Introduction
Epithelial ovarian cancer (EOC) is the most
aggressive malignant gynecological disease, and the 5-year survival
rate of patients less than 30% (1).
It is estimated that 22,280 new diagnoses and 14,240 deaths from
this neoplasm were reported in 2016 in the US (2). Currently, there is no effective approach
for the detecting of EOC in early stage, because patients in early
stage have no obvious symptoms. Almost 70% patients were diagnosed
in advanced stage with predominant ascites and wide spread implant
lesions in abdominal cavity. Ovarian cancer (OC) metastasis is
characterized by the shedding of malignant cells from the surface
of the ovary and their implantation onto the peritoneal surface
(3,4).
Unlike other solid tumors, no anatomical barrier exists to block OC
metastasis inside the peritoneal cavity. Although the metastasis of
EOC has no common patterns, some researchers thought the omentum
was the preferentially implanted site of EOC (5). The omentum, which is the largest
peritoneal fold in the peritoneal cavity, plays critical role in
several physiological process including fat deposition, immune
contribution and infection isolation. The ‘omental caking’,
indicating the thickness of omentum after invasion of OC at the
late stage, is one of a distinct pattern of advanced EOC (6). In the relative early stage of EOC, the
pathologist can also find OC cells in the omentum after surgery.
Patients without omental caking show slighter metastasis and better
prognosis (7). Thus, it was inferred
that the omentum might actually provide ‘optimal’ sign for the
migration of cancer cells. Therefore, we can know the metastatic
manner of EOC by comparing the invaded omentum with the primary
tumor. And it can provide novel potential biomarkers for screening
the early metastasis and developing new treatment strategies of the
treatment by resisting the spread of the cancer cells.
In the present study, we identified the non-coding
RNAs (ncRNAs) between primary ovarian tumors and omental metastatic
tissue using microarray analysis. The candidate altered genes,
which selected by the microarray analysis results, were further
validated by Reverse transcription quantitative polymerase chain
reaction (RT-qPCR). We also clustered the subgroup based on patient
age, International Federation of Gynecology and Obstetrics (FIGO)
stage (8), pathological grade, and
omental metastatic stage.
Materials and methods
Study samples
All the samples were collected after surgery
performed in Xiangya Hospital of Central South University
(Changsha, China) between March 2013 and May 2015. A total of 48
paired OC and omental fresh samples were stored in −80°C for
RT-qPCR analysis. The clinic pathologic data were obtained from
medical records and pathologic reports. Prior informed consent was
obtained from all recruited patients for ovarian carcinoma tissues
collection and the publication of their data. The study was
approved by the Ethics Committee of Xiangya Hospital of Central
South University (reference: 201604556).
Gene chip hybridization and data
analysis
Total RNA was extracted using the TRIzol method
(Shanghai Sangong Pharmaceutical Co., Ltd., Shanghai, China)
according to the manufacturer's instructions. Total RNA was
quantified by the Nano Drop ND-2000 (Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and the RNA integrity was assessed using
Agilent Bioanalyzer 2100 (Agilent Technologies, Inc., Santa Clara,
CA, USA). Prime Script RT-PCR kit (Takara, Dalian, China) was
utilized for reverse transcription, then total RNA were transcribed
to double strand cDNA, after that synthesized into cRNA and labeled
with Cyanine-3-CTP. The labeled cRNAs were hybridized onto the
microarray. After washing, the arrays were scanned with an Agilent
Scanner G2505C (Agilent Technologies, Inc.). Feature Extraction
software (version 10.7.1.1; Agilent Technologies, Inc.) was used to
obtain raw data from the array images. GeneSpring was employed for
basic analysis of raw data. First, raw data were normalized with
the quantile algorithm. Probes with at least 1 of 2 conditions
having flags in ‘P’ were chosen for further data analysis. The
ncRNAs were then identified through fold change as well as P-value
calculated with t-test. The threshold set for up- and downregulated
genes was a fold change ≥2.0 and a P-value ≤0.05. Finally,
hierarchical clustering of genes based on clinic pathologic data
was performed.
Quantitative real-time polymerase
chain reaction
Total RNA was extracted using the TRIzol method and
reverse transcribed to cDNA by using PrimeScript RT-PCR kit
(Takara) in accordance with the manufacturer's protocol. Real time
PCR was performed on a ViiA 7 Real-Time Fluorescent Quantitative
PCR system (Thermo Fisher Scientific, Inc.). The PCR amplification
conditions were 40 cycles of 95°C for 30 sec, 95°C for 5 sec, 60°C
for 34 sec. Primers of target genes are as follows: U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. U6
was used as the reference gene. myocardial infarction associated
transcript (MIAT) forward, 5′-GGAACAAGGATGGGAGTCG-3′ and reverse,
5′-GCACTGAGCAAATGGAGACA-3′. Small nucleolar RNA, C/D Box 114
cluster (SNORD114)-10, reverse transcription primer,
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTGGACCTC-3′, forward,
5′-ACACTCCAGCTGGGAAGATCAATGATGACT-3′ and universal primer,
5′-ACTGACTGATGCAATCTCAACTGGTGTCGTGGA-3′. SNORD114-2 reverse
transcription primer,
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTGGACCTC-3′, forward,
5′-ACACTCCAGCTGGGGGACCAATGATAATG-3′ and universal primer,
5′-ACTGACTGATGCAATCTCAACTGGTGTCGTGGA-3′. SNORD114-11, reverse
transcription primer,
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTGGACCTC-3′, forward,
5′-ACACTCCAGCTGGGTGGACCAGTGATGGTG-3′ and universal primer,
5′-ACTGACTGATGCAATCTCAACTGGTGTCGTGGA-3′. Abundance was calculated
using the 2-ΔCT method. SPSS 18.0 statistical software package
(SPSS, Inc., Chicago, IL, USA) was used for statistical analysis;
error bars represent the standard error of the mean. Statistical
analyses were performed using paired and t-test and non-parametric
test. P<0.05 was considered as statistically significant.
Results
Comprehensive analysis of ncRNAs in
primary and metastatic OC
The differentiate expressed ncRNAs in 3 paired
OC/omental metastasis (OM) samples were determined by microarray
analysis. Hierarchical clustering revealed that 179 and 56 of the
235 differentially expressed ncRNAs were up- and downregulated,
respectively (Fig. 1A). Among the
ncRNAs, MIAT (P=0.015) was upregulated, whereas SNORD114-2
(P=0.024) SNORD114-10 (P=0.0002) and SNORD114-11 (P=0.0001) were
downregulated (Fig. 1B). We further
validated the expression level of the four genes in 3 matched
primary and metastatic OC by RT-qPCR, the expression trend of the
genes were consistent with the results of chip (Fig. 2). MIAT (P=0.0363) expression increased
in the omentum, while SNORD114-2 (P=0.0246), SNORD114-10 (P=0.0025)
and SNORD114-11 (P=0.1125) were downregulated in OM tissues
compared to OC tissues.
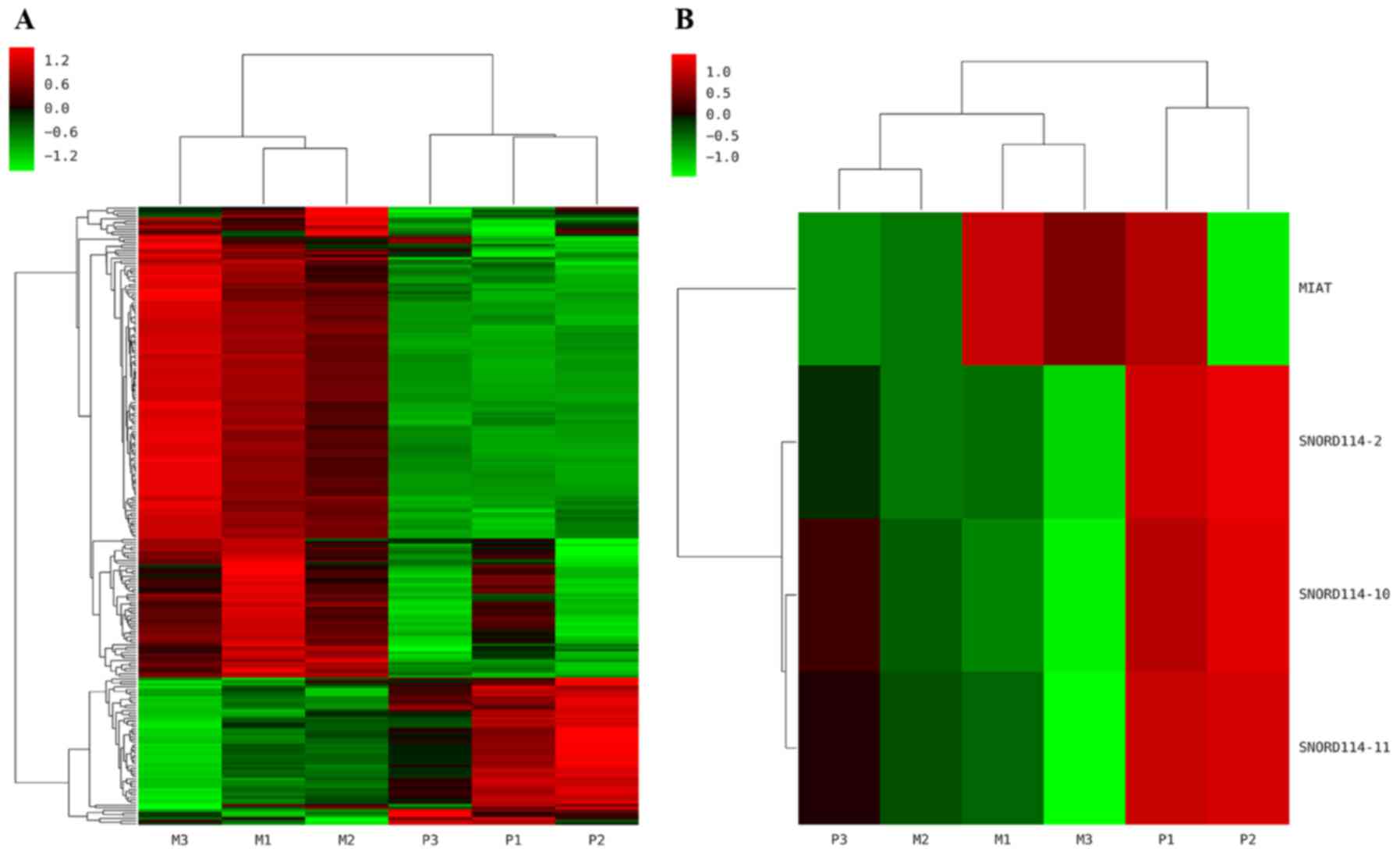 | Figure 1.Hierarchical clustering map of ncRNAs.
The heat map diagram shows the result of the two-way hierarchical
clustering of ncRNAs and samples. Each row represents a gene and
each column represents a sample. P1, P2, P3 represent 3 cases of
primary ovarian cancer sites, while M1, M2, M3 mean corresponding
omental metastasis sites respectively. (A) A total of 235 ncRNAs
were differentially expressed. (B) Of the significant ncRNAs
expressed, MIAT (P=0.015) was upregulated in the OM tissues,
whereas SNORD114-2 (P=0.024), SNORD114-10 (P=0.0002) and
SNORD114-11 (P=0.0001) were downregulated in OM tissues compared
with OC tissues. Red represents upregulated genes, and green
represents downregulated genes. ncRNA, non-coding RNA; MIAT,
myocardial infarction associated transcript; OM, omental
metastasis; SNORD114, small nucleolar RNA, C/D Box 114 cluster; OC,
ovarian cancer. |
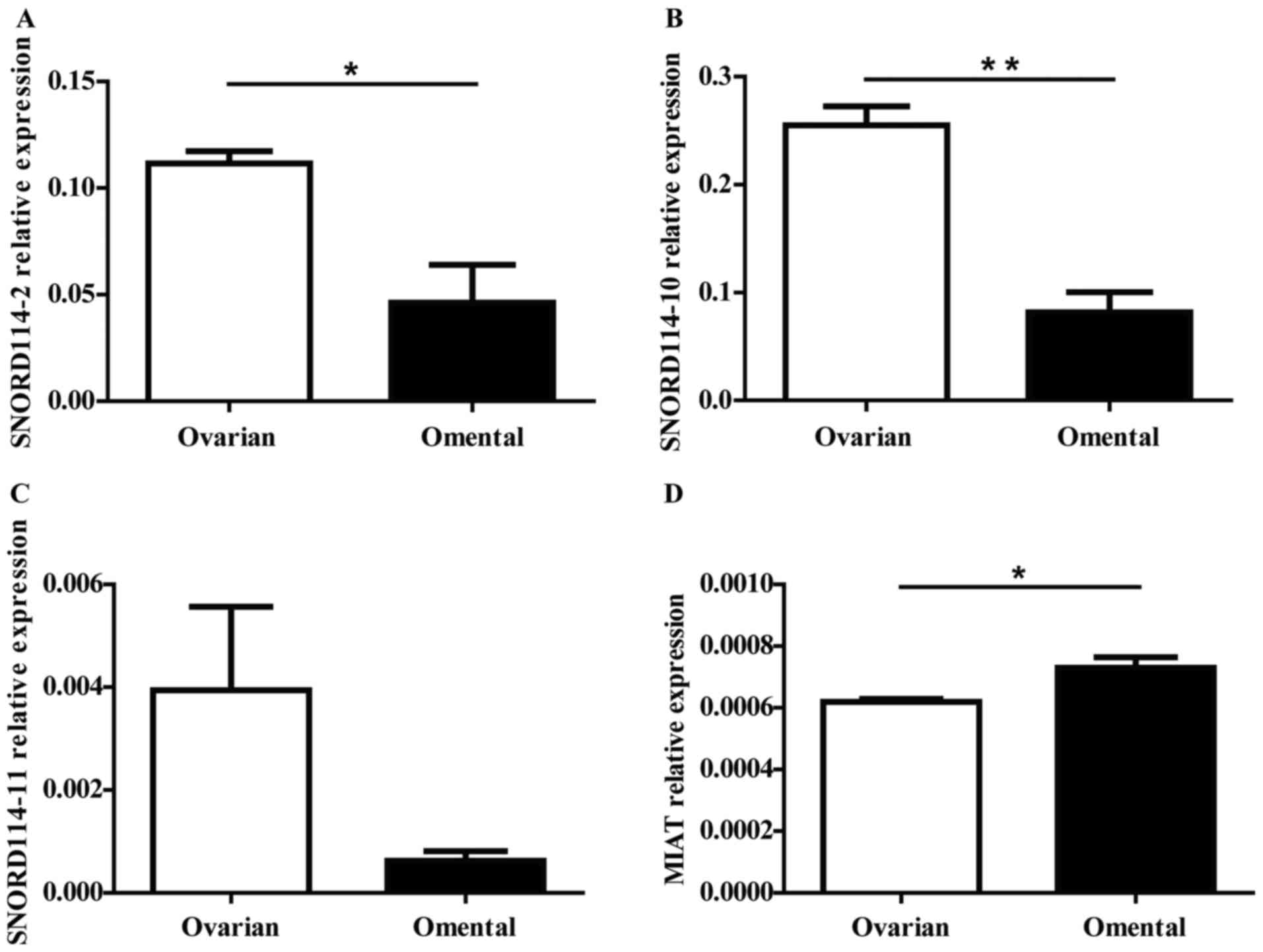 | Figure 2.The significant expressed ncRNAs were
selected for further validation by RT-qPCR. We further validated
the expression level of the (A) SNORD114-2, (B) SNORD114-10 (C)
SNORD114-11 and (D) MIAT genes in 3 matched primary and metastatic
ovarian cancer by RT-qPCR, MIAT (P=0.0363) expression increased in
the OM tissues while SNORD114-2 (P=0.0246), SNORD114-10 (P=0.0025)
and SNORD114-11 (P=0.1125) were downregulated in OM tissues
compared with OC tissues. The expression trend of the genes were
consistent with the results of the chip. *P<0.05, **P<0.01.
ncRNA, non-coding RNA; RT-qPCR, reverse transcription quantitative
polymerase chain reaction; SNORD114, small nucleolar RNA, C/D Box
114 cluster; MIAT, myocardial infarction associated transcript; OM,
omental metastasis; OC, ovarian cancer. |
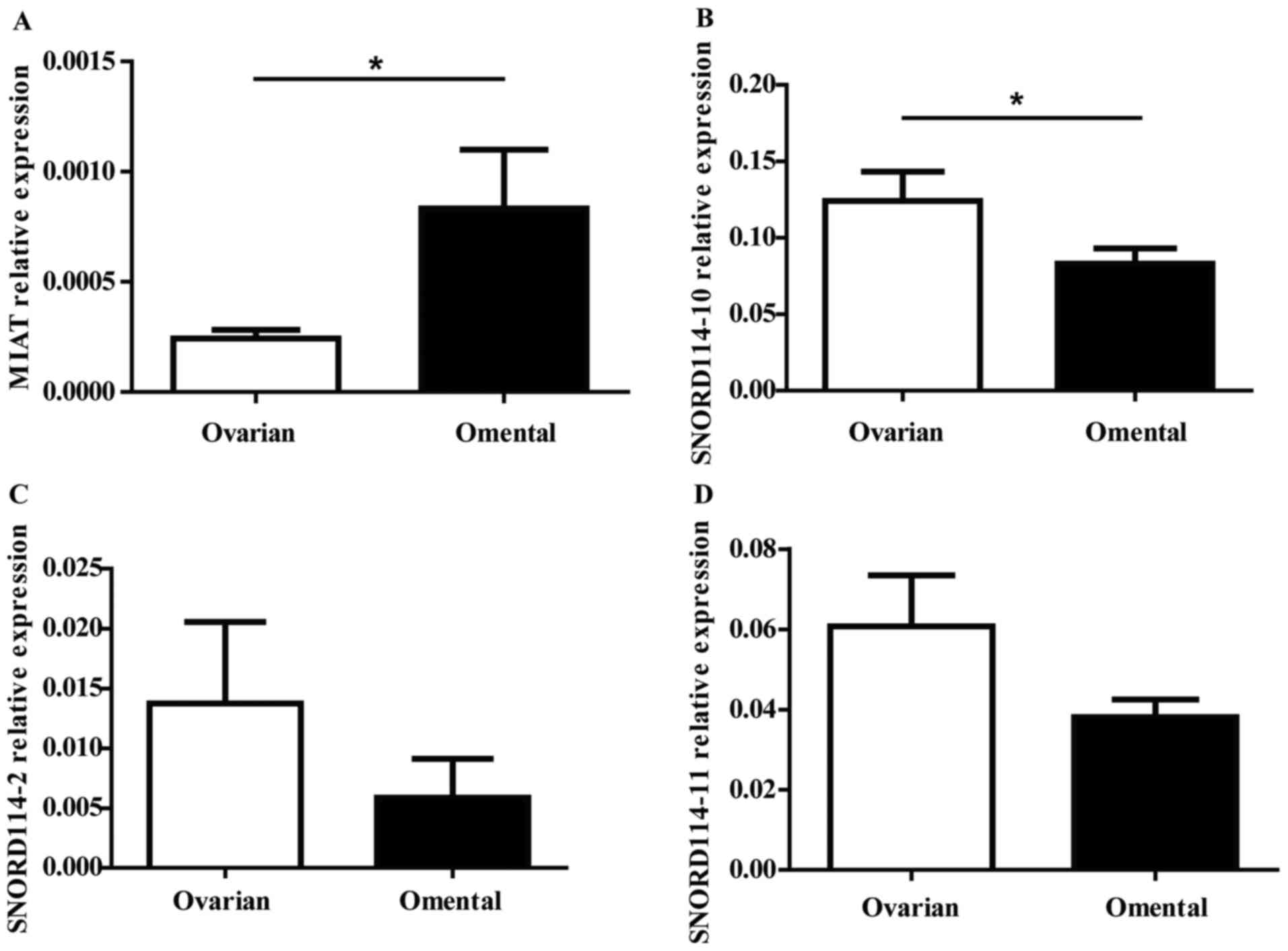
The expressions of MIAT SNORD114-10
SNORD114-2 SNORD114-11 genes in OC
To validate the array data, RT-qPCR was performed
with 27 pairs of ovarian and omentum cancer tissues. In the four
genes, MIAT (P=0.023) was statistically significant upregulated in
omentum tissues (Fig. 3A), while
SNORD114-10 (P=0.025) SNORD114-2 (P=0.082) and SNORD114-11
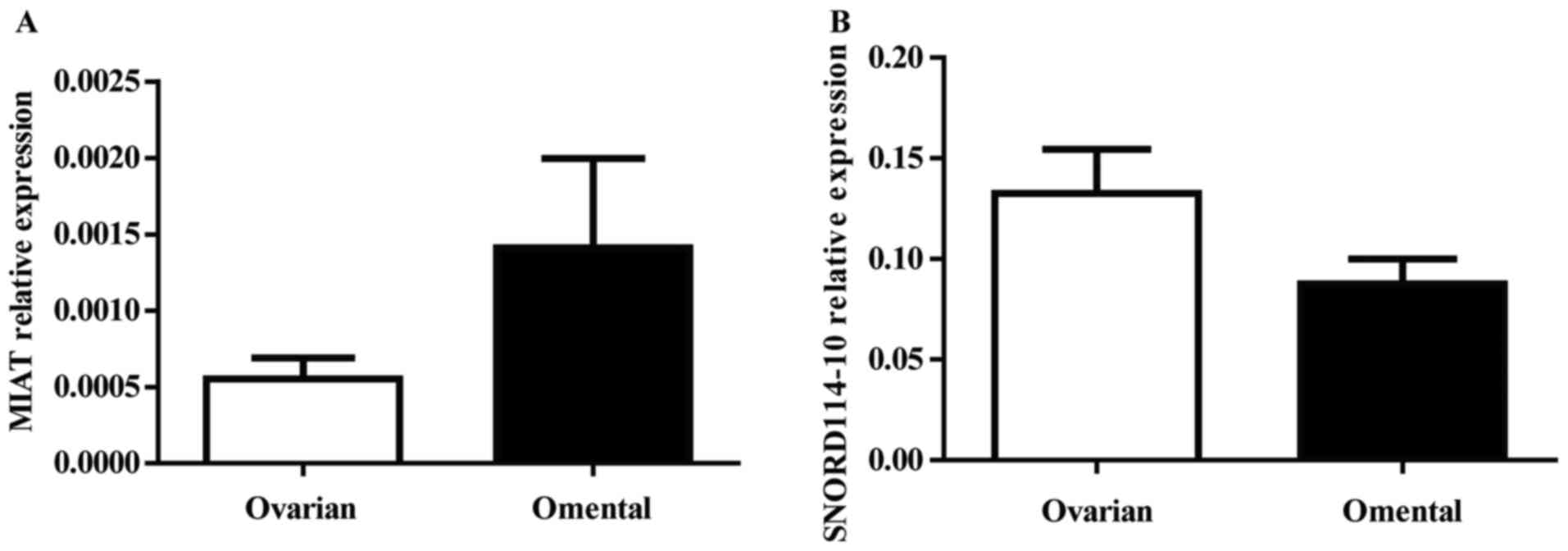
(P=0.132) were downregulated in omental tissues (Fig. 3B-D). Furthermore, to verify MIAT and
SNORD114-10 expression changes indeed originate from cancer cells,
we identified the MIAT and SNORD114-10expressionin another 21 early
stage OC samples without OM compares with the expression in paired
normal omental tissue. The expression level of these two genes
showed no statistically significant difference between ovarian and
omental tissues (P>0.05) (Fig. 4A and
B).
Association between genes expression
and prognostic factors in OC
A total of 48 paired clinical samples analyzed above
were derived from OC patients aged 21–77 years. The pathological
appearance was divided into two stages as ovarian low-grade serous
adenocarcinoma (LGSC) or high-grade serous adenocarcinoma (HGSC)
according to the FIGO. The expression of SNORD114-10 in OC samples
was not correlated with age, FIGO division, pathological
classification or the presence of omental metastases (P>0.05)
(Table I). This was essentially the
same pattern as for MIAT, with the exception that levels of this
ncRNA were significantly lower in omental samples with metastases
(P=0.042) (Table I).
 | Table I.Correlations between MIAT and
SNORD114-10 expression and clinicopathlogical features. |
Table I.
Correlations between MIAT and
SNORD114-10 expression and clinicopathlogical features.
|
|
| MIAT | SNORD114-10 |
|---|
|
|
|
|
|
|---|
| Factors | Cases | 2-ΔCT
(mean ± SD) | P-value | 2-ΔCT
(mean ± SD) | P-value |
|---|
| Age (years) |
|
| 0.597 |
| 0.205 |
|
<50 | 22 |
0.00034±0.00035 |
|
0.1079±0.0816 |
|
| ≥50 | 26 |
0.00041±0.00055 |
|
0.1446±0.1108 |
|
| FIGO stage |
|
| 0.251 |
| 0.539 |
| I–II | 13 |
0.00051±0.00039 |
|
0.1424±0.1078 |
|
|
III–IV | 35 |
0.00033±0.00049 |
|
0.1224±0.0969 |
|
| FIGO grade |
|
| 0.434 |
| 0.100 |
| Low | 9 |
0.00049±0.00033 |
|
0.0787±0.068 |
|
| High | 39 |
0.00038±0.00031 |
|
0.1391±0.1024 |
|
| Omentum
metastasis |
|
| 0.042a |
| 0.773 |
| Yes | 27 |
0.00024±0.00020 |
|
0.1241±0.0999 |
|
| No | 21 |
0.00055±0.00063 |
|
0.1326±0.1005 |
|
Discussion
The mortality rate of OC is ranked top one among all
gynecological malignancies, and as high as 75% of diagnosed
patients are already in the late stage. It is difficult to detect
omentum metastasis in OC patients due to the lacking of obvious
symptoms and specific markers at early stage. At the same time,
limited understanding about the mechanism of tumor formation also
hinders the development of new treatments. The ncRNA as a new tumor
marker, in the occurrence and development of tumors play an
important role in the treatment of cancer for people to provide a
new way of thinking.
In the present study we used microarray to determine
the ncRNAs profile between OC and OM. To the best of our knowledge,
we are the first research group to report the alterations in ncRNA
expression in OC, and to show that SNORD114-10 levels are reduced
in OM, while MIAT levels are increased when compared to OC tissue.
Consequently, we believe that the expression of MIAT and
SNORD114-10 are important factors that contribute to migration of
ovarian tumor cells from the primary site to the omentum.
OC is associated with the highest mortality rates of
all gynecologic malignancies (9). The
metastatic potential of OC cells is influenced by a series of
complex process, with the most common metastatic sites being the
peritoneum and omentum and the disease progresses rapidly when
malignant cells reach these secondary sites (4). Based on the ‘seed and soil’ hypothesis,
we decided to investigate genes that may contribute to the ‘soil’
properties of the omentum.
According to the results of RT-PCR, MIAT was
upregulated and SNORD114-10 was downregulated in the omentum in
metastatic disease cases. However, there was no statistical
difference between OC with OM and OC without OM. Thus, we speculate
that the migration in OC is not a tumor cell-intrinsic property,
but rather is dependent on the omentum ‘soil’ which provides the
conditions for tumor development and subsequent disease
acceleration.
ncRNAs usually perform catalytic or regulatory
functions (10), and can be
classified as small (<200 bps) or long ncRNAs (>200 bps)
(11). Over the past few years, many
ncRNAs have been identified including H19 (12) and Xist (13). Our present study demonstrates that the
expression of SNORD114-10 and MIAT were differentially regulated in
metastatic omentum compared to non-infiltrated tissue. Previous
studies have shown that some lncRNAs are differentially expressed
in primary and metastatic tumors (14,15).
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is
a well-known carcinogenic lncRNA that is highly expressed in lung
metastases (16). Elevated MALAT1 in
different tumors predicts shorter metastasis-free survival (MFS),
deep tissue invasion (17), higher
histological grade (18), and lower
overall survival (OS) (19). Similar
to MALAT1, HOX transcript antisense intergenic RNA (HOTAIR) is
closely related to the occurrence and development of tumor
metastasis (20–23). Zhang et al (24) found that MIR4697 host gene (MIR4697HG)
in cancerous tissues increased compared with that in adjacent
noncancerous tissues. It could promote OC growth and metastasis.
The aggressive role of MIR4697HG in OC may be related to the ERK
and AKT signaling pathways.
MIAT was described as a ncRNA expressed in mitotic
progenitors (25), and RNA-FISH
indicated that it was expressed in the inner nuclear layer (INL)
and the ganglion cell layer (GCL) in humans. The research shows
that MIAT functional abnormalities are associated with the risk of
myocardial infarction, it also plays a regulatory role in other
areas, including retinal cell growth, brain development and
fibrosis after filamentous fibrosis (26,27).
Moreover, a previous study reported that high expression of MIAT
leads to abnormal endothelial cell proliferation and cell migration
during early retinal microvascular dysfunction (28). Luan et al (29) found that MIAT downregulation inhibited
the epithelial-mesenchymal transition (EMT) and decreased migration
and invasion in MDA-MB-231 and MCF-7 breast cancer cell lines. They
also suggested that MIAT promotes breast cancer progression and
functions as competing endogenous RNA (ceRNA) to regulate dual
specificity phosphatase 7 (DUSP7) expression by sponging miR-155-5p
in breast cancer. Shen et al (30) found that MIAT acted as a ceRNA, and
formed a feedback loop with AKT and miR-150-5p to regulate Human
lens epithelial cell (HLEC) function. Our current results showing
that MIAT was almost 3-fold higher in OM tissue when compared with
OC. Therefore, we hypothesize that MIAT may promote the metastasis
of OC, which may be through the regulation of certain genes or
through the EMT or AKT pathway to promote OC metastasis. Next we
will determine whether MIAT can regulate migration in OC cells by
silencing and over-expressing MIAT. Further study are required to
determine the role of MIAT (the correlation between) in OC
metastasis/the metastssis of OC.
Emerging evidence indicates that snoRNAs can play
several non-classical roles, including reactive oxygen species
scavenging in the cytoplasm and being precursors for microRNA-like
molecules (31–33). Although the functional map of snoRNA
has not been fully elucidated, snoRNA is likely to play an
important regulatory role in the development and progression of
cancer. According to new molecular mechanisms that have been found
(34). Some snoRNAs are implicated in
tumorigenesis, such as the tumor suppressors SNOR12 and SNOR44, and
the tumor promoters SNOD33 and U70C (35). Interestingly, Mei et al
reported high expression of SNORA42 in lung cancer, and concluded
that this ncRNA might be oncogenic (36). Gee et al (37) demonstrated that SNORD48 expression was
significantly lower in low-grade breast cancers, whereas high
SNORD44 expression in breast and non-small cell lung cancer
indicated a better prognosis. Therefore, SNORD44 and SNORD48 play a
suppressing tumor function role in these cancers. Valleron et
al found SNORD112, SNORD113 and SNORD114 ectopic expression in
acute promyelocytic leukemia (APL), and further molecular studies
also shown SNORD114-1 variants through the retinoblastoma gene
(Rb)/p16 signaling pathway, allowing cells to block G0/G1 and
reduce the proportion of S phase, thereby inhibiting cell growth
(38). At the same time, it has been
found that SNORD123 was highly expressed in colorectal cancer cells
and could affect tumorigenesis by epigenetic modification (39). The levels of SNORD33, SNORD66 and
SNORD76 in the plasma of patients with non-small cell lung cancer
were significantly higher than those in healthy and chronic
obstructive pulmonary diseases, and had the value of early
diagnosis of lung cancer (40).
Overall, snoRNA was closely correlated with the pathogenesis and
progression of various tumors, but the expression pattern and
functional role of snoRNA in OC has not been elucidated.
Here, we found that SNORD114-10 levels were reduced
in metastatic omentum, suggesting that this snoRNA might have
properties that suppress migration of ovarian tumor cells to
secondary sites. SNORD114-10 maybe inhibited the metastasis of OC
by regulating the expression of certain genes or through the EMT or
AKT pathway. Next we will determine whether SNORD114-10 can
regulate migration in OC cells by silencing and over-expressing
SNORD114-10. Further study are required to determine the role of
SNORD114-10 (the correlation between) in OC metastasis/the
metastssis of OC.
Because preoperative OC omentum metastasis is
difficult to diagnose, so it is not conducive to preoperative
surgical options. Therefore, we believe that MIAT and SNORD114-10
can be used as a marker of OC OM, which can guide OC patients'
preoperative choice.
Acknowledgements
The present study was supported by the Fundamental
Research Funds of Central South University (no. 2015zzts297),
Program for Science and Technology Plan of Hunan Province (no.
2013FJ4114) and Applied of Fundamental Research Key Prjoect (no.
2016JC2036). We are grateful for all the contributions that
supported this study.
References
|
1
|
Liu CM: Cancer of the ovary. N Engl J Med.
352:1268–1269. 2005. View Article : Google Scholar
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar
|
|
3
|
Lengyel E: Ovarian cancer development and
metastasis. Am J Pathol. 177:1053–1064. 2010. View Article : Google Scholar
|
|
4
|
Naora H and Montell DJ: Ovarian cancer
metastasis: Integrating insights from disparate model organisms.
Nat Rev Cancer. 5:355–366. 2005. View
Article : Google Scholar
|
|
5
|
Pradeep S, Kim SW, Wu SY, Nishimura M,
Chaluvally-Raghavan P, Miyake T, Pecot CV, Kim SJ, Choi HJ,
Bischoff FZ, et al: Hematogenous metastasis of ovarian cancer:
Rethinking mode of spread. Cancer Cell. 26:77–91. 2014. View Article : Google Scholar
|
|
6
|
Cooper C, Jeffrey RB, Silverman PM,
Federle MP and Chun GH: Computed tomography of omental pathology. J
Comput Assist Tomogr. 10:62–66. 1986. View Article : Google Scholar
|
|
7
|
Liu T, Zhao L, Chen W, Li Z, Hou H, Ding L
and Li X: Inactivation of von Hippel-Lindau increases ovarian
cancer cell aggressiveness through the HIF1α/miR-210/VMP1 signaling
pathway. Int J Mol Med. 33:1236–1242. 2014. View Article : Google Scholar
|
|
8
|
Prat J; FIGO Committee on Gynecologic
Oncology, : Staging classification for cancer of the ovary,
fallopian tube and peritoneum: Abridged republication of guidelines
from the international federation of gynecology and obstetrics
(FIGO). Obstet Gynecol. 126:171–174. 2015. View Article : Google Scholar
|
|
9
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar
|
|
10
|
Carninci P and Hayashizaki Y: Noncoding
RNA transcription beyond annotated genes. Curr Opin Genet Dev.
17:139–144. 2007. View Article : Google Scholar
|
|
11
|
Pauli A, Rinn JL and Schier AF: Non-coding
RNAs as regulators of embryogenesis. Nat Rev Genet. 12:136–149.
2011. View
Article : Google Scholar
|
|
12
|
Keniry A, Oxley D, Monnier P, Kyba M,
Dandolo L, Smits G and Reik W: The H19 lincRNA is a developmental
reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell
Biol. 14:659–665. 2012. View
Article : Google Scholar
|
|
13
|
McHugh CA, Chen CK, Chow A, Surka CF, Tran
C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, et
al: The Xist lncRNA interacts directly with SHARP to silence
transcription through HDAC3. Nature. 521:232–236. 2015. View Article : Google Scholar
|
|
14
|
Tahira AC, Kubrusly MS, Faria MF, Dazzani
B, Fonseca RS, Maracaja-Coutinho V, Verjovski-Almeida S, Machado MC
and Reis EM: Long noncoding intronic RNAs are differentially
expressed in primary and metastatic pancreatic cancer. Mol Cancer.
10:1412011. View Article : Google Scholar
|
|
15
|
Matouk IJ, Abbasi I, Hochberg A, Galun E,
Dweik H and Akkawi M: Highly upregulated in liver cancer noncoding
RNA is overexpressed in hepatic colorectal metastasis. Eur J
Gastroenterol Hepatol. 21:688–692. 2009. View Article : Google Scholar
|
|
16
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar
|
|
17
|
Han Y, Liu Y, Nie L, Gui Y and Cai Z:
Inducing cell proliferation inhibition, apoptosis and motility
reduction by silencing long noncoding ribonucleic acid
metastasis-associated lung adenocarcinoma transcript 1 in
urothelial carcinoma of the bladder. Urology. 81:209.e1–e7. 2013.
View Article : Google Scholar
|
|
18
|
Hou Z, Xu X, Zhou L, Fu X, Tao S, Zhou J,
Tan D and Liu S: The long non-coding RNA MALAT1 promotes the
migration and invasion of hepatocellular carcinoma by sponging
miR-204 and releasing SIRT1. Tumour Biol. 39:10104283177181352017.
View Article : Google Scholar
|
|
19
|
Schmidt LH, Spieker T, Koschmieder S,
Schäffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D,
Marra A, et al: The long noncoding MALAT-1 RNA indicates a poor
prognosis in non-small cell lung cancer and induces migration and
tumor growth. J Thorac Oncol. 6:1984–1992. 2011. View Article : Google Scholar
|
|
20
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar
|
|
21
|
Xu ZY, Yu QM, Du YA, Yang LT, Dong RZ,
Huang L, Yu PF and Cheng XD: Knockdown of long non-coding RNA
HOTAIR suppresses tumor invasion and reverses
epithelial-mesenchymal transition in gastric cancer. Int J Biol
Sci. 9:587–597. 2013. View Article : Google Scholar
|
|
22
|
Nakagawa T, Endo H, Yokoyama M, Abe J,
Tamai K, Tanaka N, Sato I, Takahashi S, Kondo T and Satoh K: Large
noncoding RNA HOTAIR enhances aggressive biological behavior and is
associated with short disease-free survival in human non-small cell
lung cancer. Biochem Biophys Res Commun. 436:319–324. 2013.
View Article : Google Scholar
|
|
23
|
Li HM, Yang H, Wen DY, Luo YH, Liang CY,
DH, Ma W, Chen G, He Y and Chen JQ: Overexpression of LncRNA HOTAIR
is associated with poor prognosis in thyroid carcinoma: A study
based on TCGA and GEO data. Horm Metab Res. 49:388–399. 2017.
View Article : Google Scholar
|
|
24
|
Zhang LQ, Yang SQ, Wang Y, Fang Q, Chen
XJ, Lu HS and Zhao LP: Long noncoding RNA MIR4697HG promotes cell
growth and metastasis in human ovarian cancer. Anal Cell Pathol
(Amst). 2017:82678632017.
|
|
25
|
Tsuiji H, Yoshimoto R, Hasegawa Y, Furuno
M, Yoshida M and Nakagawa S: Competition between a noncoding exon
and introns: Gomafu contains tandem UACUAAC repeats and associates
with splicing factor-1. Genes Cells. 16:479–90. 2011. View Article : Google Scholar
|
|
26
|
Mercer TR, Dinger ME, Sunkin SM, Mehler MF
and Mattick JS: Specific expression of long noncoding RNAs in the
mouse brain. Proc Natl Acad Sci USA. 105:pp. 716–721. 2008;
View Article : Google Scholar
|
|
27
|
Sone M, Hayashi T, Tarui H, Agata K,
Takeichi M and Nakagawa S: The mRNA-like noncoding RNA gomafu
constitutes a novel nuclear domain in a subset of neurons. J Cell
Sci. 120:2498–2506. 2007. View Article : Google Scholar
|
|
28
|
Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li
YJ, Tao ZF, Song YC, Chen Q and Jiang Q: lncRNA-MIAT regulates
microvascular dysfunction by functioning as a competing endogenous
RNA. Circ Res. 116:1143–1156. 2015. View Article : Google Scholar
|
|
29
|
Luan T, Zhang X, Wang S, Song Y, Zhou S,
Lin J, An W, Yuan W, Yang Y, Cai1 H, et al: Long non-coding RNA
MIAT promotes breast cancer progression and functions as ceRNA to
regulate DUSP7 expression by sponging miR-155-5p. Oncotarget.
8:76153–76164. 2017.
|
|
30
|
Shen Y, Dong LF, Zhou RM, Yao J, Song YC,
Yang H, Jiang Q and Yan B: Role of long non-coding RNA MIAT in
proliferation, apoptosis and migration of lens epithelial cells: A
clinical and in vitro study. J Cell Mol Med. 20:537–548. 2016.
View Article : Google Scholar
|
|
31
|
Scott MS and Ono M: From snoRNA to miRNA:
Dual function regulatory non-coding RNAs. Biochimie. 93:1987–1992.
2011. View Article : Google Scholar
|
|
32
|
Brameier M, Herwig A, Reinhardt R, Walter
L and Gruber J: Human box C/D snoRNAs with miRNA like functions:
Expanding the range of regulatory RNAs. Nucleic Acids Res.
39:675–686. 2011. View Article : Google Scholar
|
|
33
|
Ono M, Scott MS, Yamada K, Avolio F,
Barton GJ and Lamond AI: Identification of human miRNA precursors
that resemble box C/D snoRNAs. Nucleic Acids Res. 39:3879–3891.
2011. View Article : Google Scholar
|
|
34
|
Mannoor K, Liao J and Jiang F: Small
nucleolar RNAs in cancer. Biochim Biophys Acta. 1826:121–128.
2012.
|
|
35
|
Thorenoor N and Slaby O: Small nucleolar
RNAs functioning and potential roles in cancer. Tumour Biol.
36:41–53. 2015. View Article : Google Scholar
|
|
36
|
Mei YP, Liao JP, Shen J, Yu L, Liu BL, Liu
L, Li RY, Ji L, Dorsey SG, Jiang ZR, et al: Small nucleolar RNA 42
acts as an oncogene in lung tumorigenesis. Oncogene. 31:2794–2804.
2012. View Article : Google Scholar
|
|
37
|
Gee HE, Buffa FM, Camps C, Ramachandran A,
Leek R, Taylor M, Patil M, Sheldon H, Betts G, Homer J, et al: The
small-nucleolar RNAs commonly used for microRNA normalisation
correlate with tumour pathology and prognosis. Br J Cancer.
104:1168–1177. 2011. View Article : Google Scholar
|
|
38
|
Valleron W, Laprevotte E, Gautier EF,
Quelen C, Demur C, Delabesse E, Agirre X, Prósper F, Kiss T and
Brousset P: Specific small nucleolar RNA expression profiles in
acute leukemia. Leukemia. 26:2052–2060. 2012. View Article : Google Scholar
|
|
39
|
Ferreira HJ, Heyn H, Moutinho C and
Esteller M: CpG island hypermethylation-associated silencing of
small nucleolar RNAs in human cancer. RNA Biol. 9:881–890. 2012.
View Article : Google Scholar
|
|
40
|
Liao J, Yu L, Mei Y, Guarnera M, Shen J,
Li R, Liu Z and Jiang F: Small nucleolar RNA signatures as
biomarkers for non-small-cell lung cancer. Mol Cancer. 9:1982010.
View Article : Google Scholar
|