Introduction
Hepatocellular carcinoma (HCC) is the fifth most
prevalent malignancy and the third-leading cause of
cancer-associated mortality worldwide (1). It was reported that there were 782,500
new HCC cases and 745,500 incidences of mortality due to HCC
worldwide during 2012, of which half were in China (2). Numerous risk factors for HCC patients
have been identified, including chronic viral hepatitis B or C
infection, chemical exposure, alcohol consumption, obesity,
hereditary hemochromatosis and hepatosteatosis (3). At present, the primary therapeutic
strategies for HCC are surgical resection, liver transplantation
and ablation, and chemotherapy (4).
For patients with early stage of HCC, the primary tumor can often
be completely resected; however, patients with advanced-stage HCC
have a poor prognosis due to the likelihood of intrahepatic
recurrence and tumor metastasis (5,6).
Furthermore, the cellular and molecular processes underlying the
recurrence and metastasis of HCC remain poorly characterized. A
complete understanding of the mechanisms that mediate HCC
progression could aid the identification of novel therapeutic
targets and consequently improve the prognosis for patients with
HCC.
The identification of microRNAs (miRNAs/miRs) has
enabled an improved understanding of carcinogenesis and HCC
progression. miRNAs are small non-coding RNAs comprised of 19–24
nucleotides (7) that negatively
regulate the expression of target genes at the post-transcriptional
level through interaction with the 3′-untranslated regions (3′UTRs)
of target mRNAs, resulting in either mRNA degradation or the
inhibition of translation (8). Since
the identification of the first miRNA, lin-4, in 1993, the
existence of a large number of miRNAs has been validated (9). At the time of writing, ~1,000 miRNAs
have been identified in humans (10).
miRNAs are believed to modulate more than a third of human genes
(11) and therefore serve notable
functions in a large number of biological processes, including cell
proliferation, cell cycle, apoptosis, differentiation, metabolism,
migration and invasion (12–14). Furthermore, increasing evidence has
demonstrated that the abnormal expression of miRNAs contributes to
the initiation and development of human malignancies, including HCC
(15–17). Although numerous miRNAs have been
identified, their expression and roles in HCC carcinogenesis and
progression, and the underlying mechanisms by which this occurs,
remain largely unidentified.
Thus, the present study aimed to determine the level
of miR-138 in HCC tissues and cells, its effects on cell
proliferation, migration and invasion, and the mechanisms for the
observed effects, including the potential targets of miR-138.
miR-138 was demonstrated to be downregulated in HCC and inhibited
cell proliferation, migration and invasion via directly targeting
SP1. These findings provide supplementation to the general
molecular mechanisms of carcinogenesis and progression in HCC and
may aid the identification of novel targets for HCC treatment.
Materials and methods
Human tissue specimens
The present study was approved by the Ethics
Committee of Qilu Hospital (Jinan, China) and each patient provided
informed written consent. A total of 32 paired HCC tissues and
corresponding adjacent non-tumor liver tissues were obtained by
surgical resection from HCC patients (male, 23; female, 9; age
range, 43–75 years; mean age 58 years) at Qilu Hospital. None of
these HCC patients were treated with chemotherapy or radiotherapy
prior to surgery. Fresh tissues were immediately frozen in liquid
nitrogen and stored at −80°C.
Cell lines, cell culture and cell
transfection
Immortalized human hepatocyte LO2 cells, and HepG2,
Hep3B, SMMC-7721, Huh7 and BEL-7402 HCC cells were purchased from
American Type Culture Collection (ATCC; Manassas, VA, USA). All
cells were cultured in Dulbecco's modified Eagle's medium (DMEM)
supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin
and 100 mg/ml streptomycin (all Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA). An miR-138 mimic, the corresponding
negative control (NC), small interfering RNA (siRNA) for SP1
(si-SP1) and si-NC were produced by Shanghai GenePharma Co., Ltd.
(Shanghai, China). The miR-138 mimics sequence was
5′-AGCUGGUGUUGUGAAUCAGGCCG-3′ and the NC sequence was
5′-UUCUCCGAACGUGUCACGUTT-3′. The si-SP1 sequence was
5′-GCAACAUGGGAAUUAUGAATT-3′ and the si-NC sequence was
5′-UUCUCCGAACGUGUCACGUTT-3′. Cells were seeded in 6-well plates at
60–70% confluence and transfected with the oligonucleotides using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturer's
instructions.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues and cell lines
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. The relative expression
levels of miR-138 were measured using a One Step SYBR®
PrimeScript™ miRNA RT-PCR kit (Takara Bio, Inc., Otsu, Japan)
according to the manufacturer's protocol. The reaction conditions
were as follows: 42°C for 5 min, 95°C for 10 sec, followed by 40
cycles of 95°C for 5 sec, 55°C for 30 sec and 70°C for 30 sec. To
assess SP1 mRNA expression levels, cDNA was synthesized using M-MLV
reverse transcriptase (Promega Corporation, Madison, WI, USA) and
qPCR was performed using SYBR green I mix (Takara Bio, Inc.). The
thermocycling conditions for qPCR were as follows: 95°C for 10 min,
followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The
primers were designed as follows: miR-138,
5′-TCCGAGCCTGACTAAGTGTTGTGGTCGA-3′ (forward) and
5′-GTGCAGGGTCCGAGGT-3′ (reverse); U6,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward) and
5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse); SP1,
5′-TGGTGGGCAGTATGTTGT-3′ (forward) and 5′-GCTATTGGCATTGGTGAA-3′
(reverse); and GAPDH, 5′-GGAGTCAACGGATTTGGT-3′ (forward) and
5′-GTGATGGGATTTCCATTGAT-3′ (reverse). U6 small nuclear RNA and
GADPH mRNA were used as internal controls for miR-138 and SP1 mRNA
expression, respectively. All experiments were performed in
triplicate, and data were analyzed with the 2−∆∆Ct
method (18).
Cell proliferation assay
At 48 h after transfection, HepG2 and Huh7 cells
were harvested and re-seeded into 96-well plates at a density of
3,000 cells per well. Cell proliferation was determined at 24, 48,
72 and 96 h after plating using Cell Counting Kit-8 (CCK8; Dojindo
Molecular Technologies, Inc., Kumamoto, Japan) according to the
manufacturer's protocol. In brief, cells were incubated with 10 µl
CCK8 solution for 2 h in a humidified atmosphere containing 5%
CO2 at 37°C. The optical density was measured at 450 nm
using an ELISA reader (Bio-Rad Laboratories, Inc., Hercules, CA,
USA). Each experiment was repeated at least three times.
Transwell cell migration and invasion
assay
Cell migration and invasion were assessed using
Transwell chambers (8-µm pore size; Corning, Inc., Corning, NY,
USA) either with or without Matrigel (BD Biosciences, San Jose, CA,
USA). At 48 h after transfection, HepG2 and Huh7 cells were
harvested and re-seeded into the upper chamber at a density of
3×104 cells in 200 µl fetal bovine serum (FBS)-free
culture medium. The bottom chamber was filled with 600 µl culture
medium containing 20% FBS. Subsequent incubation for 48 h in a
humidified atmosphere containing 5% CO2 at 37°C, the
cells remaining on the top surface of the membrane were removed
using a cotton swab. The migrated and invaded cells were fixed with
95% ethanol for 10 min at room temperature, stained with 0.5%
crystal violet for 10 min at room temperature, and 5 random fields
were counted under a light microscope (magnification, ×200).
miRNA target-gene prediction
TargetScan (http://www.targetscan.org/) and PicTar (http://pictar.bio.nyu.edu) were used to predict the
potential target genes of miR-138.
Luciferase reporter assay
The wild-type [pMIR-SP1-3′UTR Wt 1 and Wt 2] and
mutant (pMIR-SP1-3′UTR Mut 1 and Mut 2) SP1 3′UTR cloned in
pMIR-REPORT were produced by Shanghai GenePharma Co., Ltd. HEK293T
cells were seeded in a 24-well plate at a density of 40–50%
confluence. Following incubation overnight, HEK293T cells were
co-transfected with miR-138 mimics or NC, and pMIR-SP1-3′UTR Wt or
pMIR-SP1-3′UTR Mut using Lipofectamine® 2000 reagent,
following the manufacturer's recommendations. At 48 h after
transfection, cells were harvested and firefly luciferase activity
was detected using the Dual-Luciferase Reporter system (Promega
Corporation) and normalized to that of Renilla luciferase,
according to the manufacturer's protocol.
Western blot analysis
At 72 h after transfection, cells were lysed with
ice-cold lysis buffer supplemented with protease inhibitor cocktail
(Pierce; Thermo Fisher Scientific, Inc.). Total protein
concentration was determined using the Enhanced BCA Protein Assay
kit (Beyotime Institution of Biotechnology, Haimen, China). Protein
samples (20 µg) were separated on a 10% SDS-PAGE and then
transferred to a polyvinylidene fluoride membrane. Subsequent to
blocking in 5% skimmed milk at room temperature for 2 h, the
membranes were incubated overnight at 4°C with mouse anti-human SP1
monoclonal primary antibody (1:1,000, ab77441; Abcam, Cambridge,
UK) and mouse anti-human GADPH monoclonal primary antibody
(1:1,000, ab125247; Abcam) primary antibodies. Next, the membranes
were incubated with the goat anti-mouse horseradish
peroxidase-conjugated secondary antibody (1:3,000, ab6789; Abcam)
for 1 h at room temperature. The membranes were visualized with
Super Signal West Pico Chemiluminescent Substrate kit (Pierce;
Thermo Fisher Scientific, Inc.). GADPH was used as a loading
control.
Statistical analysis
Data were expressed as mean ± standard deviation and
compared with Student's t-test or one-way analysis of variance
(ANOVA) using SPSS 13.0 (SPSS Inc., Chicago, IL, USA). miR-138
expression in HCC cell lines were analyzed with ANOVA and other
results were analyzed by Student's t-test. SNK was used as a post
hoc test following ANOVA. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-138 is downregulated in HCC
tissues and cell lines
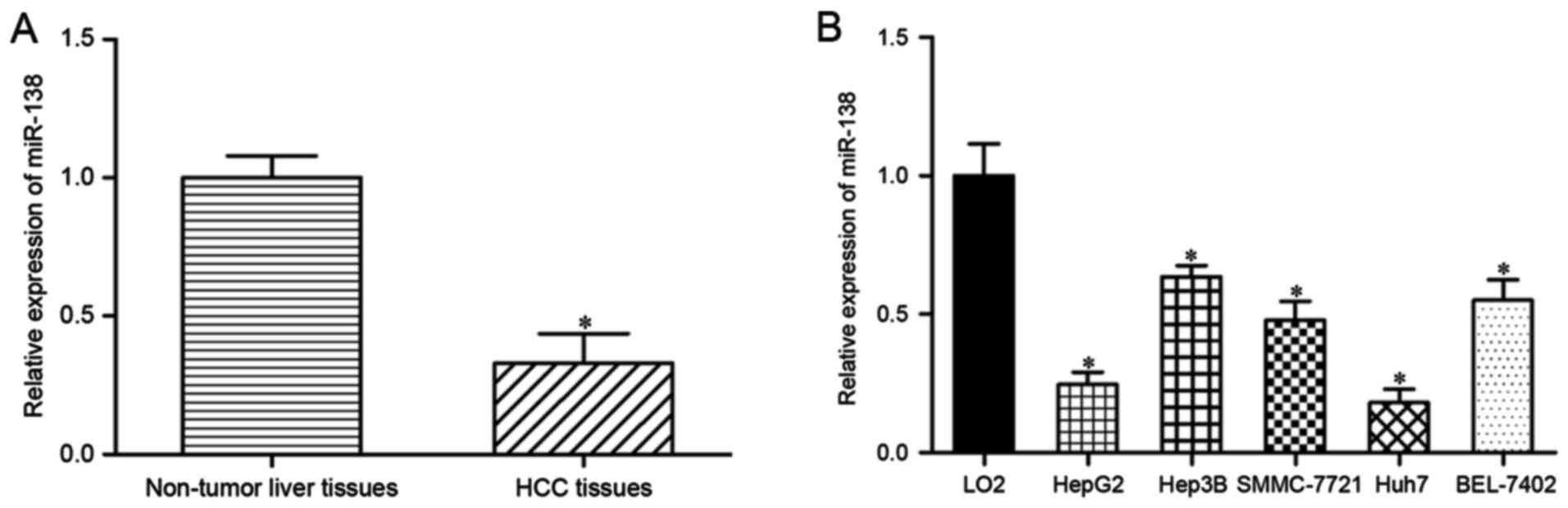
The expression levels of miR-138 were measured in
HCC tissues and their corresponding adjacent non-tumor liver
tissues using RT-qPCR. miR-138 was significantly downregulated in
HCC tissues compared with that in adjacent non-tumor liver tissue
(Fig. 1A; P<0.05). miR-138
expression levels were also assessed in HepG2, Hep3B, SMMC-7721,
Huh7 and BEL-7402 HCC cells. As demonstrated in Fig. 1B, miR-138 expression levels were
reduced in the HCC cell lines compared with the immortalized human
hepatocyte LO2 cell line (P<0.05). Therefore, these results
indicated that miR-138 was downregulated in HCC tissues and cell
lines.
miR-138 suppresses HCC cell
proliferation, migration and invasion
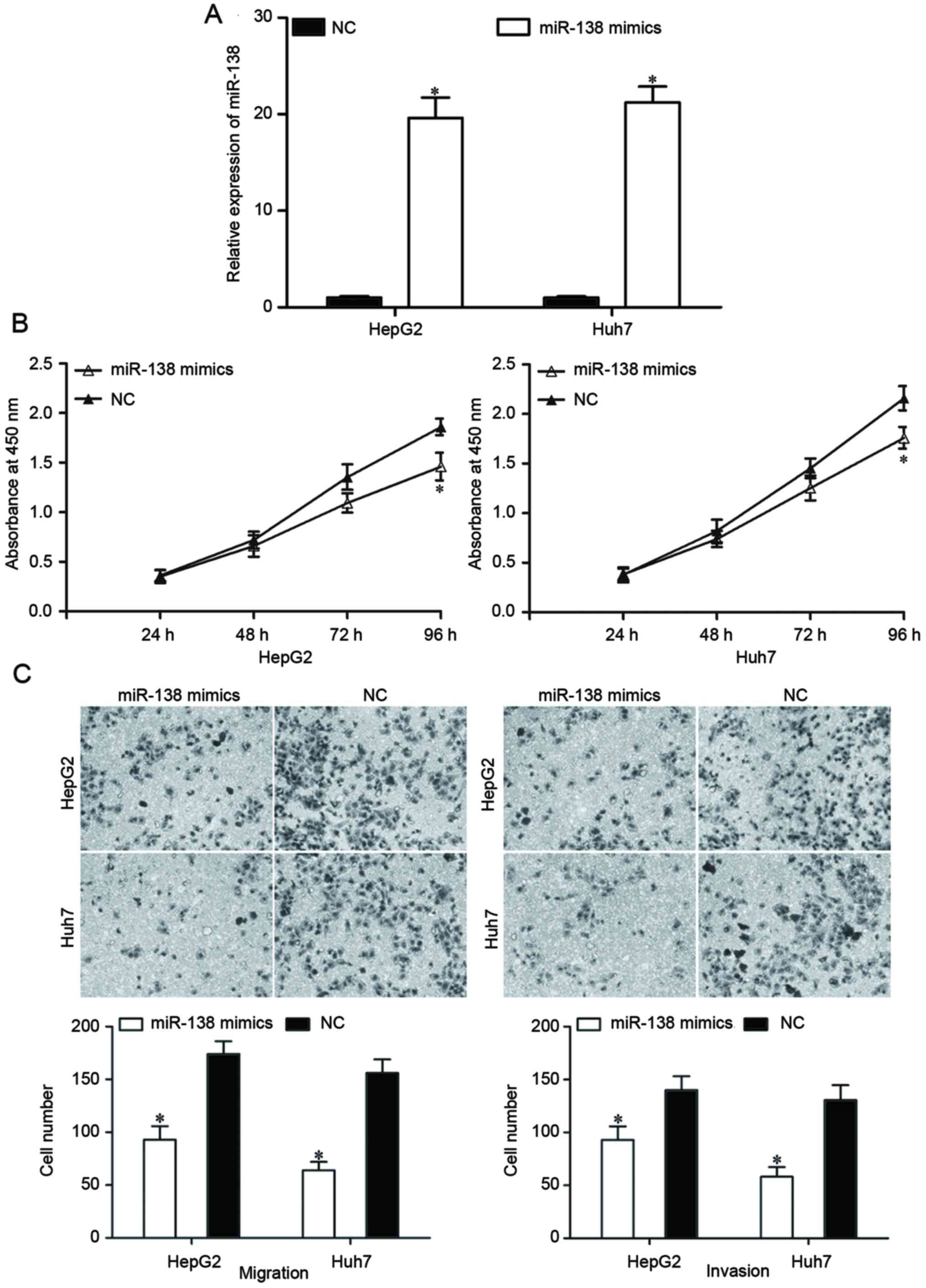
To determine the roles of miR-138 in HCC, an miR-138
mimic was transfected into HepG2 and Huh7 cells. Following
transfection with the miR-138 mimic or an NC, the expression levels
of miR-138 in HepG2 and Huh7 cells were evaluated by RT-qPCR. The
expression in the miR-138 mimic-transfected cells was significantly
increased (Fig. 2A, P<0.05).
A cell proliferation assay was performed to
investigate the effect of miR-138 on HCC cell proliferation. The
results revealed that the restoration of miR-138 expression
suppressed the proliferation of HepG2 and Huh7 cells (Fig. 2B; P<0.05). A Transwell migration
and invasion assay was performed to investigate the effects of
miR-138 on the migration and invasion capacity of HCC cells. As
demonstrated in Fig. 2C, the
migratory and invasive abilities of HepG2 and Huh7 cells
transfected with the miR-138 mimic were significantly reduced
compared with cells transfected with the NC (P<0.05). These
findings indicated that miR-138 can act as a potential tumor
suppressor in HCC.
miR-138 directly targets SP1 in
HCC
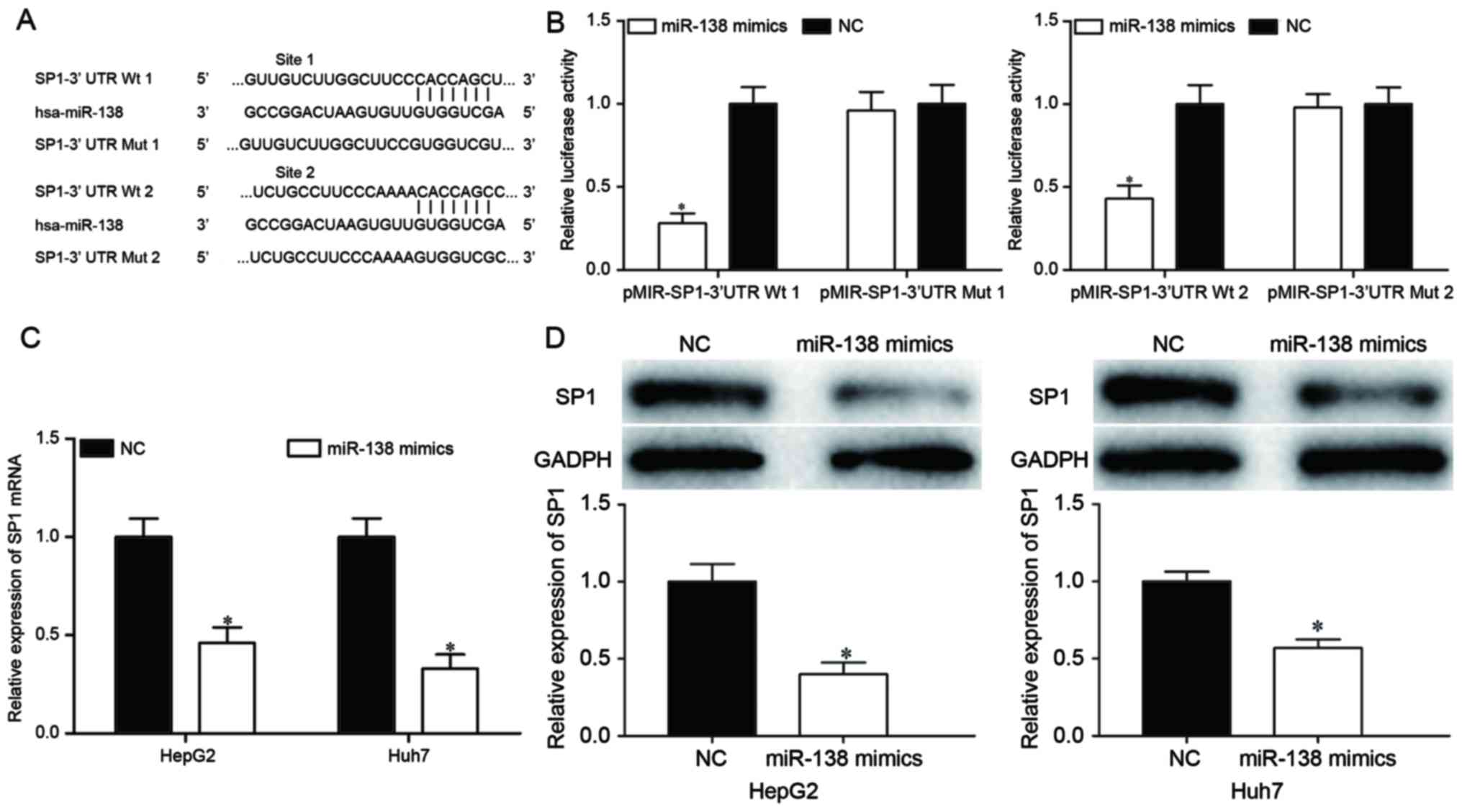
To investigate the mechanisms of miR-138-induced
cell proliferation, migration and invasion inhibition, the target
genes of miR-138 were searched for using TargetScan and PicTar.
Bioinformatics analysis demonstrated that SP1 may be a target of
miR-138, based on putative target sequences at the SP1 3′UTR
(Fig. 3A). To confirm whether SP1 was
a target of miR-138, a dual-luciferase reporter assay was
performed, which identified that miR-138 overexpression markedly
decreased the luciferase activities with pMIR-SP1-3′UTR Wt 1 and Wt
2, but not pMIR-SP1-3′UTR Mut 1 and Mut 2, indicating that miR-138
directly targeted the 3′UTR of SP1 (Fig.
3B; P<0.05). Furthermore, RT-qPCR and western blot analysis
revealed that ectopic miR-138 expression suppressed SP1 expression
compared with transfection with the NC in HepG2 and Huh7 cells at
the mRNA (Fig. 3C; P<0.05) and
protein (Fig. 3D; P<0.05) levels.
These results suggested that SP1 was a direct target gene of
miR-138 in HCC.
Knockdown of SP1 produces similar
effects to miR-138 overexpression in HCC cells
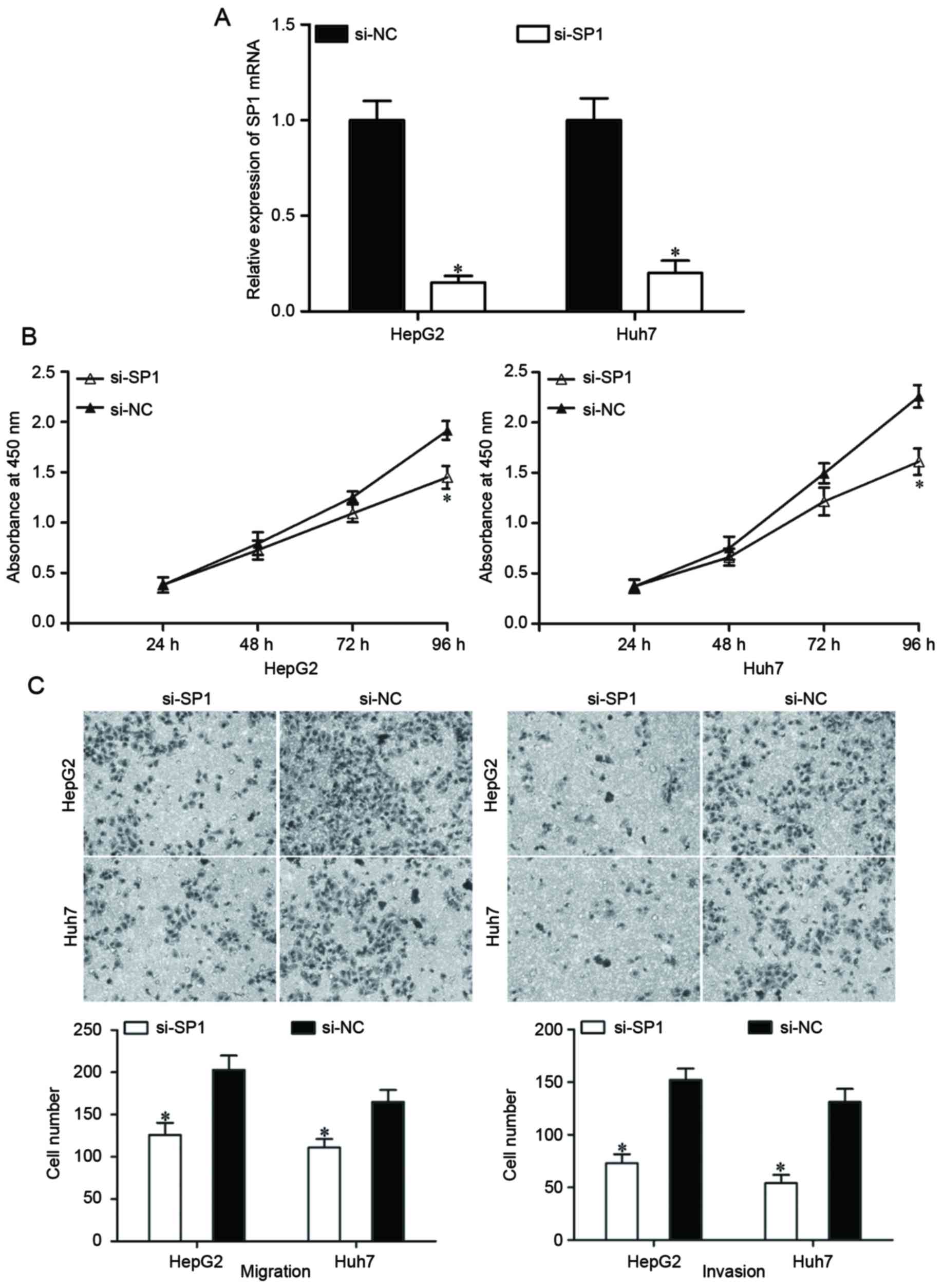
SP1 expression was knocked down using si-SP1 to
assess whether SP1 suppressed HCC cell proliferation, migration and
invasion as induced by miR-138 overexpression. At 48 h after
transfection, the expression of SP1 mRNA was determined using
RT-qPCR (Fig. 4A; P<0.05). Based
on the results of cell proliferation and Transwell migration and
invasion assays, knockdown of SP1 resulted in the suppression of
HepG2 and Huh7 cell proliferation (Fig.
4B; P<0.05), migration and invasion (Fig. 4C; P<0.05), which were similar to
the effects of miR-138 overexpression in HepG2 and Huh7 cells.
These results demonstrated that miR-138 repressed proliferation,
migration and invasion of HCC cells by downregulating SP1.
Discussion
miRNAs have emerged as critical regulators of HCC
progression and tumorigenesis (19,20). In
the present study, RT-qPCR was performed to measure miR-138
expression in HCC tissues and cell lines. The data revealed that
miR-138 expression levels were reduced in HCC tissues and cell
lines, compared with corresponding adjacent non-tumor liver
tissues, and an immortalized human hepatocyte cell line,
respectively. Further experiments demonstrated that miR-138 also
suppressed HCC cell proliferation, migration and invasion. SP1 was
validated to be a direct target gene of miR-138 in HCC.
miR-138/SP1-based targeted therapy could therefore represent a
promising therapeutic strategy for HCC patients.
Downregulation of miR-138 has been reported in
diverse types of human cancer, including anaplastic thyroid
carcinoma (21), head and neck
squamous cell carcinoma (22), clear
cell renal cell carcinoma (23),
ovarian cancer (24), glioblastoma
(25), colorectal cancer (26), non-small cell lung cancer (NSCLC)
(27), pancreatic cancer (28) and breast cancer (29). Furthermore, reduced miR-138 expression
was associated with a number of cancer characteristics. For
example, in NSCLC, low miR-138 expression levels were significantly
associated with an advanced clinical stage and positive lymph node
metastasis (27). In addition, the
overall survival of NSCLC patients with low miR-138 expression was
significantly shorter than those with high miR-138 expression
(27). Zhang et al (29) reported that low miR-138 expression
levels were associated with lymph node metastasis and invasion in
breast cancer. Another study revealed that in HCC, miR-138 was
associated with an advanced clinical stage, the presence of portal
vein invasion and lymph node metastasis (30). These studies suggested that
deregulation of miR-138 may be implicated in carcinogenesis and
progression of human cancer, and may be a promising prognostic
biomarker in these types of human cancer.
Studies have also revealed that the dysregulation of
miR-138 is associated with the carcinogenesis and progression of
diverse types of human cancer. Zhang et al (29) reported that miR-138 regulated
metastasis and the epithelial-mesenchymal transition in breast
cancer cells. In NSCLC, miR-138 suppressed cell proliferation,
migration, in vitro cisplatin sensitivity and in vivo
tumor growth (31–33). The restoration of miR-138 expression
in NSCLC cells was found to reverse gefitinib resistance (34). Xu et al (35) revealed that miR-138 overexpression
suppressed oral squamous cell carcinoma (36) and gallbladder carcinoma cell growth.
Yang et al (26) found that
miR-138 expression resulted in a significant inhibition of
colorectal cancer cell migration and invasion in vitro and
in vivo. In ovarian cancer, miR-138 overexpression repressed
cancer cell invasion and metastasis (24). In nasopharyngeal carcinoma, miR-138
markedly decreased cell proliferation and colony formation in
vitro, and tumorigenesis in vivo (37). In head and neck squamous cell
carcinoma, ectopic transfection with miR-138 inhibited cell
invasion, and enhanced cell-cycle arrest and apoptosis (22). The evidence from these prior studies,
together with the findings of the present study, indicates that
miR-138 acts as a tumor suppressor in cancer.
Evidence suggests that miRNAs can exert functions as
oncogenes or tumor suppressors in carcinogenesis and progression of
human cancer depending on the roles of their target genes (38–40).
Therefore, identification of miR-138 target genes involved in HCC
tumorigenesis and progression may provide valuable insight for the
diagnosis and therapeutic treatments of patients with HCC. Numerous
genes are directly regulated by miR-138, including vimentin in
breast cancer (29), zinc finger
E-box-binding homeobox 2 in bladder cancer (41), ARF GTPase-activating protein GIT1,
semaphoring 4C (42) and
G1/S-specific cyclin-D3 (33) in
NSCLC, transcriptional coactivator YAP1 in oral squamous cell
carcinoma (36) and BAG family
molecular chaperone regulator 1 in gallbladder carcinoma (35). In the present study, SP1 was
identified as a novel direct target gene for miR-138. Bioinformatic
analysis revealed that there are two putative binding sites of
miR-138 in the 3′UTR of SP1. A dual-luciferase reporter assay
demonstrated that miR-138 could directly bind to the 3′UTR of the
SP1 gene. RT-qPCR and western blot analysis further indicated that
the restoration of miR-138 expression decreased the expression of
SP1 mRNA and protein in HCC cells. Notably, knockdown of SP1
resulted in the suppression of HCC cell proliferation, migration
and invasion, similar to the effects of miR-138 overexpression in
HCC cells. Collectively, these findings indicate that miR-138
represses HCC cell proliferation, migration and invasion via the
negative regulation of endogenous SP1 expression at the
transcriptional and translational levels.
The present study demonstrated that miR-138 was
significantly downregulated in HCC tissues and cell lines and that
miR-138 overexpression repressed HCC cell proliferation, migration
and invasion. Furthermore, SP1 was identified as a direct target
gene of miR-138 in HCC. These results indicate that miR-138 may be
associated with the tumorigenesis and progression of HCC, and may
therefore represent a potential target for HCC treatment.
Acknowledgements
This study was supported by the Natural Science
Foundation of Shandong (grant nos. ZR2012HM064 and
ZR2009CM126).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu Y, Li Y, Wang R, Qin S, Liu J, Su F,
Yang Y, Zhao F, Wang Z and Wu Q: MiR-130a-3p regulates cell
migration and invasion via inhibition of Smad4 in gemcitabine
resistant hepatoma cells. J Exp Clin Cancer Res. 35:192016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kindrat I, Tryndyak V, de Conti A,
Shpyleva S, Mudalige TK, Kobets T, Erstenyuk AM, Beland FA and
Pogribny IP: MicroRNA-152-mediated dysregulation of hepatic
transferrin receptor 1 in liver carcinogenesis. Oncotarget.
7:1276–1287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen PJ, Furuse J, Han KH, Hsu C, Lim HY,
Moon H, Qin S, Ye SL, Yeoh EM and Yeo W: Issues and controversies
of hepatocellular carcinoma-targeted therapy clinical trials in
Asia: Experts' opinion. Liver Int. 30:1427–1438. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pelus LM and Fukuda S: Peripheral blood
stem cell mobilization: The CXCR2 ligand GRObeta rapidly mobilizes
hematopoietic stem cells with enhanced engraftment properties. Exp
Hematol. 34:1010–1020. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cortez MA and Calin GA: MicroRNA
identification in plasma and serum: A new tool to diagnose and
monitor diseases. Expert Opin Biol Ther. 9:703–711. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee RC, Feinbaum RL and Ambros V: The C.
Elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Griffiths-Jones S, Saini HK, van Dongen S
and Enright AJ: miRBase: Tools for microRNA genomics. Nucleic Acids
Res. 36(Database issue): D154–D158. 2008.PubMed/NCBI
|
|
11
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He L, Thomson JM, Hemann MT,
Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe
SW, Hannon GJ and Hammond SM: A microRNA polycistron as a potential
human oncogene. Nature. 435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang JG, Shi Y, Hong DF, Song M, Huang D,
Wang CY and Zhao G: MiR-148b suppresses cell proliferation and
invasion in hepatocellular carcinoma by targeting WNT1/β-catenin
pathway. Sci Rep. 5:80872015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zheng B, Liang L, Wang C, Huang S, Cao X,
Zha R, Liu L, Jia D, Tian Q, Wu J, et al: MicroRNA-148a suppresses
tumor cell invasion and metastasis by downregulating ROCK1 in
gastric cancer. Clin Cancer Res. 17:7574–7583. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen X, Bo L, Zhao X and Chen Q:
MicroRNA-133a inhibits cell proliferation, colony formation
ability, migration and invasion by targeting matrix
metallopeptidase 9 in hepatocellular carcinoma. Mol Med Rep.
11:3900–3907. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang W, Liu K, Liu S, Ji B, Wang Y and
Liu Y: MicroRNA-133a functions as a tumor suppressor by targeting
IGF-1R in hepatocellular carcinoma. Tumour Biol. 36:9779–9788.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dong Y, Zou J, Su S, Huang H, Deng Y, Wang
B and Li W: MicroRNA-218 and microRNA-520a inhibit cell
proliferation by downregulating E2F2 in hepatocellular carcinoma.
Mol Med Rep. 12:1016–1022. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ji J, Shi J, Budhu A, Yu Z, Forgues M,
Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, et al: MicroRNA
expression, survival, and response to interferon in liver cancer. N
Engl J Med. 361:1437–1447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang X, Chen J, Li F, Lin Y, Zhang X, Lv Z
and Jiang J: MiR-214 inhibits cell growth in hepatocellular
carcinoma through suppression of β-catenin. Biochem Biophys Res
Commun. 428:525–531. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mitomo S, Maesawa C, Ogasawara S, Iwaya T,
Shibazaki M, Yashima-Abo A, Kotani K, Oikawa H, Sakurai E, Izutsu
N, et al: Downregulation of miR-138 is associated with
overexpression of human telomerase reverse transcriptase protein in
human anaplastic thyroid carcinoma cell lines. Cancer Sci.
99:280–286. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu X, Jiang L, Wang A, Yu J, Shi F and
Zhou X: MicroRNA-138 suppresses invasion and promotes apoptosis in
head and neck squamous cell carcinoma cell lines. Cancer Lett.
286:217–222. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liang J, Zhang Y, Jiang G, Liu Z, Xiang W,
Chen X, Chen Z and Zhao J: MiR-138 induces renal carcinoma cell
senescence by targeting EZH2 and is downregulated in human clear
cell renal cell carcinoma. Oncol Res. 21:83–91. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yeh YM, Chuang CM, Chao KC and Wang LH:
MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis
by targeting SOX4 and HIF-1α. Int J Cancer. 133:867–878. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qiu S, Huang D, Yin D, Li F, Li X, Kung HF
and Peng Y: Suppression of tumorigenicity by microRNA-138 through
inhibition of EZH2-CDK4/6-pRb-E2F1 signal loop in glioblastoma
multiforme. Biochim Biophys Acta. 1832:1697–1707. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Long L, Huang G, Zhu H, Guo Y, Liu Y and
Huo J: Down-regulation of miR-138 promotes colorectal cancer
metastasis via directly targeting TWIST2. J Transl Med. 11:2752013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Han L, Zhang G, Zhang N, Li H, Liu Y, Fu A
and Zheng Y: Prognostic potential of microRNA-138 and its target
mRNA PDK1 in sera for patients with non-small cell lung cancer. Med
Oncol. 31:1292014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yu C, Wang M, Li Z, Xiao J, Peng F, Guo X,
Deng Y, Jiang J and Sun C: MicroRNA-138-5p regulates pancreatic
cancer cell growth through targeting FOXC1. Cell Oncol (Dordr).
38:173–181. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang J, Liu D, Feng Z, Mao J, Zhang C, Lu
Y, Li J, Zhang Q, Li Q and Li L: MicroRNA-138 modulates metastasis
and EMT in breast cancer cells by targeting vimentin. Biomed
Pharmacother. 77:135–141. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang B, Li H, Huang L, Luo C and Zhang Y:
Clinical significance of microRNA 138 and cyclin D3 in
hepatocellular carcinoma. J Surg Res. 193:718–723. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang H, Zhang H, Zhao M, Lv Z, Zhang X,
Qin X, Wang H, Wang S, Su J, Lv X, et al: MiR-138 inhibits tumor
growth through repression of EZH2 in non-small cell lung cancer.
Cell Physiol Biochem. 31:56–65. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ye XW, Yu H, Jin YK, Jing XT, Xu M, Wan ZF
and Zhang XY: miR-138 inhibits proliferation by targeting
3-phosphoinositide-dependent protein kinase-1 in non-small cell
lung cancer cells. Clin Respir J. 9:27–33. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Han LP, Fu T, Lin Y, Miao JL and Jiang QF:
MicroRNA-138 negatively regulates non-small cell lung cancer cells
through the interaction with cyclin D3. Tumour Biol. 37:291–298.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gao Y, Fan X, Li W, Ping W, Deng Y and Fu
X: miR-138-5p reverses gefitinib resistance in non-small cell lung
cancer cells via negatively regulating G protein-coupled receptor
124. Biochem Biophys Res Commun. 446:179–186. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ma F, Zhang M, Gong W, Weng M and Quan Z:
MiR-138 Suppresses Cell Proliferation by Targeting Bag-1 in
Gallbladder Carcinoma. PLoS One. 10:e01264992015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu R, Zeng G, Gao J, Ren Y, Zhang Z, Zhang
Q, Zhao J, Tao H and Li D: miR-138 suppresses the proliferation of
oral squamous cell carcinoma cells by targeting Yes-associated
protein 1. Oncol Rep. 34:2171–2178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu X, Lv XB, Wang XP, Sang Y, Xu S, Hu K,
Wu M, Liang Y, Liu P, Tang J, et al: MiR-138 suppressed
nasopharyngeal carcinoma growth and tumorigenesis by targeting the
CCND1 oncogene. Cell Cycle. 11:2495–2506. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sarver AL, Li L and Subramanian S:
MicroRNA miR-183 functions as an oncogene by targeting the
transcription factor EGR1 and promoting tumor cell migration.
Cancer Res. 70:9570–9580. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gong C, Yao Y, Wang Y, Liu B, Wu W, Chen
J, Su F, Yao H and Song E: Up-regulation of miR-21 mediates
resistance to trastuzumab therapy for breast cancer. J Biol Chem.
286:19127–19137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang Z, Liu S, Shi R and Zhao G: miR-27
promotes human gastric cancer cell metastasis by inducing
epithelial-to-mesenchymal transition. Cancer Genet. 204:486–491.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sun DK, Wang JM, Zhang P and Wang YQ:
MicroRNA-138 regulates metastatic potential of bladder cancer
through ZEB2. Cell Physiol Biochem. 37:2366–2374. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li J, Wang Q, Wen R, Liang J, Zhong X,
Yang W, Su D and Tang J: MiR-138 inhibits cell proliferation and
reverses epithelial-mesenchymal transition in non-small cell lung
cancer cells by targeting GIT1 and SEMA4C. J Cell Mol Med.
19:2793–2805. 2015. View Article : Google Scholar : PubMed/NCBI
|