Introduction
Glioma originates from the neural epithelium, which
is the most common primary malignant tumor in the brain,
responsible for 40–65% of these tumors (1). The incidence of glioma is on the
increase among youth in China (2).
Pathological types of gliomas can be divided into 4 grades; the
higher the grade, the higher the degree of malignancy (3). Glioma develops faster, according to the
location, structure and tumor size. Additionally, it presents
different clinical symptoms such as elevated intracranial pressure
(headache, vomiting, papilloedema and consciousness disturbance)
and focal symptoms and signs (movement disorders, sensory
impairment and epilepsy) (4).
Surgery remains the main method of treatment in
combination with various chemotherapeutic regimens. These treatment
modalities extend survival time to some extent, but the quality of
life remains unsatisfactory (5). The
high postoperative recurrence rate is the main cause of death in
patients (6). Aggressive growth of
tumors constitutes the underlying causes of poor prognosis
(7). Cellular and molecular
biological characteristics have shown that abnormal gene
expression, which regulates tumor growth, proliferation, migration,
differentiation and apoptosis, is an important factor in glioma
(8). MicroRNA (miRNA) is involved in
90% of gene transcription and translation process, affecting the
expression of protein, and activation of the cell signaling pathway
(9). Previous findings showed that
miRNA is an important tumor control factor (10). There are 18 types of miRNA expression
upregulation in gliomas, such as miRNA-9-2, miRNA-21, 13 types of
miRNA expression downregulation, such as miRNA-128-1, and miRNA-181
(11). miRNA-184 is a newly
identified miRNA abnormally expressed in many malignant tumors,
such as liver cancer, lung cancer, and nasopharyngeal carcinoma
(12). It appears to regulate the
c-Myc and BCL2 signaling pathways, act as a cancer gene regulatory
factor, or upregulate SND1 signal to promote tumor occurrence
(13–15). Based on these prior finings, the
present study analyzed the expression of miRNA-184 in different
pathological grades of glioma, and the relationship with survival
prognosis, to provide a reference for clinical diagnosis and
treatment.
Materials and methods
Object data
Forty patients diagnosed as having glioma for the
first time were selected from January 2013 to January 2016, of
which 26 cases were male and 14 were female. The participants were
42–76 years of age, with an average age of (56.8±14.6) years.
Grading of the gliomas as per WHO revealed 10 grade I cases, all of
which were hair cell astrocytoma; 8 grade II cases, of which 2
cases of astrocytoma were of the original type, 4 cases were
ependymoma, and 2 cases were diffuse astrocytoma; 16 grade III
cases, of which 4 cases were oligodendroglial tumors, 4 cases were
central neurocytoma, 2 cases were anaplastic cell tumor, 4 cases
were anaplastic astrocytoma, 2 cases were anaplastic room tube
membrane tumor; and 6 grade IV cases, including 4 cases of
glioblastoma, and 2 cases of medulloblastoma. Ten cases of normal
brain tissue were selected (obtained by decompression because of
traumatic brain injury), including 6 males and 4 females aged 40–78
years with an average age of 55.7±13.5 years. Sex and age of the
two groups were comparable. Exclusion criteria included a history
of chemotherapy, brain trauma, meningitis, with underlying diseases
such as serious heart, liver, lung, kidney and other organ
dysfunctions, autoimmune diseases, infection diseases and other
types of benign and malignant tumors.
The present study was approved by the Ethics
Committee of Shandong Provincial Hospital (Shandong, China).
Written and signed informed consent was obtained from the patients
and their families.
Methods
RT-PCR and immunohistochemical methods were used to
detect the expression level and intensity of miRNA-184 in different
grades of glioma tissues. The length of survival of
miRNA-184-positive expressed patients was followed up and
analyzed.
RT-PCR
Conventional TRIzol reagent and miRNA extraction kit
were used to extract total RNA as per manufacturer instructions.
Determination of miRNA concentration was performed using a
spectrophotometer (Hitachi, Tokyo, Japan) and completion was
detected by agarose gel electrophoresis. The reverse transcriptase
kit was used to produce cDNA, and design primers. The primers used
were miR-184: forward, 5′-GCATGCCTAAATGTTGACAGCC-3′ and reverse,
5′-CACAUGUAUGAAUUGACAGCC-3′; reference U6: forward,
5′-GCGCGTCGTGAAGCGTTC-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′. The
reaction system was cDNA 4 µl + SYBR Master Mix 10 µl + upstream
and downstream primers each 0.2 µl, with 20 µl of water. The
reaction conditions were 95°C for 5 min, 95°C for 15 sec, 60°C for
l min, a total of 35 cycles, stored at 4°C. A standard curve and
melting curve were constructed. The results were expressed by
2−ΔΔCq method of cycle number.
Immunohistochemical method
Conventional paraffin sections were prepared. Xylene
dewaxing and gradient ethanol hydration were performed. Antigen
repair and close endogenous peroxidase were used. The section were
incubated at room temperature for 20 min in 3%
H2O2, and PBS was used for washing for 5 min
× 3 times with oscillation. Closed non-specific binding was
performed and normal goat serum was then added to the sections, in
the wet box at room temperature, followed by incubation for 30 min.
Primary antibody was added as follows: goat serum on the surface of
the sections was discarded, the mouse anti-human miR-184 monoclonal
antibody (cat. no. SY786520; Wuhan Sanying Co., Wuhan, China) was
added at a dilution of 1:200. The sections were then incubated in
the wet box at 4°C overnight, and normal mice IgG replaced the
primary antibody as the negative control. Then the goat anti-rabbit
secondary antibody (dilution, 1:2,000; cat. no. ab6721; Abcam,
Cambridge, MA, USA) was added as follows: the primary antibody on
the surface of the section was discarded, PBS was used for washing
for 5 min × 3 times with oscillation, the biotin-labeled secondary
anti working liquid in the SP immunohistochemistry kit was added to
the section, and they were incubated in the wet box at room
temperature for 20 min. HRP streptavidin solution was added by
discarding the secondary antibody on the surface of the section,
and washing with PBS for 5 min × 3 times with oscillation, adding
HRP streptavidin solution to the section, followed by incubation in
the wet box at room temperature for 20 min, and washing with PBS
for 5 min × 3 times with oscillation. DAB color rendering was then
performed, and the sections were counterstained with hematoxylin,
hydrochloric acid alcohol differentiation was conducted, ammonia
back to blue, gradient ethanol dehydration, xylene transparent, and
neutral gum mounting were applied. The sections were then dried at
room temperature.
The results were shown according to the staining
intensity and positive cell number: 0, no staining; 1–3, light to
dark brown. Positive cell rate according to random selection of 5
visual fields was: 0, positive cells; 1, 25% positive cells, 2,
25–50% positive cells and 3, >50% positive cells. The score was
the product of both, indicated as: 0, negative (−); 1–2, weak
positive (+); 3–4, positive (+ +); and 5–9, strong positive (+ +
+).
Statistical methods
IBM SPSS 20.0 (Armonk, NY, USA) software was used
for statistical analysis. Measurement data were expressed as mean ±
standard deviation. The comparison of two groups was tested by
independent sample t-test and the multiple group comparison was
analyzed by single-factor ANOVA analysis. Pairwise comparison was
tested by LSD method. Countable data were expressed by cases or
percentage. Inter-group comparison was tested by (correction)
χ2 or Fisher's exact probability and ranked data were
tested by rank sum. Survival time was compared by Kaplan-Meier
method with log-rank χ2 test. P<0.05 was considered
to indicate a statistically significant difference.
Results
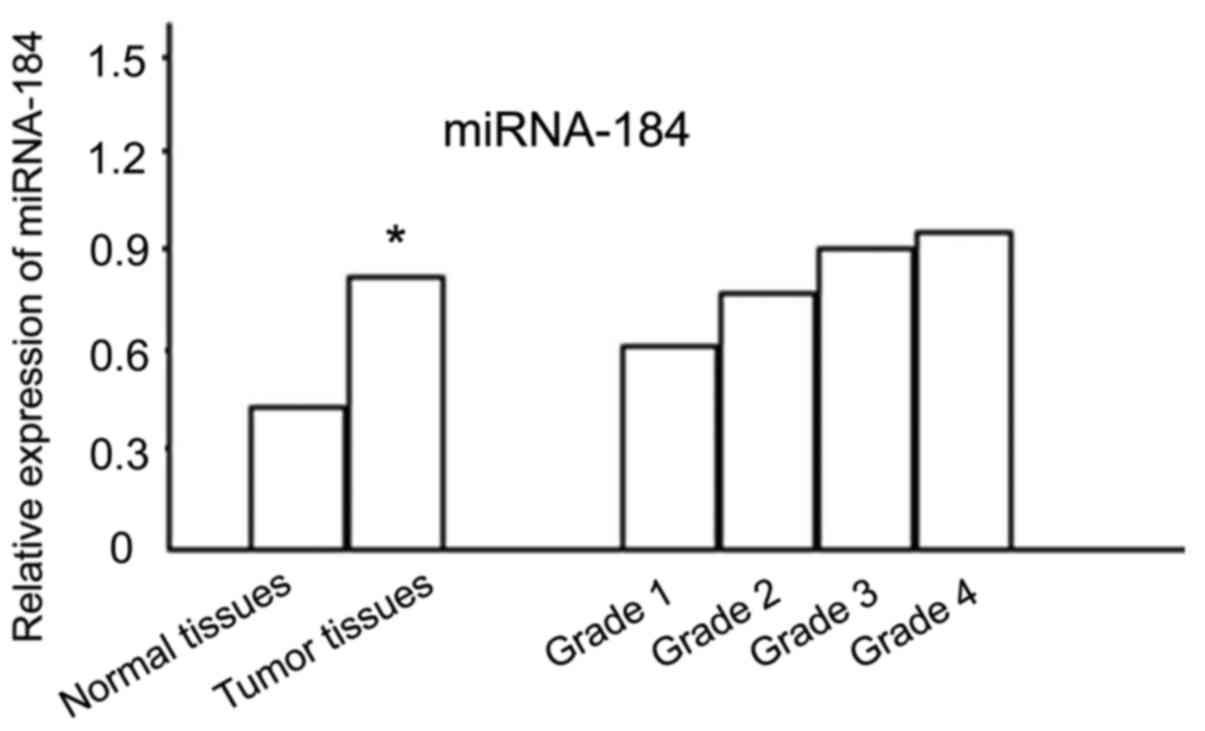
Quantitative comparison of miRNA-184
mRNA
miRNA-184 mRNA expression was found in normal
tissues and tumor tissues, and the expression significantly
increased in tumor tissues, and the difference was statistically
significant (P<0.05). miRNA-184 mRNA expression in different
grades was compared, and the difference was statistically
significant (P<0.05) with a higher grade being associated with a
higher expression level (Fig. 1).
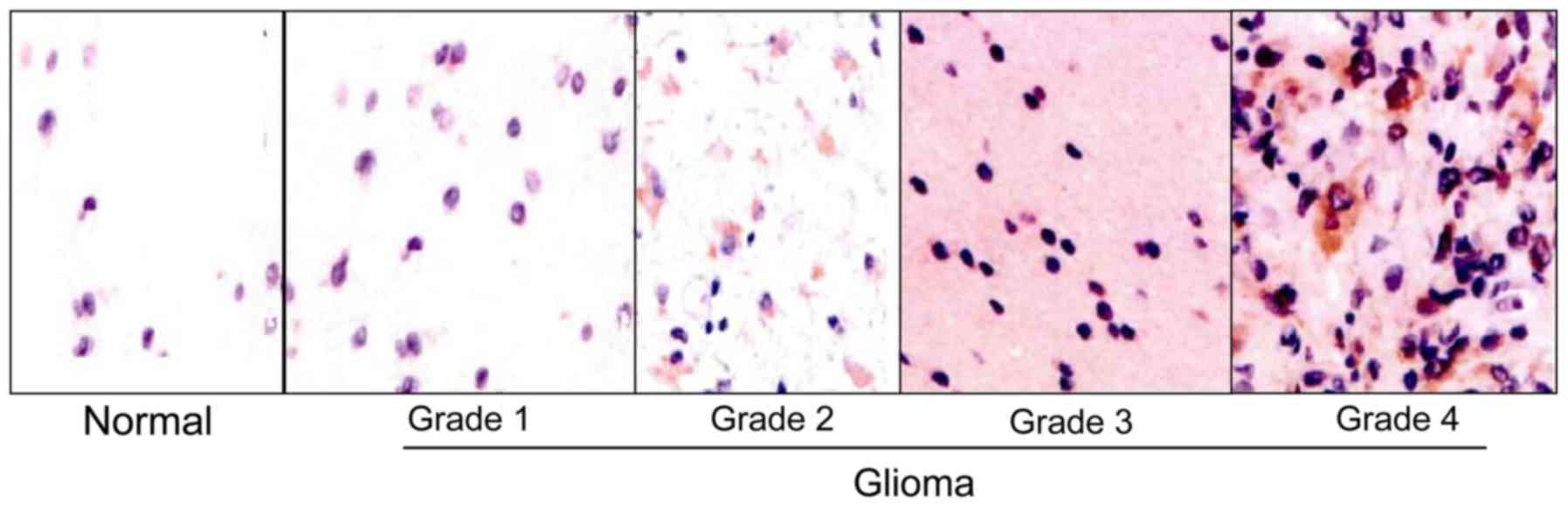
Comparison of miRNA-184-positive
expression rate
miRNA-184-positive expression rate increased with
the increase of tumor grade; the higher the level, the higher the
positive expression intensity. The differences were statistically
significant (P<0.05). Positive expression was not associated
with the type of glioma cells (Table
I and Fig. 2).
 | Table I.Comparison of microRNA-184-positive
expression rate [cases (%)]. |
Table I.
Comparison of microRNA-184-positive
expression rate [cases (%)].
| Group | No. of cases | Negative | Weak positive | Positive | Strong positive | Positive rate |
|---|
| Normal brain
tissue | 10 | 8 (80.0) | 2 (20.0) | 0 | 0 | 2
(20.0) |
| Grade I gliomas | 10 | 6 (60.0) | 2 (20.0) | 1 (10.0) | 1 (10.0) | 4
(40.0) |
| Grade II | 8 | 3 (37.5) | 2 (25.0) | 2 (25.0) | 1 (12.5) | 5
(62.5) |
| Grade III | 16 | 3 (18.8) | 4 (25.0) | 6 (37.5) | 3 (18.8) | 13 (81.3) |
| Grade IV | 6 | 1 (16.7) | 1 (16.7) | 1 (16.7) | 3 (50.0) | 5
(83.3) |
| χ2 |
| 26.537 |
|
|
| 13.127 |
| P-value |
|
0.000 |
|
|
|
0.011 |
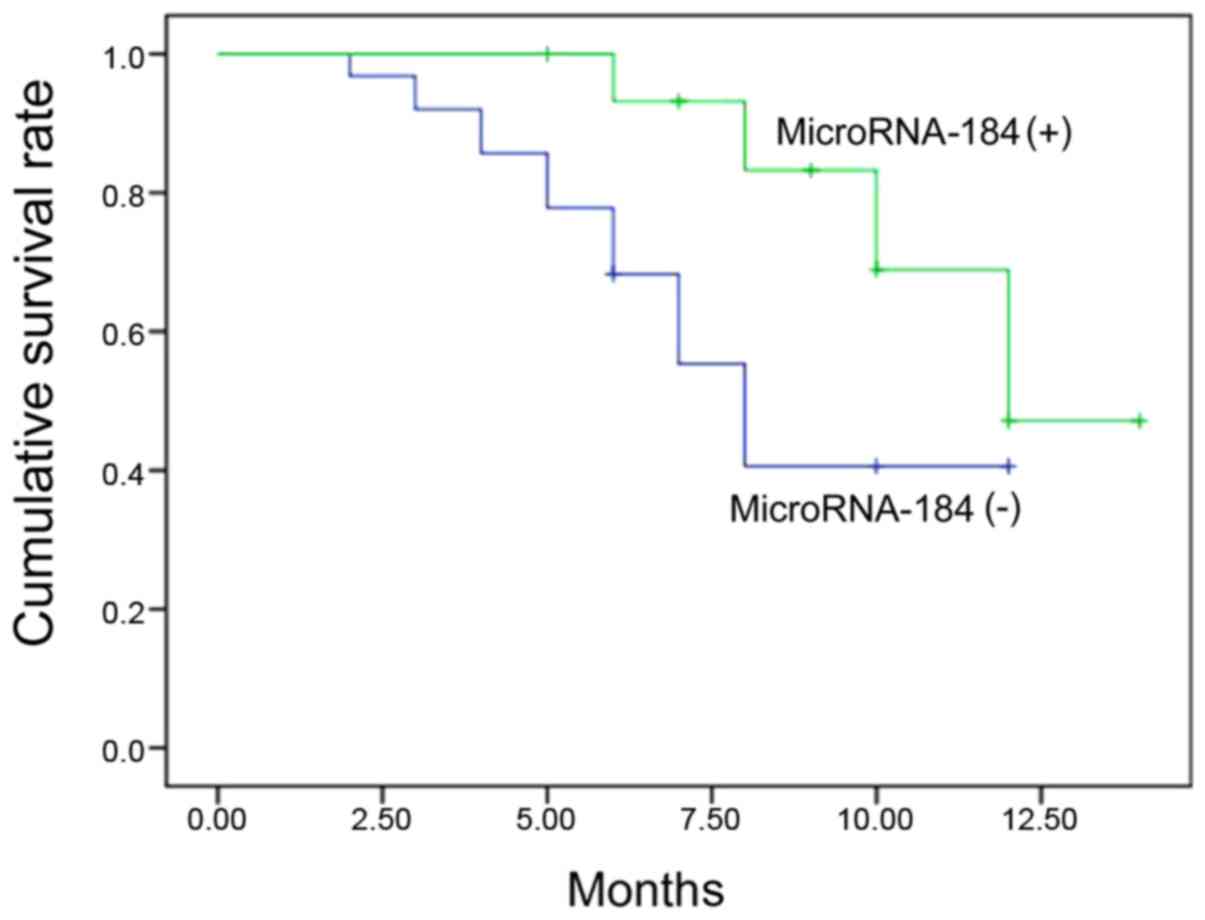
Comparison of survival time
The median survival time of miRNA-184-positive
expression was significantly shorter than that of the negative
expression group [compare (8.0±0.4) with (12.0±0.1),
χ2=12.480, P<0.001] (Fig.
3).
Discussion
Although there are many miRNAs, the common molecular
biological characteristics, including similar frame structure
having no open reading frame can be found in all eukaryotic
genomes, and code for proteins that are similar in length,
approximately the size of 18–22 nT. They are small RNA molecules of
a short sequence (16). miRNA is an
important regulatory factor, involved in cell growth,
proliferation, differentiation, metabolism and apoptosis,
angiogenesis, cell invasion and metastasis, including stem cell
regulation (17).
The present study demonstrates that miRNA-184 mRNA
expression is expressed at a statistically significant greater
level in glioma tissue than normal tissue. miRNA-184 mRNA
expression for each tumor grade also showed statistically
significant differences with increasing expression as the grade
increased. The present findings also show that miRNA-184 expression
is upregulated in the occurrence or progression of glioma, which
may promote the occurrence and development of gliomas. The
miRNA-184-positive expression rate increased with the increase of
the tumor grade and the higher the level of expression, the higher
the positive expression intensity. Differences for each grade level
were statistically significant. A positive expression is not
related to the type of glioma cells. miRNA-184 does not express in
neurons, and is only expressed in glial cells, and mainly expressed
in plasma cells, but is not expressed in normal brain tissue. The
presence of glioma indicates a positive expression of miR-184, and
an increase of staining suggess a correlation with the degree of
glioma pathological grades. An increase of pathological grade leads
to stronger staining, which is not related to the type of glioma
cells. Thus, miR-184 can be used as a marker for the pathological
grade of gliomas. Furthermore, the follow up showed that the median
survival time of miRNA-184-positive expression was significantly
shorter than that of the negative expression group.
In conclusion, miRNA-184 is highly expressed in
gliomas, which is positively correlated with pathological grade,
but is not correlated with pathological type, and negatively
correlated with survival time, and may be an important molecular
marker. Future studies aim to examine the miRNA-184 modulation
mechanisms of glioma cell proliferation, migration and apoptosis,
such as cell signaling pathways and regulators and identify
appropriate targets for intervention and provide clues for clinical
diagnosis, treatment and prognostic evaluation.
References
|
1
|
Gramatzki D, Dehler S, Rushing EJ, Zaugg
K, Hofer S, Yonekawa Y, Bertalanffy H, Valavanis A, Korol D,
Rohrmann S, et al: Glioblastoma in the Canton of Zurich,
Switzerland Revisited: 2005 to 2009. Cancer. 122:2206–2215. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hu J, Wu W, Zhu B, Wang H, Liu R, Zhang X,
Li M, Yang Y, Yan J, Niu F, et al: Cerebral glioma grading using
bayesian network with features extracted from multiple modalities
of magnetic resonance imaging. PLoS One. 11:e1533692016.
|
|
3
|
Pessina F, Navarria P, Cozzi L, Ascolese
AM, Simonelli M, Santoro A, Tomatis S, Riva M, Fava E, Scorsetti M,
et al: Value of surgical resection in patients with newly diagnosed
grade III glioma treated in a multimodal approach: Surgery,
chemotherapy and radiotherapy. Ann Surg Oncol. 23:3040–3046. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sehm T, Fan Z, Ghoochani A, Rauh M,
Engelhorn T, Minakaki G, Dörfler A, Klucken J, Buchfelder M,
Eyüpoglu IY, et al: Sulfasalazine impacts on ferroptotic cell death
and alleviates the tumor microenvironment and glioma-induced brain
edema. Oncotarget. 7:36021–36033. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bush NA, Chang SM and Berger MS: Current
and future strategies for treatment of glioma. Neurosurg Rev.
16:7–8. 2016.
|
|
6
|
Yin Q, Zhou YY, Wang P, Ma LI, Li P, Wang
XG, She CH and Li WL: CD44 promotes the migration of bone
marrow-derived mesenchymal stem cells toward glioma. Oncol Lett.
11:2353–2358. 2016.PubMed/NCBI
|
|
7
|
Garcia MA, Solomon DA and Haas-Kogan DA:
Exploiting molecular biology for diagnosis and targeted management
of pediatric low-grade gliomas. Future Oncol. 12:1493–1506. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lam FC and Yaffe MB: Kicking Genomic
Profiling to the Curb: How Re-wiring the Phosphoproteome Can
Explain Treatment Resistance in Glioma. Cancer Cell. 29:435–436.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun G, Yan SS, Shi L, Wan ZQ, Jiang N, Fu
LS, Li M and Guo J: MicroRNA-15b suppresses the growth and invasion
of glioma cells through targeted inhibition of cripto-1 expression.
Mol Med Rep. 13:4897–4903. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Belter A, Rolle K, Piwecka M,
Fedoruk-Wyszomirska A, Naskręt-Barciszewska MZ and Barciszewski J:
Inhibition of miR-21 in glioma cells using catalytic nucleic acids.
Sci Rep. 6:245162016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Goodwin CR, Woodard CL, Zhou X, Pan J,
Olivi A, Xia S, Bettegowda C, Sciubba DM, Pevsner J, Zhu H, et al:
Microarray-based phospho-proteomic profiling of complex biological
systems. Transl Oncol. 9:124–129. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang X, Ding H, Han Y, Sun D, Wang H and
Zhai XU: The significance of microRNA-184 on JAK2/STAT3 signaling
pathway in the formation mechanism of glioblastoma. Oncol Lett.
10:3510–3514. 2015.PubMed/NCBI
|
|
13
|
Liu Z, Mai C, Yang H, Zhen Y, Yu X, Hua S,
Wu Q, Jiang Q, Zhang Y, Song X, et al: Candidate tumour suppressor
CCDC19 regulates miR-184 direct targeting of C-Myc thereby
suppressing cell growth in non-small cell lung cancers. J Cell Mol
Med. 18:1667–1679. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stringer BW, Bunt J, Day BW, Barry G,
Jamieson PR, Ensbey KS, Bruce ZC, Goasdoué K, Vidal H, Charmsaz S,
et al: Nuclear factor one B (NFIB) encodes a subtype-specific
tumour suppressor in glioblastoma. Oncotarget. 7:29306–29320. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Emdad L, Janjic A, Alzubi MA, Hu B,
Santhekadur PK, Menezes ME, Shen XN, Das SK, Sarkar D and Fisher
PB: Suppression of miR-184 in malignant gliomas upregulates SND1
and promotes tumor aggressiveness. Neuro Oncol. 17:419–429. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fu TG, Wang L, Li W, Li JZ and Li J:
miR-143 inhibits oncogenic traits by degrading NUAK2 in
glioblastoma. Int J Mol Med. 37:1627–1635. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Feng R and Dong L: Inhibitory effect of
miR-184 on the potential of proliferation and invasion in human
glioma and breast cancer cells in vitro. Int J Clin Exp Pathol.
8:9376–9382. 2015.PubMed/NCBI
|