumi One L (Nacalai Tesque, Inc.,
Kyoto, Japan). Protein levels were quantified with ImageJ (version
1.51k) (36), and β-actin was used as
the internal control.
Immunocytochemistry
Caco-2 cells were seeded on Nunc Lab-Tek chamber
slides (Thermo Fisher Scientific, Inc.) and incubated at 37°C until
confluent. Cells were subsequently washed twice with PBS and fixed
with 3.7% formalin at room temperature for 15 min. The cells were
then permeabilized with 0.25% Triton X-100 in PBS for 10 min and
blocked with 2% bovine serum albumin (Wako Pure Chemical
Industries, Ltd.) in PBS for 1 h. This was followed by incubations
at room temperature for 2 h with anti-claudin-3 (1:100),
anti-claudin-7 (1:100), mouse monoclonal anti-β-catenin (cat. no.
M3539; 1:200; Dako; Agilent Technologies, Inc., Santa Clara, CA,
USA) and mouse monoclonal anti-E-cadherin (cat. no. M3612; 1:200;
Dako; Agilent Technologies, Inc.) antibodies. Cells were rinsed
three times with PBS and incubated at room temperature for 15 min
with secondary antibodies provided in the Histofine Simple Stain
MAX PO (MULTI) detection reagent (universal immunoperoxidase
polymer, anti-mouse and anti-rabbit; Nichirei Biosciences, Inc.,
Tokyo, Japan) according to the manufacturer's protocol. Cells were
then stained with 3,3′-diaminobenzidine tetrahydrochloride (DAB)
using the Histofine DAB substrate kit (Nichirei Biosciences, Inc.,
Tokyo, Japan) at room temperature for 1 min. Sections were
counterstained with Mayer's hematoxylin at room temperature for 4
min, dehydrated in a graded series of ethanol (Muto Pure Chemicals,
Co., Ltd., Tokyo, Japan) from 80 to 100%, cleared with 99% xylene
(Muto Pure Chemicals, Co., Ltd.) for 15 min and mounted in malinol
(Muto Pure Chemicals, Co., Ltd.). The expression of claudin-3 and
claudin-7 in cells was observed at magnification, ×200 using a
light microscope (BX53; Olympus Corporation, Tokyo, Japan) and
images were captured with a microscopic camera (DP20-5; Olympus
Corporation).
Patients
Tissues were obtained from 100 patients with
colorectal adenocarcinoma who underwent surgical resection between
January 2010 and June 2014 at Kagawa University Hospital (Kagawa,
Japan). The histological findings of these patients, in addition to
their lymph node metastases, venous invasion and tumor node
metastasis stages, were evaluated based on the Japanese
Classification of Colorectal Carcinoma (8th edition) (37). All subjects provided written informed
consent. The present study was conducted with the approval of the
Institutional Research Ethics Committee of Kagawa Prefectural
University of Health Sciences (Kagawa, Japan).
Immunohistochemistry
Tissues were obtained from primary tumors
(4-µm-thick), deparaffinized in 99% xylene for 15 min, and then
rehydrated in a graded series of ethanol, followed by antigen
retrieval by microwave heating for 15 min at 2 kW in 0.01 M citrate
buffer (pH 6.0). Endogenous peroxidase activity was blocked using
3% hydrogen peroxide (Wako Pure Chemical Industries, Ltd.) at room
temperature for 10 min, and non-specific binding was blocked with
PBS containing 0.1% skimmed milk powder (Wako Pure Chemical
Industries, Ltd.) at room temperature for 10 min. Sections were
incubated at room temperature for 2 h with the aforementioned
anti-claudin-3 (1:200) and anti-claudin-7 (1:300) primary
antibodies. Slides were rinsed three times with PBS and incubated
at room temperature for 15 min with the secondary antibodies of
Histofine Simple Stain MAX PO (MULTI), according to the
manufacturer's protocol, and subsequently stained with DAB using
the aforementioned DAB substrate kit (Nichirei Biosciences, Inc.,
Tokyo, Japan) at room temperature for 1 min. Sections were
counterstained with Mayer's hematoxylin at room temperature for 4
min, dehydrated in a graded series of ethanol from 80 to 100%,
cleared with 99% xylene for 15 min and then mounted in malinol.
Normal colorectal mucosa samples were used as positive controls.
The expression of claudin-3 and claudin-7 in tissues was observed
at ×200 magnification using a light microscope (BX53; Olympus
Corporation), and images were captured with a microscopic camera
(DP20-5; Olympus Corporation).
Immunoscore
The classification of membranous claudin expression
was based on the criteria of Jung et al (38). Briefly, immunostaining for claudin-3
or claudin-7 was assessed using the following scoring: No staining,
0; <25% cells positive and incomplete membranous staining, 1+;
25–50% cells positive and incomplete membranous staining, 2+;
50–75% cells positive and complete or incomplete membranous
staining, 3+; and >75% cells positive and complete membranous
staining, 4+. In the evaluation, the expression levels of claudin-3
and claudin-7 were grouped into negative (0 and 1+) and positive
(2+, 3+ and 4+) groups.
The classification of nuclear claudin expression was
scored by a similar method of membranous staining: Negative
expression, 0 (0%) and 1+ (<25%); positive expression, 2+
(25–50%), 3+ (50–75%) and 4+ (>75%).
Statistical analysis
Data was expressed as the mean ± standard deviation.
Differences between two groups were evaluated with the paired
Student's t-test. Univariate analysis was performed using the
χ2 or Fisher's exact test for categorical data. All
statistical analyses were performed using SPSS 23.0 software (IBM
SPSS, Armonk, NY, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Claudin-3 and claudin-7 are expressed
in human colorectal adenocarcinoma cell lines
In order to study the expression of claudin-3 and
claudin-7 in human colorectal adenocarcinoma cell lines, RT-PCR was
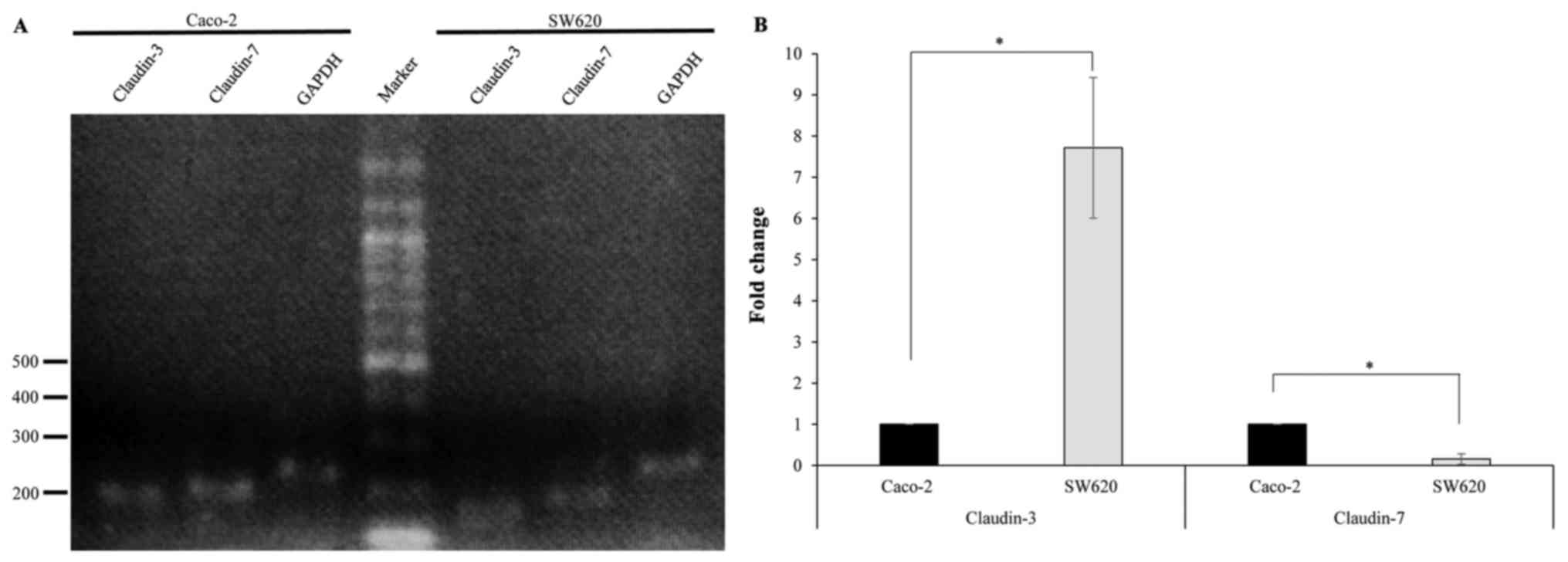
performed using specific primers. RT-PCR amplification yielded DNA
fragments of the expected size (~200 bp), indicating the presence
of claudin-3 and claudin-7 in Caco-2 and SW620 cells (Fig. 1A). qPCR revealed that claudin-3 mRNA
levels were significantly lower in Caco-2 cells compared with those
in SW620 cells, while claudin-7 mRNA levels were significantly
increased in Caco-2 cells compared with those in SW620 cells
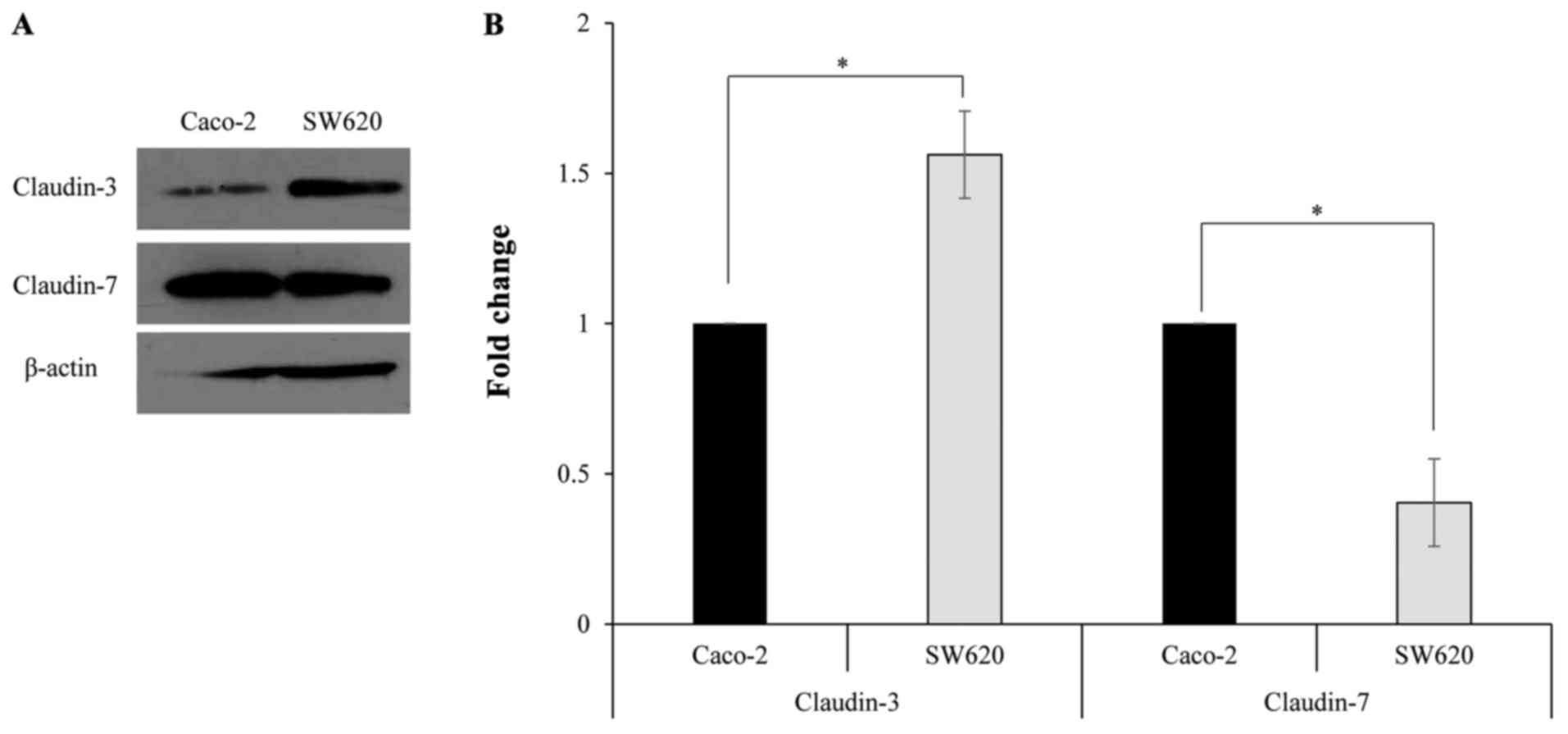
(Fig. 1B). Western blotting was
performed using a primary antibody against claudin-3 or claudin-7
to determine claudin protein expression in Caco-2 and SW620 cells.
The specific band corresponding to claudin-3 or claudin-7 was
detected in Caco-2 and SW620 cells (Fig.
2A). Furthermore, the protein expression levels of claudin-3
were significantly lower in Caco-2 cells compared with those in
SW620 cells, while the protein expression levels of claudin-7 were
significantly increased in Caco-2 cells compared with those in
SW620 cells (Fig. 2B).
Localization of claudin-3 and
claudin-7 in Caco-2 and SW620 cells
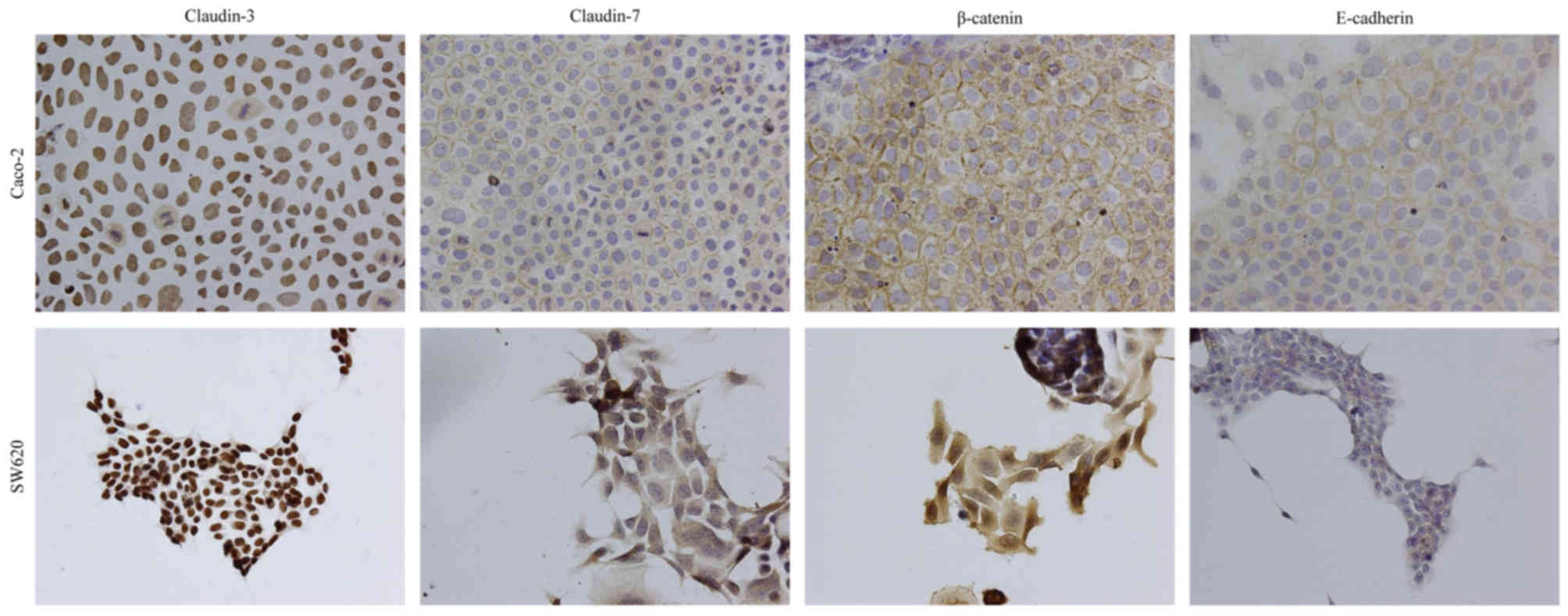
An immunocytochemical analysis was performed to
determine the subcellular localization of claudin-3 and claudin-7
in Caco-2 and SW620 cells. Staining with antibodies against
E-cadherin, β-catenin and claudin-7 revealed immunoreactivity
predominantly at the site of cell-cell contact for claudin-7,
whereas claudin-3 immunoreactivity was localized in the nucleus in
Caco-2 cells (Fig. 3). Similar
results were obtained for staining with an anti-claudin-3 antibody
in SW620 cells (Fig. 3). However,
claudin-7 and β-catenin were largely localized at the cytoplasm,
and E-cadherin was lowly expressed in SW620 cells (Fig. 3).
Expression and localization of
claudin-3 and claudin-7 proteins in colorectal adenocarcinoma
tissues
The expression of claudin-3 and claudin-7 was
investigated in colorectal adenocarcinoma tissues. The protein
expression levels of claudin-3 and claudin-7 were determined using
immunohistochemical staining. Table I
summarizes the clinical parameters of patients with colorectal
adenocarcinoma. A total of 59 (59.0%) patients were men and 41
(41.0%) were women, with a median age of 69 years (range, 39–95
years). An analysis of tumor samples revealed that 1 (1.0%) patient
had stage 0, 15 (15.0%) patients had stage I, 49 (49.0%) patients
had stage II, 34 (34.0%) patients had stage III and 1 (1.0%)
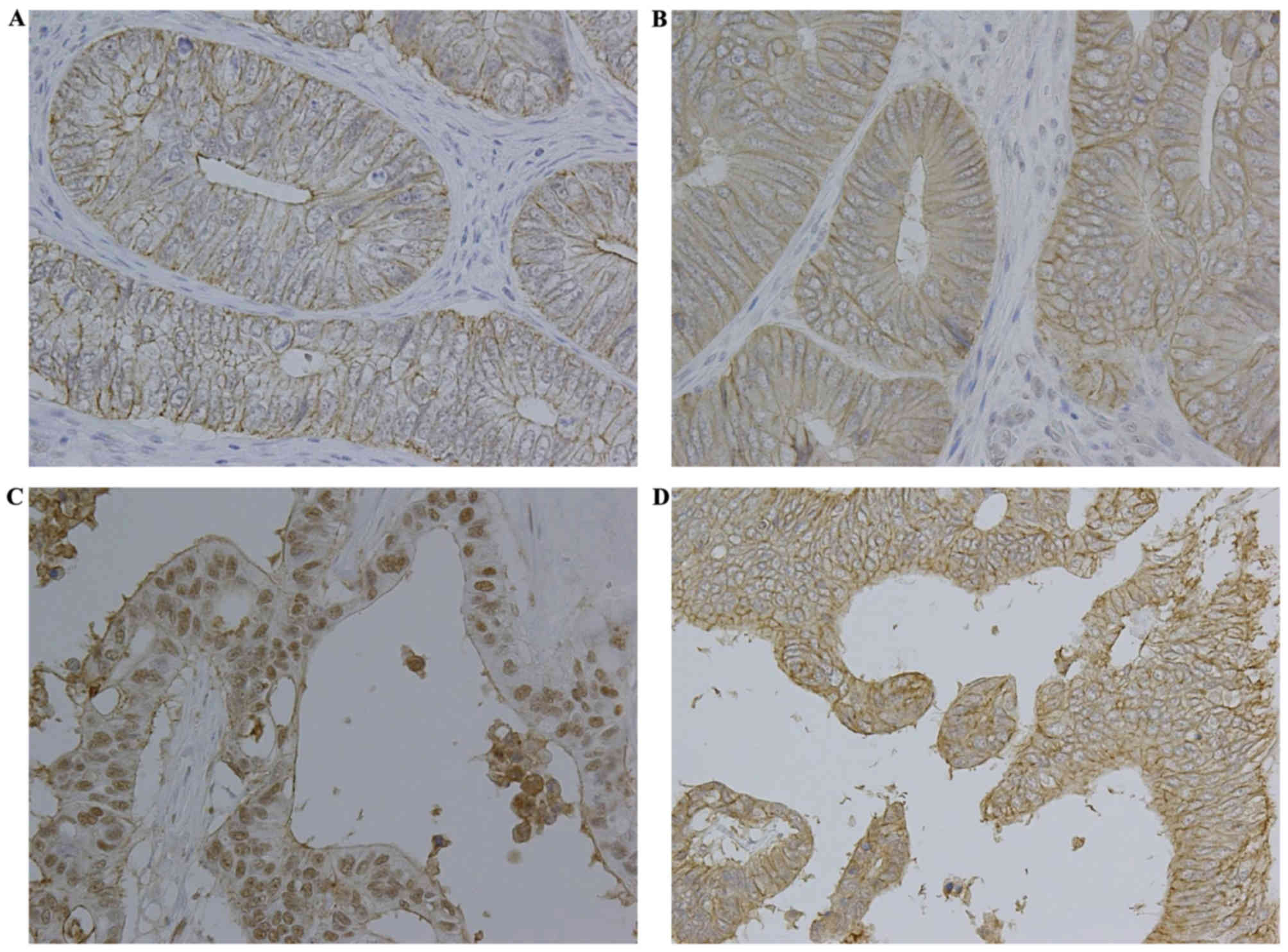
patient had stage IV colorectal adenocarcinoma. Claudin-3 and
claudin-7 were primarily expressed in the cell membrane of
colorectal adenocarcinoma cells; certain samples exhibited nuclear
claudin expression and a low level of cytoplasmic staining
(Fig. 4). The membranous expression
rates of claudin-3 and claudin-7 were 58.0 and 50.0%, respectively
(Table II). The nuclear expression
rates of claudin-3 and claudin-7 were 22.0 and 2.0%, respectively
(Table II). Therefore, membranous
and nuclear claudin expression in colorectal adenocarcinoma tissues
was evaluated.
 | Table I.Clinical characteristics of 100
patients with colorectal adenocarcinoma. |
Table I.
Clinical characteristics of 100
patients with colorectal adenocarcinoma.
|
Characteristics | Patients, n |
|---|
| Mean age ± standard
deviation, years | 68.5±11.4 |
| Sex |
|
|
Male | 59 |
|
Female | 41 |
| Histological
type |
|
| Well-
to moderately-differentiated | 81 |
|
Mucinous | 19 |
| Lymphatic
invasion |
|
|
Negative | 31 |
|
Positive | 69 |
| Venous
invasion |
|
|
Negative | 20 |
|
Positive | 80 |
| Lymph node
metastasis |
|
| N0 | 65 |
| N1 | 28 |
| N2 | 6 |
| N3 | 1 |
| Depth of tumor
invasion |
|
|
Tis | 1 |
| T1 | 10 |
| T2 | 6 |
| T3 | 52 |
| T4 | 31 |
| Stage |
|
| 0 | 1 |
| I | 15 |
| II | 49 |
|
III | 34 |
| IV | 1 |
 | Table II.Immunohistochemical staining for
positive expression rate (membrane or nucleus)a of claudins in 100 colorectal
adenocarcinoma tissue samples. |
Table II.
Immunohistochemical staining for
positive expression rate (membrane or nucleus)a of claudins in 100 colorectal
adenocarcinoma tissue samples.
| Claudin | Membranous
expression, n (%) | Nuclear expression,
n (%) |
|---|
| Claudin-3 | 58 (58.0) | 22 (22.0) |
| Claudin-7 | 50 (50.0) | 2 (2.0) |
The membranous expression of claudin-3 or claudin-7
was not associated with any clinicopathological factors (Fig. 4 and Table
III). As shown in Fig. 4 and
Table IV, the nuclear expression of
claudin-3 was associated with histological type and was
significantly increased in colorectal mucinous adenocarcinomas
compared with that in well- to moderately-differentiated colorectal
adenocarcinomas (P<0.01). However, no associations were observed
between the nuclear expression of claudin-3 and age, gender,
lymphatic invasion, venous invasion, depth of tumor invasion, lymph
node metastasis or stage of colorectal adenocarcinoma in the
present cohort of patients (Table
IV). The nuclear expression rate of claudin-7 in colorectal
mucinous adenocarcinomas was similar to that in well- to
moderately-differentiated colorectal adenocarcinomas; therefore,
claudin-7 was not observed to be associated with any
clinicopathological factor (Table
IV).
 | Table III.Association between membranous
claudin expression and clinicopathological characteristics of
colorectal adenocarcinoma in 100 tissue samples from patients. |
Table III.
Association between membranous
claudin expression and clinicopathological characteristics of
colorectal adenocarcinoma in 100 tissue samples from patients.
|
| Claudin-3
expression, n | Claudin-7
expression, n |
|---|
|
|
|
|
|---|
|
Characteristics | (−) | (+) | P-value | (−) | (+) | P-value |
|---|
| Age, years |
|
| 0.389 |
|
| 1.000 |
|
<60 | 11 | 11 |
| 11 | 11 |
|
|
≥60 | 31 | 47 |
| 39 | 39 |
|
| Sex |
|
| 0.185 |
|
| 0.542 |
|
Male | 28 | 31 |
| 31 | 28 |
|
|
Female | 14 | 27 |
| 19 | 22 |
|
| Histological
type |
|
| 0.432 |
|
| 0.308 |
| Well-
to moderately-differentiated | 32 | 49 |
| 38 | 43 |
|
|
Mucinous | 10 | 9 |
| 12 | 7 |
|
| Lymphatic
invasion |
|
| 0.386 |
|
| 0.130 |
|
Negative | 15 | 16 |
| 12 | 19 |
|
|
Positive | 27 | 42 |
| 38 | 31 |
|
| Venous
invasion |
|
| 0.648 |
|
| 0.453 |
|
Negative | 7 | 13 |
| 8 | 12 |
|
|
Positive | 35 | 45 |
| 42 | 38 |
|
| Lymph node
metastasis |
|
| 0.426 |
|
| 0.295 |
| N0 | 25 | 40 |
| 29 | 36 |
|
| N1 | 13 | 15 |
| 17 | 11 |
|
| N2 | 4 | 2 |
| 4 | 2 |
|
| N3 | 0 | 1 |
| 0 | 1 |
|
| Depth of tumor
invasion |
|
| 0.740 |
|
| 0.563 |
|
Tis | 0 | 1 |
| 0 | 1 |
|
| T1 | 3 | 7 |
| 3 | 7 |
|
| T2 | 3 | 3 |
| 3 | 3 |
|
| T3 | 24 | 28 |
| 27 | 25 |
|
| T4 | 12 | 19 |
| 17 | 14 |
|
| Stage |
|
| 0.586 |
|
| 0.323 |
| 0 | 0 | 1 |
| 0 | 1 |
|
| I | 5 | 10 |
| 6 | 9 |
|
| II | 20 | 29 |
| 23 | 26 |
|
|
III | 17 | 17 |
| 21 | 13 |
|
| IV | 0 | 1 |
| 0 | 1 |
|
 | Table IV.Association between nuclear claudin
expression and clinicopathological characteristics of colorectal
adenocarcinoma in 100 tissue samples from patients. |
Table IV.
Association between nuclear claudin
expression and clinicopathological characteristics of colorectal
adenocarcinoma in 100 tissue samples from patients.
|
| Claudin-3
expression | Claudin-7
expression |
|---|
|
|
|
|
|---|
|
Characteristics | (−) | (+) | P-value | (−) | (+) | P-value |
|---|
| Age, years |
|
| 0.563 |
|
| 1.000 |
|
<60 | 16 | 6 |
| 22 | 0 |
|
|
≥60 | 62 | 16 |
| 76 | 2 |
|
| Sex |
|
| 0.088 |
|
| 0.511 |
|
Male | 50 | 9 |
| 57 | 2 |
|
|
Female | 28 | 13 |
| 41 | 0 |
|
| Histological
type |
|
| 0.001 |
|
| 0.345 |
| Well-
to moderately-differentiated | 69 | 12 |
| 80 | 1 |
|
|
Mucinous | 9 | 10 |
| 18 | 1 |
|
| Lymphatic
invasion |
|
| 0.867 |
|
| 0.526 |
|
Negative | 25 | 6 |
| 30 | 1 |
|
|
Positive | 53 | 16 |
| 68 | 1 |
|
| Venous
invasion |
|
| 0.370 |
|
| 1.000 |
|
Negative | 14 | 6 |
| 20 | 0 |
|
|
Positive | 64 | 16 |
| 78 | 2 |
|
| Lymph node
metastasis |
|
| 0.794 |
|
| 0.777 |
| N0 | 52 | 13 |
| 63 | 2 |
|
| N1 | 21 | 7 |
| 28 | 0 |
|
| N2 | 4 | 2 |
| 6 | 0 |
|
| N3 | 1 | 0 |
| 1 | 0 |
|
| Depth of tumor
invasion |
|
| 0.181 |
|
| 0.319 |
|
Tis | 1 | 0 |
| 1 | 0 |
|
| T1 | 8 | 2 |
| 9 | 1 |
|
| T2 | 4 | 2 |
| 6 | 0 |
|
| T3 | 45 | 7 |
| 52 | 0 |
|
| T4 | 20 | 11 |
| 30 | 1 |
|
| Stage |
|
| 0.350 |
|
| 0.662 |
| 0 | 1 | 0 |
| 1 | 0 |
|
| I | 11 | 4 |
| 14 | 1 |
|
| II | 40 | 9 |
| 48 | 1 |
|
|
III | 26 | 8 |
| 34 | 0 |
|
| IV | 0 | 1 |
| 1 | 0 |
|
Discussion
The present study examined the expression of
claudin-3 and claudin-7 in human colorectal adenocarcinoma cell
lines and in 100 patients with colorectal adenocarcinoma. The
expression of claudin-3 was localized in the nuclei of Caco-2 and
SW620 cells, and the nuclear expression of claudin-3 was
significantly increased in colorectal mucinous adenocarcinomas
compared with that in well- to moderately-differentiated colorectal
adenocarcinomas. However, no associations were observed between the
expression of claudin-7 in colorectal adenocarcinomas and
clinicopathological parameters. Therefore, the present results
provided evidence for the development of a useful marker for
predicting cancer progression and prognosis in colorectal
adenocarcinoma, since the nuclear expression of claudin-3 may be a
phenotypic feature of colorectal mucinous adenocarcinomas.
An important result of the present study is the
nuclear localization of claudin-3 in human colorectal
adenocarcinoma cell lines (Caco-2 and SW620) and tissues (Figs. 3 and 4C). Although claudins are members of the
tetraspanin family of proteins, which are integral to the structure
and function of TJs, changes in the cellular localization of
claudins serve an important role during tumorigenesis (19). Previous studies reported that the
expression of claudin-1 increased in human colon cancers with
mislocalization from the cell membrane to the nucleus and cytoplasm
(19), the predominant cytoplasmic
and nuclear localization of claudin-1 reduced apoptosis in
nasopharyngeal carcinoma cells (28),
and claudin-1 showed loss of membrane expression and increased
nuclear localization in follicular thyroid carcinoma (39). The cytoplasmic expression of claudin-1
in metastatic melanoma cells was also shown to be critical for
increased malignancy (27,40). In addition, nuclear distribution of
claudin-2 resulted in enhanced proliferation in human lung
adenocarcinoma cells (34). As
aforementioned, the cellular distribution of claudins has been
reported in several types of cancer (41); however, the pathophysiological
functions of nuclear claudins have not yet been determined. The
present results indicated that claudin-3 is mainly distributed in
the nucleus of Caco-2 and SW620 cells. Therefore, it was
hypothesized that nuclear claudin-3, but not claudin-7, serves an
important function in enhancing cell proliferation in human
colorectal adenocarcinoma cell lines.
The nuclear localization of several cell junction
proteins, including ZO-1, ZO-2 and β-catenin, is known to be
associated with oncogenic transformation and cell proliferation
(42,43). In addition, ZO-1 and ZO-2 contain a
nuclear localization signal (NLS), and exhibit shuttle localization
between the cytoplasm and nucleus (42,43).
Therefore, the amino acid sequence of claudin-3 was analyzed using
an NLS prediction program (cNLS Mapper; http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi)
(44), but no putative NLS was
detected. Although NLS is a well-known sequence of nuclear import
elements, other sequences, including the PDZ domain, are known to
be important for nuclear localization (45). A previous study reported that the
nuclear localization of armadillo-repeat gene deleted in
velocardiofacial syndrome (ARVCF), an armadillo-repeat protein of
the p120ctn family, may be mediated by the PDZ domain of ZO-2
(46). One of the potential
mechanisms by which claudin-3 may translocate to the nucleus is
through interaction with the PDZ domain of ZO-2 in a manner similar
to that of ARVCF. Several studies have indicated that tumor
suppressors, including adenomatous polyposis coli and breast cancer
gene 1, translocate to the nucleus via an NLS-independent pathway
(47). Additionally, β-catenin, which
has no NLS, was imported into the nucleus by binding directly to
the nuclear pore machinery, similarly to importin-β/β-karyopherin
or other importin-β-like import factors (48). Another potential mechanism is that
claudin-3 relocates from the cellular membrane to the nucleus using
an NLS-independent import pathway similar to that used by these
proteins.
The membranous localization of β-catenin is
necessary for cadherin-mediated cell adhesion (49), and its cytoplasmic and nuclear
localization have been associated with oncogenesis (50). Furthermore, β-catenin regulates the
expression of claudin-1 in colorectal cancer cells (26) and that of claudin-3 in primary brain
endothelial cells (51). Although the
precise mechanism through which β-catenin regulates the expression
of claudin-1 and claudin-3 has not yet been defined, Wnt signaling
may be involved in the expression and localization of claudins. As
presented in Fig. 3, β-catenin was
largely localized at the cytoplasm, and E-cadherin was weakly
expressed in SW620 cells, whereas β-catenin and E-cadherin were
primarily localized at the cell membrane in Caco-2 cells. The
expression patterns of these proteins in SW620 cells were
consistent with those reported in previous studies (19). Additionally, overexpression of
claudin-3 in Caco-2 cells may induce changes in the localization of
β-catenin, since overexpression of claudin-1 in the primary colon
adenocarcinoma SW480 cell line was previously reported to induce
changes in the localization of β-catenin from the cell membrane to
the cytosol (19). The interactions
between nuclear claudin-3 and β-catenin in colorectal
adenocarcinoma cell proliferation and metastasis will be further
investigated by our group.
The present study detected mRNA and protein
expression of claudin-3 and claudin-7 in Caco-2 and SW620 cells,
and revealed that the expression levels of claudin-3 were
significantly increased in SW620 cells compared with those in
Caco-2 cells, whereas the expression levels of claudin-7 were lower
in SW620 cells compared with those in Caco-2 cells (Figs. 1 and 2).
In addition, in SW620 cells, the localization of claudin-7
decreased at the membrane and exhibited diffuse staining at the
cytoplasm (Fig. 3). One of the
reasons to explain the differential expression of claudin-3 and
claudin-7 exhibited by these two cell lines may be their functional
differences. Caco-2 cells were isolated from a primary colonic
tumor, while SW620 cells were established from the lymph nodes of a
patient with colorectal adenocarcinoma (52,53).
Furthermore, SW620 cells are known to exhibit highly tumorigenic
and metastatic features (54).
Additionally, previous studies demonstrated that upregulation of
claudin-3 was associated with increased tumorigenic potential in
ovarian epithelial cells (55,56), and
that loss of claudin-7 was associated with increased cellular
discohesion in breast carcinoma (30). Therefore, high expression of claudin-3
and low expression of claudin-7 in the metastatic colon cancer
SW620 cell line may be associated with tumor cell proliferation and
metastasis.
Several studies reported that patients with
colorectal mucinous adenocarcinoma had a poorer prognosis compared
with that of patients with non-mucinous adenocarcinoma (57–59).
Differences in metastatic patterns between histological subtypes of
colorectal cancer and a high number of peritoneal metastases in
colorectal mucinous adenocarcinomas have also been reported
(60–63). However, the underlying mechanisms for
the differences in metastatic patterns between various histological
subtypes remain unclear. Molecular and biological differences may
be associated with the histological features and behaviors of
different subtypes of colorectal cancer, and thus, the nuclear
localization of claudin-3 may contribute to a more aggressive
behavior. The present results indicated that claudin-3 serves a
role in histological transformation, since the nuclear expression
rate of claudin-3 was significantly increased in colorectal
mucinous adenocarcinomas (52.6%, 10/19) compared with that in well-
to moderately-differentiated colorectal adenocarcinomas (14.8%,
12/81) (Table IV), and was also
significantly increased (P=0.029; data not shown) in advanced stage
T4 (35.5%, 11/31) compared with that in stages Tis-T3 (15.9%,
11/69) (Table IV). Thus, additional
studies are required in order to establish whether claudin-3
contributes to the regulation of tumor cell proliferation and
metastasis in colorectal adenocarcinomas.
The nuclear localization of claudin-3 in colorectal
adenocarcinoma cells is involved in the biological transformation
of tumor behavior. The present results indicated that claudin-3
serves an important role in determining the tumor histological type
and has potential as a prognostic marker. Although the mechanisms
underlying the translocation of claudin-3 to the nucleus in
tumorigenesis have not yet been elucidated in detail, the present
study indicated the potential of claudin-3 as a histopathological
biomarker for colorectal adenocarcinomas.
Acknowledgements
The present study was supported by the Japan Society
for the Promotion of Science KAKENHI (grant no. JP15K08381).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Weitz J, Koch M, Debus J, Höhler T, Galle
PR and Büchler MW: Colorectal cancer. Lancet. 365:153–165. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fearon ER and Vogelstein B: A genetic
model for colorectal tumorigenesis. Cell. 61:759–767. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huber MA, Kraut N and Beug H: Molecular
requirements for epithelial-mesenchymal transition during tumor
progression. Curr Opin Cell Biol. 17:548–558. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thiery JP: Epithelial-mesenchymal
transitions in development and pathologies. Curr Opin Cell Biol.
15:740–746. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Laskowski P, Klim B, Ostrowski K,
Szkudlarek M, Litwiejko-Pietryńczak E, Kitlas K, Nienartowicz S and
Dzięcioł J: Local inflammatory response in colorectal cancer. Pol J
Pathol. 67:163–171. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Matter K and Balda MS: Signalling to and
from tight junctions. Nat Rev Mol Cell Biol. 4:225–236. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tsukita S, Furuse M and Itoh M:
Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol.
2:285–293. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mitic LL, Van Itallie CM and Anderson JM:
Molecular physiology and pathophysiology of tight junctions I.
Tight junction structure and function: Lessons from mutant animals
and proteins. Am J Physiol Gastrointest Liver Physiol.
279:G250–G254. 2000.PubMed/NCBI
|
|
10
|
Mitic LL and Anderson JM: Molecular
architecture of tight junctions. Annu Rev Physiol. 60:121–142.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Langbein L, Pape UF, Grund C, Kuhn C,
Praetzel S, Moll I, Moll R and Franke WW: Tight junction-related
structures in the absence of a lumen: Occludin, claudins and tight
junction plaque proteins in densely packed cell formations of
stratified epithelia and squamous cell carcinomas. Eur J Cell Biol.
82:385–400. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Itoh M and Bissell MJ: The organization of
tight junctions in epithelia: Implications for mammary gland
biology and breast tumorigenesis. J Mammary Gland Biol Neoplasia.
8:449–462. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Martin TA and Jiang WG: Tight junctions
and their role in cancer metastasis. Histol Histopathol.
16:1183–1195. 2001.PubMed/NCBI
|
|
14
|
Martin TA and Jiang WG: Loss of tight
junction barrier function and its role in cancer metastasis.
Biochim Biophys Acta. 1788:872–891. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Morita K, Furuse M, Fujimoto K and Tsukita
S: Claudin multigene family encoding four-transmembrane domain
protein components of tight junction strands. Proc Natl Acad Sci
USA. 96:pp. 511–516. 1999; View Article : Google Scholar : PubMed/NCBI
|
|
16
|
McCarthy KM, Skare IB, Stankewich MC,
Furuse M, Tsukita S, Rogers RA, Lynch RD and Schneeberger EE:
Occludin is a functional component of the tight junction. J Cell
Sci. 109:2287–2298. 1996.PubMed/NCBI
|
|
17
|
Matter K, Aijaz S, Tsapara A and Balda MS:
Mammalian tight junctions in the regulation of epithelial
differentiation and proliferation. Curr Opin Cell Biol. 17:453–458.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tsukita S, Yamazaki Y, Katsuno T, Tamura A
and Tsukita S: Tight junction-based epithelial microenvironment and
cell proliferation. Oncogene. 27:6930–6938. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dhawan P, Singh AB, Deane NG, No Y, Shiou
SR, Schmidt C, Neff J, Washington MK and Beauchamp RD: Claudin-1
regulates cellular transformation and metastatic behavior in colon
cancer. J Clin Invest. 115:1765–1776. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Itoh M, Furuse M, Morita K, Kubota K,
Saitou M and Tsukita S: Direct binding of three tight
junction-associated MAGUKs, ZO-1, ZO-2 and ZO-3, with the COOH
termini of claudins. J Cell Biol. 147:1351–1363. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mineta K, Yamamoto Y, Yamazaki Y, Tanaka
H, Tada Y, Saito K, Tamura A, Igarashi M, Endo T, Takeuchi K and
Tsukita S: Predicted expansion of the claudin multigene family.
FEBS Lett. 585:606–612. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Turksen K and Troy TC: Barriers built on
claudins. J Cell Sci. 117:2435–2447. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Swisshelm K, Macek R and Kubbies M: Role
of claudins in tumorigenesis. Adv Drug Deliv Rev. 57:919–928. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Morin PJ: Claudin proteins in human
cancer: Promising new targets for diagnosis and therapy. Cancer
Res. 65:9603–9606. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Soini Y, Tommola S, Helin H and
Martikainen P: Claudins 1, 3, 4 and 5 in gastric carcinoma, loss of
claudin expression associates with the diffuse subtype. Virchows
Arch. 448:52–58. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Miwa N, Furuse M, Tsukita S, Niikawa N,
Nakamura Y and Furukawa Y: Involvement of claudin-1 in the
beta-catenin/Tcf signaling pathway and its frequent upregulation in
human colorectal cancers. Oncol Res. 12:469–476. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Leotlela PD, Wade MS, Duray PH, Rhode MJ,
Brown HF, Rosenthal DT, Dissanayake SK, Earley R, Indig FE,
Nickoloff BJ, et al: Claudin-1 overexpression in melanoma is
regulated by PKC and contributes to melanoma cell motility.
Oncogene. 26:3846–3856. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee JW, Hsiao WT, Chen HY, Hsu LP, Chen
PR, Lin MD, Chiu SJ, Shih WL and Hsu YC: Upregulated claudin-1
expression confers resistance to cell death of nasopharyngeal
carcinoma cells. Int J Cancer. 126:1353–1366. 2010.PubMed/NCBI
|
|
29
|
Ikari A, Sato T, Watanabe R, Yamazaki Y
and Sugatani J: Increase in claudin-2 expression by an
EGFR/MEK/ERK/c-Fos pathway in lung adenocarcinoma A549 cells.
Biochim Biophys Acta. 1823:1110–1118. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kominsky SL, Argani P, Korz D, Evron E,
Raman V, Garrett E, Rein A, Sauter G, Kallioniemi OP and Sukumar S:
Loss of the tight junction protein claudin-7 correlates with
histological grade in both ductal carcinoma in situ and invasive
ductal carcinoma of the breast. Oncogene. 22:2021–2033. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
de Oliveira SS, de Oliveira IM, De Souza W
and Morgado-Diaz JA: Claudins upregulation in human colorectal
cancer. FEBS Lett. 579:6179–6185. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kinugasa T, Huo Q, Higashi D, Shibaguchi
H, Kuroki M, Tanaka T, Futami K, Yamashita Y, Hachimine K, Maekawa
S, et al: Selective up-regulation of claudin-1 and claudin-2 in
colorectal cancer. Anticancer Res. 27:3729–3734. 2007.PubMed/NCBI
|
|
33
|
Gröne J, Weber B, Staub E, Heinze M,
Klaman I, Pilarsky C, Hermann K, Castanos-Velez E, Röpcke S, Mann
B, et al: Differential expression of genes encoding tight junction
proteins in colorectal cancer: Frequent dysregulation of claudin-1,
−8 and −12. Int J Colorectal Dis. 22:651–659. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ikari A, Watanabe R, Sato T, Taga S,
Shimobaba S, Yamaguchi M, Yamazaki Y, Endo S, Matsunaga T and
Sugatani J: Nuclear distribution of claudin-2 increases cell
proliferation in human lung adenocarcinoma cells. Biochim Biophys
Acta. 1843:2079–2088. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schneider CA, Rasband WS and Eliceiri KW:
NIH Image to ImageJ: 25 years of image analysis. Nat Methods.
9:671–675. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Japanese Society for Cancer of the Colon
and Rectum: Japanese Classification of Colorectal Carcinoma. 8th.
Kanehara, Tokyo: 2013
|
|
38
|
Jung H, Jun KH, Jung JH, Chin HM and Park
WB: The expression of claudin-1, claudin-2, claudin-3, and
claudin-4 in gastric cancer tissue. J Surg Res. 167:e185–e191.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zwanziger D, Badziong J, Ting S, Moeller
LC, Schmid KW, Siebolts U, Wickenhauser C, Dralle H and Fuehrer D:
The impact of CLAUDIN-1 on follicular thyroid carcinoma
aggressiveness. Endocr Relat Cancer. 22:819–830. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
French AD, Fiori JL, Camilli TC, Leotlela
PD, O'Connell MP, Frank BP, Subaran S, Indig FE, Taub DD and
Weeraratna AT: PKC and PKA phosphorylation affect the subcellular
localization of claudin-1 in melanoma cells. Int J Med Sci.
6:93–101. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Rabinsky EF, Joshi BP, Pant A, Zhou J,
Duan X, Smith A, Kuick R, Fan S, Nusrat A, Owens SR, et al:
Overexpressed claudin-1 can be visualized endoscopically in colonic
adenomas in vivo. Cell Mol Gastroenterol Hepatol. 2:222–237. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Islas S, Vega J, Ponce L and
Gonzalez-Mariscal L: Nuclear localization of the tight junction
protein ZO-2 in epithelial cells. Exp Cell Res. 274:138–148. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gottardi CJ, Arpin M, Fanning AS and
Louvard D: The junction-associated protein, zonula occludens-1,
localizes to the nucleus before the maturation and during the
remodeling of cell-cell contacts. Proc Natl Acad Sci USA. 93:pp.
10779–10784. 1996; View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kosugi S, Hasebe M, Tomita M and Yanagawa
H: Systematic identification of cell cycle-dependent yeast
nucleocytoplasmic shuttling proteins by prediction of composite
motifs. Proc Natl Acad Sci USA. 106:pp. 10171–10176. 2009;
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Du G, Gu Y, Hao C, Yuan Z, He J, Jiang WG
and Cheng S: The cellular distribution of Na+/H+ exchanger
regulatory factor 1 is determined by the PDZ-I domain and regulates
the malignant progression of breast cancer. Oncotarget.
7:29440–29453. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kausalya PJ, Phua DC and Hunziker W:
Association of ARVCF with zonula occludens (ZO)-1 and ZO-2: binding
to PDZ-domain proteins and cell-cell adhesion regulate plasma
membrane and nuclear localization of ARVCF. Mol Biol Cell.
15:5503–5515. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Fabbro M and Henderson BR: Regulation of
tumor suppressors by nuclear-cytoplasmic shuttling. Exp Cell Res.
282:59–69. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fagotto F, Glück U and Gumbiner BM:
Nuclear localization signal-independent and
importin/karyopherin-independent nuclear import of beta-catenin.
Curr Biol. 8:181–190. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kemler R: From cadherins to catenins:
Cytoplasmic protein interactions and regulation of cell adhesion.
Trends Genet. 9:317–321. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Munemitsu S, Albert I, Souza B, Rubinfeld
B and Polakis P: Regulation of intracellular beta-catenin levels by
the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc
Natl Acad Sci USA. 92:pp. 3046–3050. 1995; View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liebner S, Corada M, Bangsow T, Babbage J,
Taddei A, Czupalla CJ, Reis M, Felici A, Wolburg H, Fruttiger M, et
al: Wnt/beta-catenin signaling controls development of the
blood-brain barrier. J Cell Biol. 183:409–417. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Fogh J, Wright WC and Loveless JD: Absence
of HeLa cell contamination in 169 cell lines derived from human
tumors. J Natl Cancer Inst. 58:209–214. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Leibovitz A, Stinson JC, McCombs WB III,
McCoy CE, Mazur KC and Mabry ND: Classification of human colorectal
adenocarcinoma cell lines. Cancer Res. 36:4562–4569.
1976.PubMed/NCBI
|
|
54
|
Hewitt RE, McMarlin A, Kleiner D, Wersto
R, Martin P, Tsokos M, Stamp GW and Stetler-Stevenson WG:
Validation of a model of colon cancer progression. J Pathol.
192:446–454. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Agarwal R, D'Souza T and Morin PJ:
Claudin-3 and claudin-4 expression in ovarian epithelial cells
enhances invasion and is associated with increased matrix
metalloproteinase-2 activity. Cancer Res. 65:7378–7385. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kwon MJ, Kim SS, Choi YL, Jung HS, Balch
C, Kim SH, Song YS, Marquez VE, Nephew KP and Shin YK: Derepression
of CLDN3 and CLDN4 during ovarian tumorigenesis is associated with
loss of repressive histone modifications. Carcinogenesis.
31:974–983. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Symonds DA and Vickery AL: Mucinous
carcinoma of the colon and rectum. Cancer. 37:1891–1900. 1976.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Umpleby HC, Ranson DL and Williamson RC:
Peculiarities of mucinous colorectal carcinoma. Br J Surg.
72:715–718. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Minsky BD, Mies C, Rich TA, Recht A and
Chaffey JT: Colloid carcinoma of the colon and rectum. Cancer.
60:3103–3112. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Hugen N, van de Velde CJ, de Wilt JH and
Nagtegaal ID: Metastatic pattern in colorectal cancer is strongly
influenced by histological subtype. Ann Oncol. 25:651–657. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Catalano V, Loupakis F, Graziano F,
Torresi U, Bisonni R, Mari D, Fornaro L, Baldelli AM, Giordani P,
Rossi D, et al: Mucinous histology predicts for poor response rate
and overall survival of patients with colorectal cancer and treated
with first-line oxaliplatin- and/or irinotecan-based chemotherapy.
Br J Cancer. 100:881–887. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chen JS, Hsieh PS, Hung SY, Tang R, Tsai
WS, Changchien CR, Lin PY, Wang JY and Yeh CY: Clinical
significance of signet ring cell rectal carcinoma. Int J Colorectal
Dis. 19:102–107. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Mekenkamp LJ, Heesterbeek KJ, Koopman M,
Tol J, Teerenstra S, Venderbosch S, Punt CJ and Nagtegaal ID:
Mucinous adenocarcinomas: Poor prognosis in metastatic colorectal
cancer. Eur J Cancer. 48:501–509. 2012. View Article : Google Scholar : PubMed/NCBI
|