Introduction
Lung cancer is one of the most commonest malignant
neoplasms among no matter men or women, and has the highest
mortality (1). About 80 percent of
all lung cancers are non-small cell lung cancer. Of those, ~50% are
lung adenocarcinoma (LUAD) (2).
Hence, it is important to study the biological mechanism of LUAD,
which are valuable to targeted therapy.
miRNAs play important roles in initiation and
development of cancers, which suggest that they have potential to
be targets for therapy in cancers (3). With the genetic sequencing technology
developing, differentially expression miRNAs (DEMs) in LUAD tissues
vs. noncancerous lung tissues have been detected (4). miR-373-3p is one of the members of
miRNAs-371-372-373 family, which is transcribed from chromosome
19q13.42. Deregulation of miR-373-3p has been declared in many
cancers, whether it acts as an oncogene or a tumor suppressor gene
(5). Researches had proved that
miR-373-3p has significant regulating function in breast cancer
(6,7),
testicular germ cell tumors (8) and
so on. Nevertheless, mechanism of miR-373-3p in LUAD has not been
much clarified.
Amyloid precursor protein (APP) is a highly
conserved type 1 transmembrane glycoprotein with a receptor-like
structure and was considered to be closely related to the
occurrence and progress of Alzheimer's disease (9). Meanwhile, APP has been shown
up-regulated pathophysiologically in various kinds of cancers,
including breast cancer (10),
melanoma (11), and lung cancer
(12). Bioinformatics analysis
suggested that APP may be a target of miR-373-3p. In this study,
the expression levels of APP and miR-373-3p in LUAD tissues vs.
adjacent tissues were first time to be detected. Furthermore, using
human LUAD cell line A549, the tumor-promoting effect of miR-373-3p
in A549 was delineated then interaction between miRNA-373 and APP
was identified to explore the in-depth mechanism.
Materials and methods
Bioinformatics analysis
The information of miRNA microarray was obtained
from the NCBI GEO (Gene Expression Omnibus) under Platform GPL4717
and Series GSE18692. Detailed experimental process and design of
the microarray was previously described by Puissegur et al
(4). The differential expression of
miRNAs (DEMs) was analyzed with the limma package. Under the
condition of adjusted P-value <0.01 and |logFC|>2, DEMs were
considered to have significant differential expression between
experiment groups and controls. The statistical tests were done by
the R program version 3.2.2 (http://www.r-project.org/). To predict the target gene
of miRNAs, 4 miRNA-target gene databases were searched including
miRanda, RNA22Sites, TargetScan and picTarSites.
Clinical specimen collection
50 pairs of LUAD tissues and the adjacent nontumor
lung tissues (2 cm from the margin of the tumor), were collected
from patients who operated at the First Affiliated Hospital of
China Medical University (Shenyang, China) and we got approval from
the Ethics Committee of the First Affiliated Hospital of China
Medical University (IRB Approval 2012-40-2).
Cell culture and Cell
transfection
Human LUAD cell lines A549, purchased from Shanghai
Cell Bank of Chinese Academy of Sciences (Shanghai, China), were
cultured in RPMI-1640 (Invitrogen, Carlsbad, CA, USA) medium
supplemented with 10% fetal bovine serum (FBS; Invitrogen), 100
µg/ml streptomycin and 100 IU/ml penicillin. All the cells were
maintained in a 37°C, 5% CO2 incubator.
All the miRNA inhibitors, miRNA mimics, negative
control (NC) of miRNA and siRNAs were chemically synthesized by
Genepharma (Shanghai, China). All transfections in our study were
transient and JetPRIME reagent (Polyplus-transfection) was added
following the protocol. The cells were harvested for subsequent
assays after RNA oligonucleotides successfully transfected for 48
h.
RNA isolation and quantitative reverse
transcription-PCR (qRT-PCR)
Total RNA was extracted from LUAD tissues, adjacent
normal tissues and A549 by miRNeasy Mini kit (Qiagen, Hilden,
Germany) according to the manufacturer's instructions.
Complementary DNA (cDNA) of miR-373-3p was obtained with
application of the QuantiMir RT kit (SBI). To verify the result of
bioinformatics analysis, mRNA and miRNA of specimen were quantified
by SYBR-Green qPCR Master Mix (Takara) through ABI 7500 Fast System
thermocycler (Applied Biosystems Life Technologies, Foster City,
CA, USA). Then, the qRT-PCR was applied to verify the transfection
efficiency of oligonucleotides. The detection of miR-373-3p in
detail was described by Zhang et al (13) and RUN6B (U6) was considered as an
endogenous control. The primers of APP and β-actin are as followed:
APP forward, GGA AGC GAT GAT AAG GTG GTA GAA GAA CAA and reverse,
CAT CAC CAT CAT CAT CGT CAT CAT CAT CAG; β-actin forward, CCT TGC
ACA TGC CGG AG and reverse, GCA CAG AGC CTC GCC TT. All the
experiments were performed in triplicate and data were calculated
through 2−ΔΔCt method.
Protein extraction and western
blotting
48 h after transfection, proteins of the cell were
extracted. Protein of the clinical sample and the cultured cell
were separated by SDS-PAGE and transferred to nitrocellulose
membranes, which were blocked with 5% fat-free milk for 1 h. Then,
the membranes incubated with anti-APP antibody (1:1,000; Abcam,
Cambridge, UK) at 4°C overnight. Anti-β-actin antibody was served
as the internal reference. The membranes were incubated with
secondary antibodies for 30 min at room temperature after washing
thoroughly. We detected the results by enhanced chemiluminescence
technique (Amersham, Piscataway, NJ, USA) and quantified the level
of expression of these proteins by application of Image J
software.
Dual-luciferase assays
3′UTR of the APP mRNA containing the potential
target region for miR-373-3p were amplified by PCR. Overlap
extension PCR was applied to amplify the mutant region of 3′UTR of
the APP mRNA. Then, the region was cloned into the pmirGLO
Dual-Luciferase miRNA Target Expression Vector (Promega Corp.,
Madison, WI, USA) and identified as dual-luciferase reporter
vectors. The insertions were confirmed by Sangon Biotech (Shanghai,
China) with commercial sequencing. The dual-luciferase reporter
plasmids named pmirGLO-wt-APP and pmirGLO-mut-APP were
co-transfected with miRNA mimics (50 nM) or NC (50 nM) using
JetPRIME reagent (Polyplus-transfection). The cells were harvested
after transfection for 48 h. The luciferase activity was detected
by the Dual-Luciferase® Reporter Assay System (Promega
Corp.), according to the manufacturer's instructions.
Cell counting kit-8 (CCK-8)
assays
After transfection 24 h, A549 cells were seeded into
96-well plates which density was 3–5×103 cells/well. At
0, 24, 48, or 72 h, CCK-8 solution (Beyotime Biotech, Jiangsu,
China) was added. The cells were cultured in a 37°C incubator for
another 3 h. To evaluate the number of viable cells, OD values at
450 nm wave-length (OD 450) were assessed daily.
Statistical analysis
All the statistical analyses except bioinformatics
analysis were performed with SPSS 23.0 statistical software package
(IBM SPSS, Armonk, NY, USA) and Graphpad prism 5 (GraphPad
Software, Inc., La Jolla, CA, USA). Results were presented as mean
± SD and P<0.05 was considered to indicate a statistically
significant difference.
Results
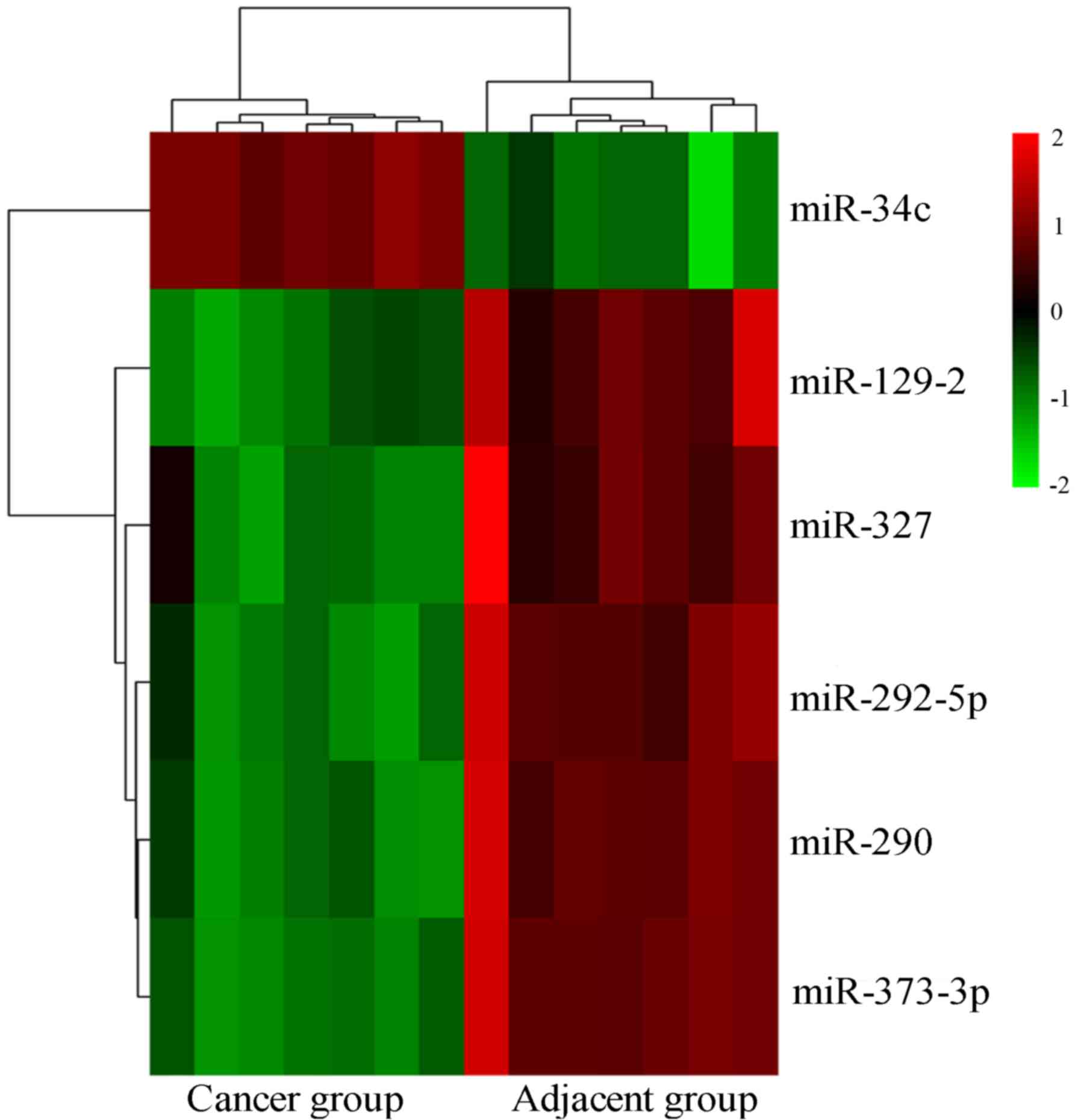
Identification of DEMs
A total of 6 DEMs were identified from GSE18692
datasets (GPL4717) on the basis of adj. P<0.01 and |logFC|>2
(Fig. 1). Among them, 5 DEMs were
significantly downregulation in LUAD, we selected miR-373-3p as our
target.
miR-373-3p is downregulated and APP is
overexpressed in LUAD tissues
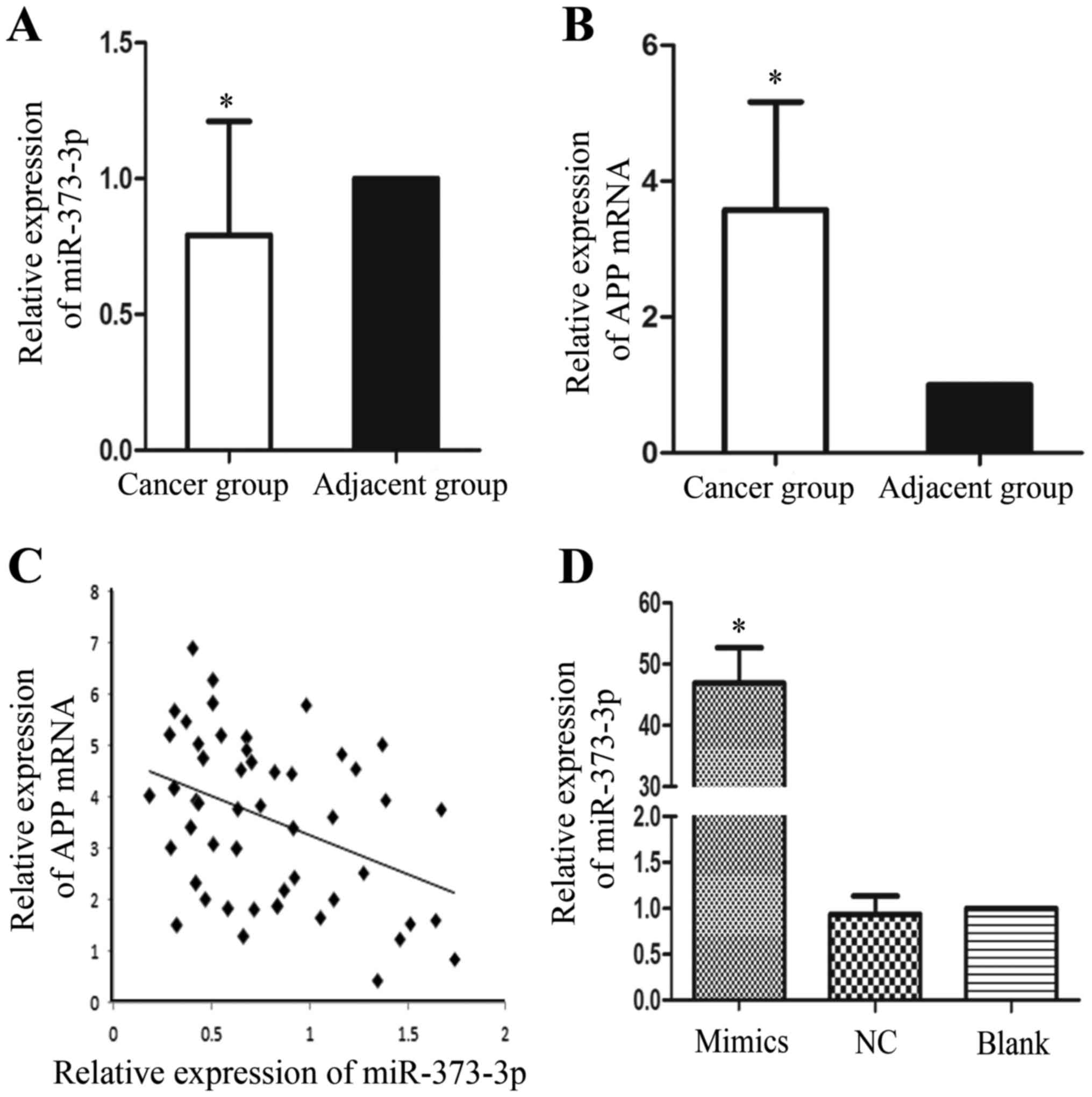
In order to detect the expression levels of
miR-373-3p and APP mRNA, qRT-PCR was used after the total RNA was
extracted (Fig. 2). In comparison to
the adjacent tissues, miR-373-3p expression was significantly
downregulation in the LUAD tissue samples (Fig. 2A). In sharp contrast, the levels of
APP mRNA were upregulation in the LUAD tissues compared with those
in the adjacent normal tissues (Fig.
2B). Furthermore, we found that there is a negative
relationship between miR-373-3p and APP mRNA (r=−0.4; P<0.05)
(Fig. 2C). Briefly, our results
suggested that miR-373-3p may participate in the oncogenesis of
LUAD partly through regulating APP, which has been reported as an
oncogene in NSCLC.
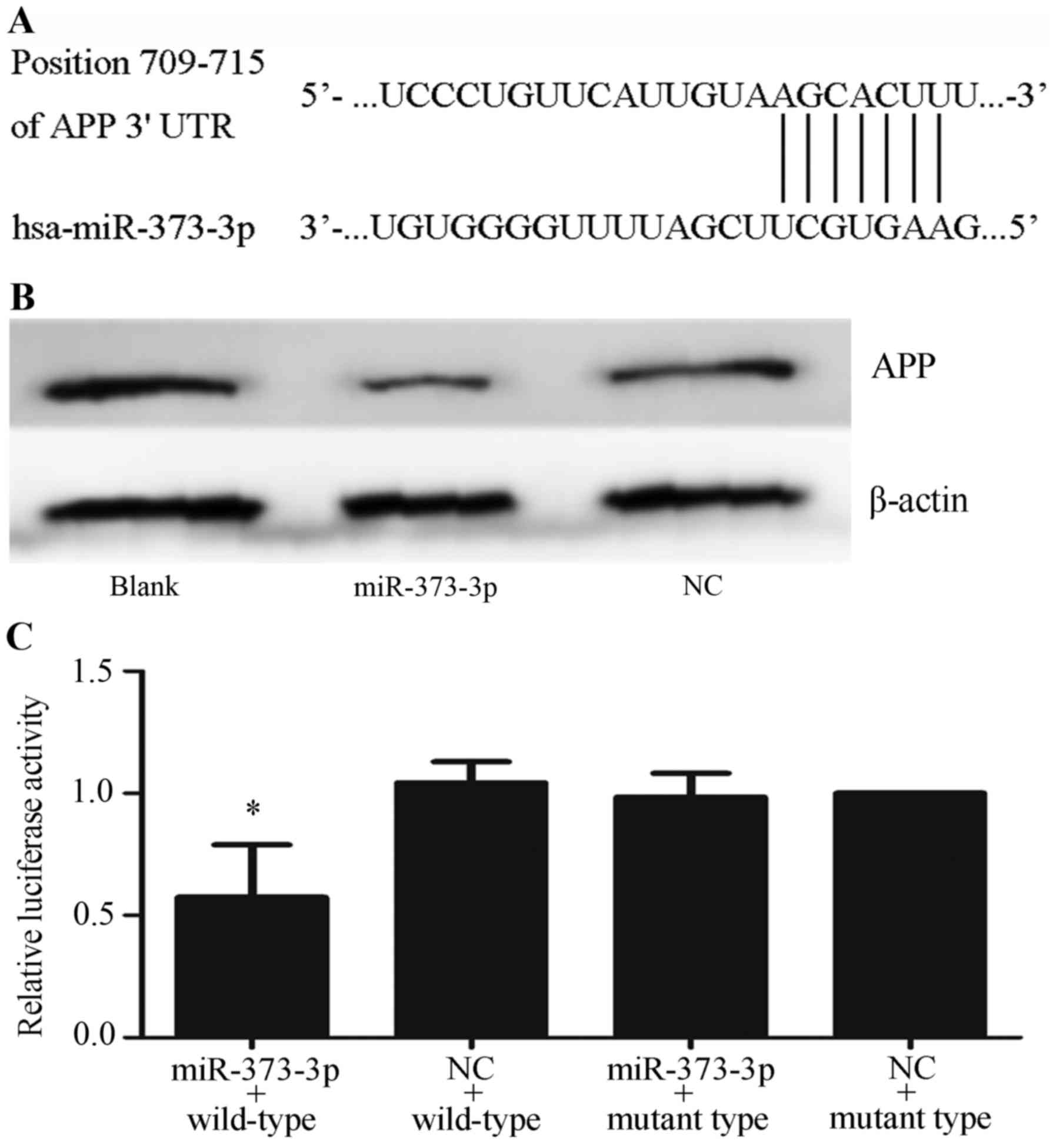
miR-373-3p targets APP mRNA
A bioinformatic analysis using 4 miRNA-target gene
databases showed that APP mRNA is a presumed target of miR-373-3p.
The potential target region for miR-373-3p in the APP mRNA 3′UTR
are showed in Fig. 3A. To verify that
miR-373-3p can regulate APP mRNA, we investigated APP protein
expression in A549 cells transfected with the miR-373-3p mimic.
Successful transfection was confirmed by qRT-PCR (Fig. 2D). Western blotting results
demonstrated that the expression of APP was markedly lower in the
miR-373-3p group than in the blank or NC groups (Fig. 3B). Dual-luciferase reporter assays
were implemented to validate if regulation exists. Dual-luciferase
reporter vectors containing either the mutant or wild-type 3′UTR of
APP mRNA was then constructed, and cotransfected into A549 cells
together with the miR-373-3p mimic or NC. The results found that
pmirGLO-wt-APP group was specifically responsive to miR-373-3p
overexpression, but no significant difference was noticed between
the relative luciferase activity of the miR-373-3p mimics group and
that of cells cotransfected with NC in pmirGLO-mut-APP groups
(P<0.05) (Fig. 3C). The results
implied that miR-373-3p acts directly on the 3′-UTR of APP mRNA,
and negatively regulates APP expression.
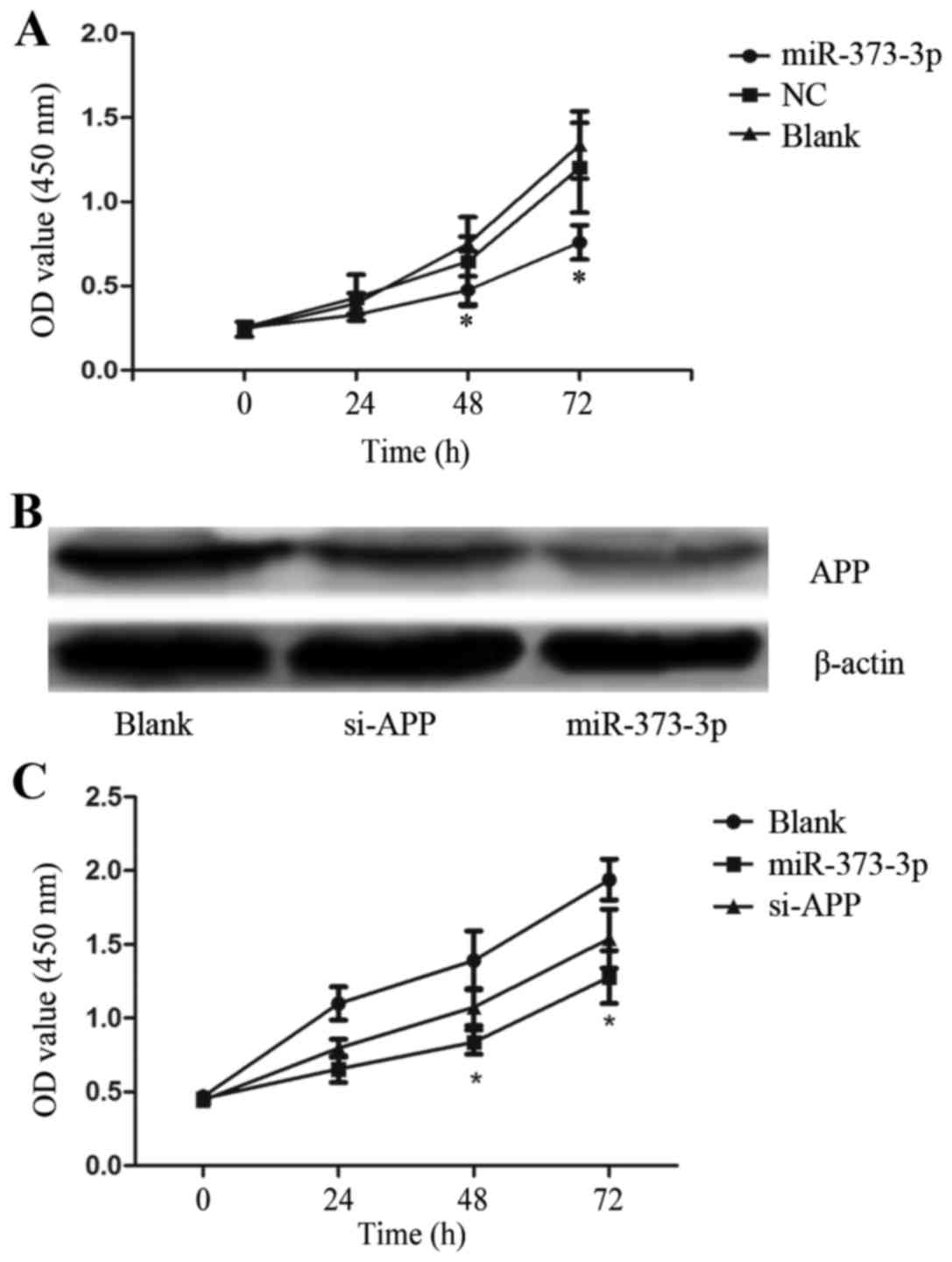
Forced expression of miR-373-3p
inhibits the proliferation of LUAD cell line A549
To confirm the antitumor effect of miR-373-3p on
LUAD, we synthesized a NC and a miR-373-3p mimic then introduced
them respectively into the human LUAD cell line A549. The effects
of miR-373-3p overexpression on LUAD cells were explored by CCK-8.
In the CCK-8 assays, the miR-373-3p transfected cells showed slower
proliferation trend than other groups (Fig. 4A). This indicates that the
upregulation of miR-373-3p inhibits LUAD cell proliferation in
vitro.
Overexpression of miR-373-3p more
effectively inhibits the proliferation of A549 than APP si-RNA
(si-APP)
To compare the biological effect of miR-373-3p with
that of si-APP on A549 behaviors, we used si-APP or miR-373-3p to
silence APP expression, and the efficacy was verified with western
blotting assays (Fig. 4B). The CCK-8
assays were performed to investigate the impact of miR-373-3p and
si-APP in altering the growth trend of A549 cells. Both miR-373-3p
and si-APP group showed proliferation-suppressing trend, but the
proliferation trend was lower in miR-373-3p group (Fig. 4C). These data indicate that miR-373-3p
had a stronger effect on A549 cells than si-APP, and miR-373-3p is
a promising target for LUAD treatment since it affects more than
one target mRNA and may involve in many regulatory network.
Discussion
Epidermal growth factor receptor (EGFR) inhibitors
and ALK inhibitors have been applied in clinical practice for
several years (14,15), however, the patients with EGFR
mutations or ALK gene translocations only account for a small part,
which leads to the prognosis of LUAD patients has not been
dramatically improved (16). Hence,
it's necessary to develop more effective targeted therapies. With
the help of genetic sequencing technology, we selected miR-373-3p,
which is repressed in LUAD as our target. The expression of
miR-373-3p is abnormal in various kinds of tumors, effecting
process of proliferation, invasion and metastasis. miR-373-3p has
been proved to be served as an oncogenic miRNA in testicular germ
cell tumors by inhibiting LATS2, the tumor suppressor directly
(8). A research suggested that
expression of miR-373-3p are up-regulated in breast cancer, while
invasion and metastasis of cancer cells could be promoted through
down-regulation of CD44 expression (7). But the role of miR-373-3p remains
controversial because later then, the miR-520/373 family has been
reported to serve as tumor suppressor in ER(−) breast cancer
through linking the NF-κB pathways with TGF-β pathways, and result
in several consequences such as inflammation, tumor progression and
dissemination (6). Recent reports
demonstrated that miR-373-3p expression is silenced by histone
modification in lung cancer cells, then promote proliferation,
migration, and invasion of cancer cells via up-regulation of IRAK2
and LAMP1 target genes (17).
Besides, there is very little study known about the relationship
between miR-373-3p and LUAD.
A bioinformatic analysis suggested that APP mRNA may
be a target of miR-373-3p, which aroused our interest to exploit
the roles of these two molecules in LUAD. APP is a highly
transmembrane glycoprotein and its expression is related with
carcinogenesis of several tumors. For example, upexpression of APP
is associated with greater metastatic tendencies as well as high
motility and proliferation in breast cancer (10). A recent study conducted by Sobol et
al (12) showed that decreased
expression of APP leads to several consequences such as G0/G1 cell
cycle arrest, decreased pRb phosphorylation, cell size
abnormalities and necrosis in NSCLC. Furthermore, according to the
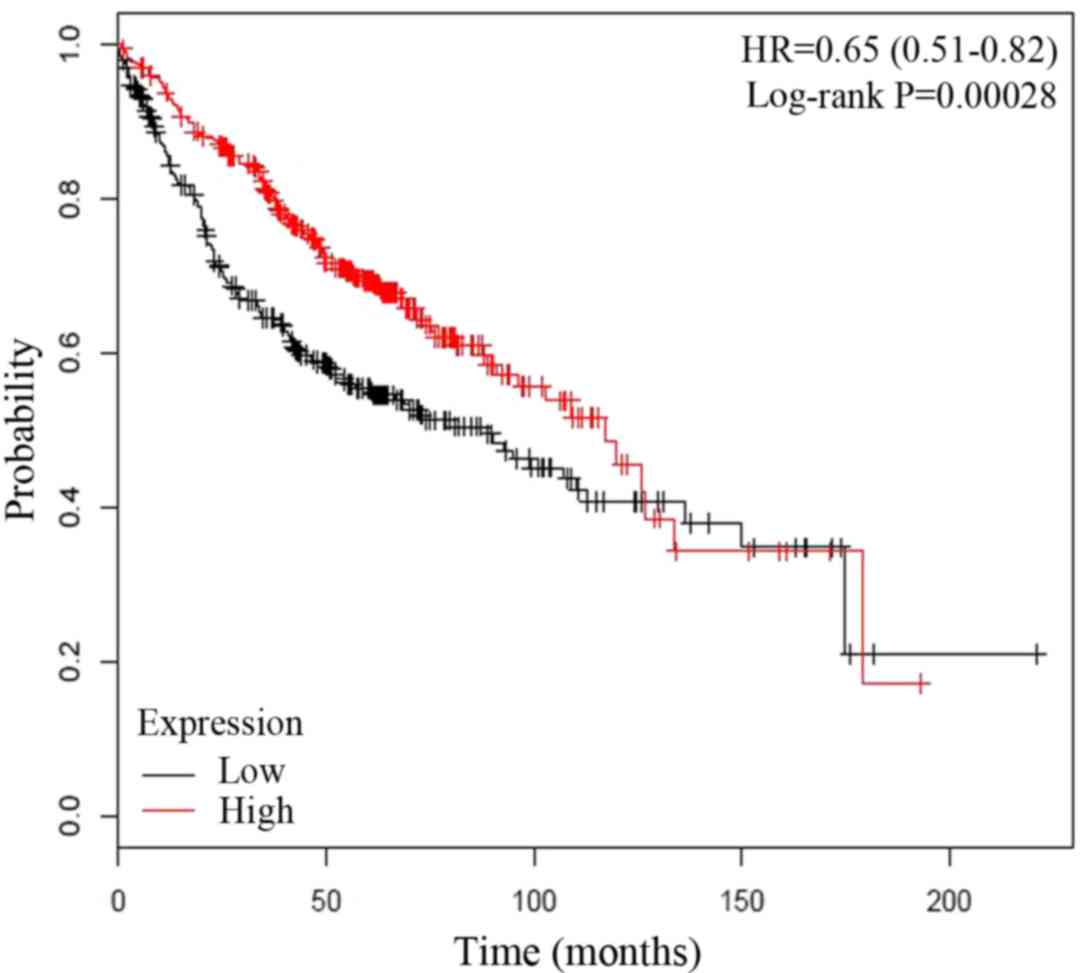
analysis of 866 LUAD patients' overall survival rate by
Kaplan-Meier plotter (http://www.kmplot.com/lung/), the elevated expression
of APP may be relevant with poor prognosis (Fig. 5) (18).
Taking these researches into consideration, we
detected the miR-373-3p and APP mRNA expression in 50 paired
tissues of LUAD patients. miR-373-3p was downregulation and APP
mRNA was overexpression in their LUAD tissues compared with the
adjacent normal tissues. Moreover, the expression of APP mRNA is
negatively related with the expression of miR-373-3p. Through dual
luciferase assays, the interaction between miR-373-3p and APP mRNA
was verified. We then studied the effects of miR-373-3p on the cell
growth of A549, and found that the upregulation of miR-373-3p
inhibited cell proliferation by directly targeting APP mRNA.
Furthermore, we observed that force expression of miR-373-3p had a
stronger effect than APP siRNA on the behavior of LUAD cell line
A549, which implies that miR-373-3p may target other oncogenes and
is participate in more than one signaling pathway apart from this.
Therefore, miR-373-3p could be a valuable target in LUAD treatment,
and to achieve the goal, more LUAD patients and systematic
follow-up should be included to make up for this study's
deficiency. In addition, in vivo experiments and deeper
exploration of the mechanisms of miR-373-3p/APP axis will also need
to be exploited in the future.
In general, these data indicated miR-373-3p
regulates the proliferation of LUAD partly via APP, and the
expression of APP may be significantly related to the clinical
outcomes of LUAD patients. These conclusions suggested that
miR-373-3p may be a valuable target for potential anticancer
strategy to treat LUAD.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Diaz-Garcia CV, Agudo-López A, Pérez C,
López-Martín JA, Rodríguez-Peralto JL, de Castro J, Cortijo A,
Martínez-Villanueva M, Iglesias L, García-Carbonero R, et al:
DICER1, DROSHA and miRNAs in patients with non-small cell lung
cancer: Implications for outcomes and histologic classification.
Carcinogenesis. 34:1031–1038. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim VN and Nam JW: Genomics of microRNA.
Trends in genetics. 22:165–173. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Puissegur MP, Mazure NM, Bertero T,
Pradelli L, Grosso S, Robbe-Sermesant K, Maurin T, Lebrigand K,
Cardinaud B, Hofman V, et al: miR-210 is overexpressed in late
stages of lung cancer and mediates mitochondrial alterations
associated with modulation of HIF-1 activity. Cell death and
differentiation. 18:465–478. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wei F, Cao C, Xu X and Wang J: Diverse
functions of miR-373 in cancer. J Transl Med. 13:1622015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Keklikoglou I, Koerner C, Schmidt C, Zhang
JD, Heckmann D, Shavinskaya A, Allgayer H, Gückel B, Fehm T,
Schneeweiss A, et al: MicroRNA-520/373 family functions as a tumor
suppressor in estrogen receptor negative breast cancer by targeting
NF-kappaB and TGF-beta signaling pathways. Oncogene. 31:4150–4163.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang Q, Gumireddy K, Schrier M, le Sage
C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, et al:
The microRNAs miR-373 and miR-520c promote tumour invasion and
metastasis. Nat Cell Biol. 10:202–210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bing Z, Master SR, Tobias JW, Baldwin DA,
Xu XW and Tomaszewski JE: MicroRNA expression profiles of seminoma
from paraffin-embedded formalin-fixed tissue. Virchows Arch.
461:663–668. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pandey P, Sliker B, Peters HL, Tuli A,
Herskovitz J, Smits K, Purohit A, Singh RK, Dong J, Batra SK, et
al: Amyloid precursor protein and amyloid precursor-like protein 2
in cancer. Oncotarget. 7:19430–19444. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lim S, Yoo BK, Kim HS, Gilmore HL, Lee Y,
Lee HP, Kim SJ, Letterio J and Lee HG: Amyloid-beta precursor
protein promotes cell proliferation and motility of advanced breast
cancer. BMC cancer. 14:9282014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Botelho MG, Wang X, Arndt-Jovin DJ, Becker
D and Jovin TM: Induction of terminal differentiation in melanoma
cells on downregulation of beta-amyloid precursor protein. J Invest
Dermatol. 130:1400–1410. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sobol A, Galluzzo P, Weber MJ, Alani S and
Bocchetta M: Depletion of amyloid precursor protein (APP) causes G0
arrest in non-small cell lung cancer (NSCLC) cells. J Cell Physiol.
230:1332–1341. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y, Zhao FJ, Chen LL, Wang LQ, Nephew
KP, Wu YL and Zhang S: MiR-373 targeting of the Rab22a oncogene
suppresses tumor invasion and metastasis in ovarian cancer.
Oncotarget. 5:12291–12303. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xing RC, Zheng J, Zheng WH, Qin ZP, Liu W
and Yao RC: Relevance of E-cadherin expression to EGFR-TKI
molecular targeted therapy sensitivity/resistance and its clinical
significance. Genet Mol Res. 14:5785–5792. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Steuer CE, Khuri FR and Ramalingam SS: The
next generation of epidermal growth factor receptor tyrosine kinase
inhibitors in the treatment of lung cancer. Cancer. 121:E1–E6.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ellis PM, Coakley N, Feld R, Kuruvilla S
and Ung YC: Use of the epidermal growth factor receptor inhibitors
gefitinib, erlotinib, afatinib, dacomitinib and icotinib in the
treatment of non-small-cell lung cancer: A systematic review. Curr
Oncol. 22:e183–e215. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Seol HS, Akiyama Y, Shimada S, Lee HJ, Kim
TI, Chun SM, Singh SR and Jang SJ: Epigenetic silencing of
microRNA-373 to epithelial-mesenchymal transition in non-small cell
lung cancer through IRAK2 and LAMP1 axes. Cancer Lett. 353:232–241.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gyorffy B, Surowiak P, Budczies J and
Lanczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PloS one. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|