Introduction
Notable progress has been achieved in previous years
in the treatment of metastatic gastric cancer (MGC). For instance,
trastuzumab combined with chemotherapy markedly prolongs overall
survival (OS) in comparison with chemotherapy alone for patients
with human epidermal growth factor receptor 2 (HER2)-positive MGC
(1). However, HER2-positive disease
accounts for only 7–34% of all gastric cancer (GC) (1); for patients with the HER2-negative
disease, chemotherapy alone remains the standard treatment. EOF
[oxaliplatin and 5-fluorouracil (5-FU) combined with epirubicin] is
one of the classic first-line chemotherapeutic treatments for MGC.
Nevertheless, <50% of all patients respond to this treatment
(2), driving the demand for
predictive biomarkers in order to improve the selection of patients
likely to respond to EOF therapy.
Tumor hypoxia develops in the majority of solid
tumors due to the imbalance between the tumor growth and blood
supply (3). In GC, previous studies
have revealed that hypoxia may lead to drug resistance and
stimulate the epithelial-mesenchymal transition (EMT) of GC cells
(4,5),
and the presence of low levels of oxygenation in the tumor tissues
of patients with GC has been associated with poor survival
(6). Prior studies have additionally
revealed that hypoxia-inducible factor (HIF)-1, the major
transcription factor significantly activated by hypoxia, may confer
hypoxia-induced drug resistance via the inhibition of drug-induced
apoptosis, a decrease in intracellular drug accumulation and by
prompting multidrug resistance (MDR) (7). Thus, the study of genes associated with
tumor hypoxia may be of value in predicting the treatment response
and prognosis of patients with MGC.
Myoglobin (MB) is an oxygen-binding respiratory
protein that has been identified in various non-muscle tissues
(8). Previous studies have revealed
that mRNA and/or protein levels of MB may be induced by hypoxia and
were correlated with the expression of numerous hypoxia biomarkers,
including HIF-1α, HIF-2α, and carbonic anhydrase IX (CAIX) in
various types of cancer (9–13).
The ATP-binding cassette sub-family G member 2
(ABCG2) promoter has been revealed to involve three hypoxia
response elements (14). Under
conditions of hypoxia, ABCG2 may be induced, thus the tumor cells
would be provided with a survival advantage by reducing the
accumulation of heme or porphyrin (15).
MutL homolog 1 (MLH1), a key DNA mismatch repair
(MMR) gene, has been revealed to be specifically reduced in tumor
cells and stem cells under hypoxia (16,17), which
eventually leads to genetic instability, tumor progression and
resistance to chemotherapeutic agents including oxaliplatin, 5-FU,
and irinotecan (18–21).
Poly(ADP-ribose) polymerase-1 (PARP-1) has been
demonstrated to serve important functions in carcinogenesis
(22). Previous studies have revealed
that PARP-1 interacts with HIF-1α and HIF-2α (23,24), and
that pre-exposure to hypoxia followed by oxidative stress may lead
to the over activation of PARP-1 in human lung cancer cells
(25).
In the present study, a retrospective analysis was
conducted to evaluate the effect of 6 hypoxia-associated genetic
polymorphisms (rs7292, rs7293, rs2231142, rs1800734, rs9852810, and
rs1136410) on the treatment response and survival of Chinese
patients with MGC receiving EOF chemotherapy.
Materials and methods
Patients
The present study retrospectively enrolled 108
Chinese patients with untreated MGC from May 2008 to June 2012 at
the Fudan University Shanghai Cancer Center (Shanghai, China). All
patients had pathologically diagnosed gastric adenocarcinoma with
metastasis confirmed by magnetic resonance imaging or computed
tomography, and were receiving the EOF treatment (intravenous
epirubicin 50 mg/m2 combined with a 2 h intravenous
infusion of oxaliplatin 130 mg/m2 on day 1, followed by
a 24 h continuous infusion of 5-FU 375–425 mg/m2/day for
5 days) as first-line chemotherapy. The chemotherapy was repeated
every 3 weeks until disease progression, intolerable toxicity, or
withdrawal of consent. The response evaluation criteria in solid
tumors (1.0) guidelines were used to evaluate tumor responses
(26). Patients who achieved complete
remission, partial remission, or stable disease were defined as
‘controlled,’ and patients with progressive disease were defined as
‘uncontrolled.’ Follow-up was conducted every 3–6 months, and OS
was defined as the interval between the dates of the beginning of
treatment and the first documentation of mortality from any cause,
while progression-free survival (PFS) was defined as the interval
between the dates of the beginning of treatment and the first
documentation of disease progression or mortality from any
cause.
The present study was approved by the Ethics
Committee of Fudan University Shanghai Cancer Center (Shanghai,
China) and was conducted in accordance with the Declaration of
Helsinki. The blood samples obtained prior to treatment were
acquired from the tissue bank of Fudan University Shanghai Cancer
Center with written informed consent from the patients.
Genotyping
A total of six single nucleotide polymorphisms (SNP)
in four hypoxia-related genes from the National Center for
Biotechnology Information dbSNP database: MB (rs7292, rs7293),
ABCG2 (rs2231142), MLH1 (rs1800734, rs9852810), and PARP-1
(rs1136410) were genotyped (Table I).
Genomic DNA extraction and SNP genotyping were performed as
described previously (27).
Specifically, genomic DNA was extracted from the peripheral blood
using the standard phenol-chloroform method. All probes and primers
were designed by the Assay-on-Design service. The primer sequences
were as follows: 1) rs7292, forward primer (5′-3′): ACG TTG GAT GCT
CAT GAT GCC CCT TCT TCT, reverse primer (5′-3′): ACG TTG GAT GGA
GGA CTT AAA GAA GCA TGG, extension primer (5′-3′): tct gAA AGA AGC
ATG GTG CCA C. 2) rs7293, forward primer (5′-3′): ACG TTG GAT GGT
TTG ACA AGT TCA AGC ACC, reverse primer (5′-3′): ACG TTG GAT GTG
GCA CCA TGC TTC TTT AAG, extension primer (5′-3′): GCT TCT TTA AGT
CCT CAG A. 3) rs2231142, forward primer (5′-3′): ACG TTG GAT GTG
ATG TTG TGA TGG GCA CTC, reverse primer (5′-3′): ACG TTG GAT GGT
CAT AGT TGT TGC AAG CCG, extension primer (5′-3′): ccc tCA AGC CGA
AGA GCT GCT GAG AAC T. 4) rs1800734, forward primer (5′-3′): ACG
TTG GAT GAT CAA TAG CTG CCG CTG AAG, reverse primer (5′-3′): ACG
TTG GAT GAG TGC CTC GTG CTC ACG TTC, extension primer (5′-3′): gGC
TCA CGT TCT TCC TT. 5) rs9852810, forward primer (5′-3′): ACG TTG
GAT GTT TAT GGA GCA TCT ACG GTG, reverse primer (5′-3′): ACG TTG
GAT GAC TTT CCC TGC AGG GAT AAG, extension primer (5′-3′): atG GAT
AAG AGC ATT AAA TGA GAT AA. 6) rs1136410, forward primer (5′-3′):
ACG TTG GAT GTG AGC AGA CTG TAG GCC AC, reverse primer (5′-3′): ACG
TTG GAT GGC TTT CTT TTG CTC CTC CAG, extension primer (5′-3′): cct
taCT TTT GCT CCT CCA GGC CAA GG.
 | Table I.Summary of the six SNPs in the four
hypoxia-related genes in the present study. |
Table I.
Summary of the six SNPs in the four
hypoxia-related genes in the present study.
| Gene | SNP IDa |
Chromosomeb | Function | Allele | HW test
P-value |
|---|
| MB | rs7292 | ch.22:35610998 | Synonymous | T>C | 0.912 |
| MB | rs7293 | ch.22:35611028 | Synonymous | G>A | 0.911 |
| ABCG2 | rs2231142 | ch.4:88131171 | Missense | C>A | 0.479 |
| MLH1 | rs1800734 | ch.3:36993455 | 5′UTR | G>A | 0.505 |
| MLH1 | rs9852810 | ch.3:37027478 | Intron variant | G>A | 1.000 |
| PARP-1 | rs1136410 | ch.1:226367601 | Missense | T>C | 0.878 |
All SNPs were genotyped with the TaqMan assay method
using the ABI 7900 DNA Detection System (Applied Biosystems, Thermo
Fisher Scientific, Inc., Waltham, MA, USA). The polymerase chain
reaction (PCR) was performed using the TaqMan Universal PCR Master
Mix reagent (Applied Biosystems; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. PCR was performed at 95°C
for 2 min, followed by 45 cycles at 95°C for 30 sec, 56°C for 30
sec and 72°C for 1 min, with a final incubation at 72°C for 5 min;
the extension reactions were conducted at 94°C for 30 sec and then
94°C for 5 sec, followed by 5 cycles at 52°C for 5 sec and at 80°C
for 5 sec for a total of 40 cycles and finally at 72°C for 3
min.
Statistical analysis
Hardy-Weinberg equilibrium and pairwise linkage
disequilibrium (LD), as well as allele and genotype distributions,
were analyzed using SHEsis (http://analysis.bio-x.cn/myAnalysis.php) (28). The discrepancies in allelic and
genotype frequency between the controlled and uncontrolled groups
were analyzed using χ2 or Fisher's exact tests. OS and
PFS were analyzed using the Kaplan-Meier method with the log-rank
test using SPSS software (version 19.0; IBM Corp., Armonk, NY,
USA). Risk factors with P<0.1 were further analyzed using
multivariate Cox regression models to assess the influence of
genotypes on treatment-association survival. The software Testing
Haplotype Effects in Association Studies (version 3.1) was used to
perform haplotype analysis (29). A
two-tailed P<0.05 was considered to indicate a statistically
significant difference.
Results
Study population and SNPs
The study population included 108 Chinese patients
with MGC receiving EOF chemotherapy. Baseline demographic and
clinical data were described in a previous study (30). In brief, 41.0% (44/108) of the
patients were female, and the age of the patients ranged from 23–74
years. Within the study population, 95.4% (103/108) of patients had
an Eastern Cooperative Oncology Group (ECOG) score (31) of 0–1. All the patients had at least
one unresectable lesion including lung metastasis, liver
metastasis, ascites, pleural effusion or retroperitoneal lymph node
invasion. According to the best treatment response, 89 (82.4%)
patients were regarded as controlled and 19 (17.6%) patients were
uncontrolled. There was no difference between the controlled and
uncontrolled patients in terms of sex, age, ECOG score, tumor
differentiation, synchronous metastasis status and number of
lesions. The median follow-up duration was 28.4 (range, 7.7–69.0)
months.
Table I presents the 6
SNPs analyzed in the present study. Each of the SNPs was in
Hardy-Weinberg equilibrium. Patients for whom data was lacking for
certain SNPs (for instance, 1/108 patients for rs2231142) were
excluded from the corresponding analysis.
LD analysis
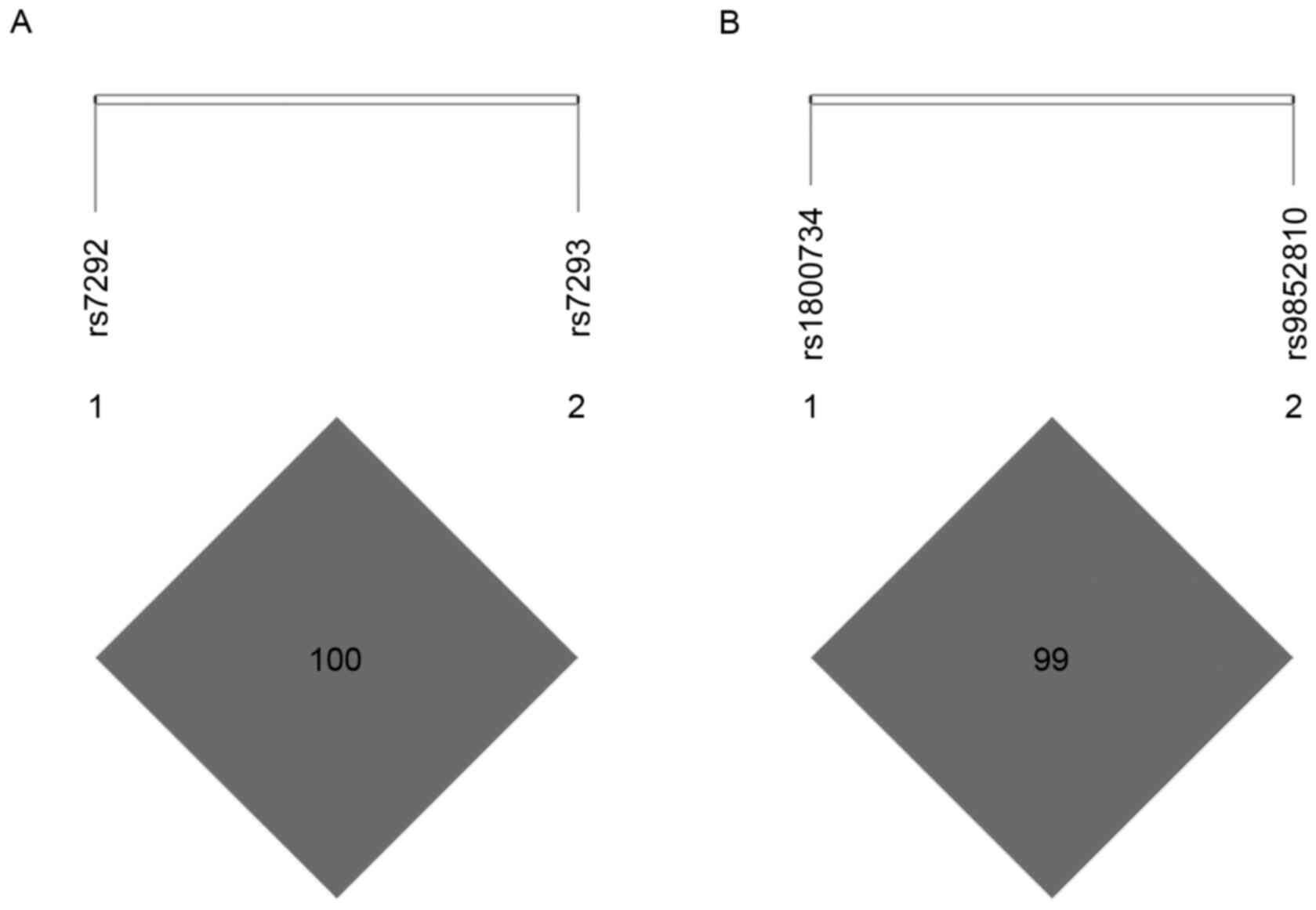
Pairwise LD analysis revealed strong LD in MB SNPs
rs7292 and rs7293 (D'=1.000, r2=1.000; Fig. 1A), as well as MLH1 SNPs rs1800734 and
rs9852810 (D'=1.000, r2=0.084; Fig. 1B). Since rs7292 and rs7293 were in
complete LD (D'=1.000, r2=1.000), only rs7292 was
included in the subsequent analysis to avoid unnecessary
duplication.
Association of alleles and genotype
frequency with disease control
Differences between the four SNPs analyzed in the
present study and disease control are presented in Table II. No allele or genotype exhibited a
significant association with treatment response.
 | Table II.Allele and genotype distribution in
controlled and uncontrolled patients. |
Table II.
Allele and genotype distribution in
controlled and uncontrolled patients.
| Gene | SNP ID | Total No. | Genotype frequency
(%) |
P-valuea | Allele frequency
(%) | χ2 |
P-valuea | Odds ratio (95%
CI) |
|---|
| MB | rs7292 |
| CC | CT | TT | 0.136 | C | T | 0.043 | 0.837 | 0.919
(0.414–2.043) |
|
| Controlled | 89 | 5 (5.6) | 34 (38.2) | 50 (56.2) |
| 44 (24.7) | 134 (75.3) |
|
|
|
|
| Uncontrolled | 19 | 3 (15.8) | 4 (21.1) | 12 (63.2) |
| 10 (26.3) | 28 (73.7) |
|
|
|
| ABCG2 | rs2231142 |
| AA | AC | CC | 0.172 | A | C | 2.081 | 0.136 | 1.915
(0.790–4.640) |
|
| Controlled | 89 | 11 (12.4) | 34 (38.2) | 44 (49.4) |
| 56 (31.5) | 122 (68.5) |
|
|
|
|
| Uncontrolled | 18 | 2 (11.1) | 3 (16.7) | 13 (72.2) |
| 7 (19.4) | 29 (80.6) |
|
|
|
| MLH1 | rs1800734 |
| AA | AG | GG | 0.085 | A | G | 0.639 | 0.424 | 1.331
(0.660–2.684) |
|
| Controlled | 89 | 28 (31.5) | 41 (46.1) | 20 (22.5) |
| 97 (54.5) | 81 (45.5) |
|
|
|
|
| Uncontrolled | 19 | 7 (36.8) | 4 (21.1) | 8 (42.1) |
| 18 (47.4) | 20 (52.6) |
|
|
|
| MLH1 | rs9852810 |
| AA | AG | GG | 0.275 | A | G | 0.877 | 0.313 | 0.567
(0.170–1.886) |
|
| Controlled | 88 | 0 (0) | 11 (12.5) | 77 (87.5) |
| 11 (6.3) | 165 (93.8) |
|
|
|
|
| Uncontrolled | 19 | 1 (5.2) | 2 (10.5) | 16 (84.2) |
| 4 (10.5) | 34 (89.5) |
|
|
|
| PARP-1 | rs1136410 |
| CC | CT | TT | 0.289 | C | T | 0.056 | 0.813 | 1.094
(0.520–2.302) |
|
| Controlled | 89 | 13 (14.6) | 42 (47.2) | 34 (38.2) |
| 68 (2.8) | 110 (97.2) |
|
|
|
|
| Uncontrolled | 18 | 4 (22.2) | 5 (27.8) | 9 (50) |
| 13 (0.0) | 23 (100.0) |
|
|
|
Survival analysis
In addition to pathological grade and number of
lesions, as reported in a previous study (27), the genotypes of MB rs7292 and MLH1
rs9852810 were revealed to be significantly associated with PFS
(Table III). T carriers of MB
rs7292 had significantly more favorable PFS compared with CC
carriers (median PFS: 179.0 and 71.0 days, respectively P=0.001;
Fig. 2A). Similarly, GG carriers of
MLH1 rs9852810 had significantly longer PFS in comparison to A
carriers (median PFS: 178.0 and 147.0 days, respectively; P=0.040;
Fig. 2B). In the Cox regression
models, the T allele of MB rs7292 and the GG genotype of MLH1
rs9852810 were associated with decreased risks of disease
progression [rs7292: hazard ratio (HR)=0.135, 95% confidence
interval (CI)=0.057–0.321; P<0.001; rs9852810: HR=0.494, 95%
CI=0.267–0.913; P=0.024, respectively], indicating that, along with
pathological grade (P=0.005) and number of lesions (P=0.020), MB
rs7292 and MLH1 rs9852810 were independent predictive factors of
PFS for patients with MGC receiving EOF chemotherapy (Table IV). However, none of the SNPs were
revealed to have an association with OS using Kaplan-Meier analysis
(Table III).
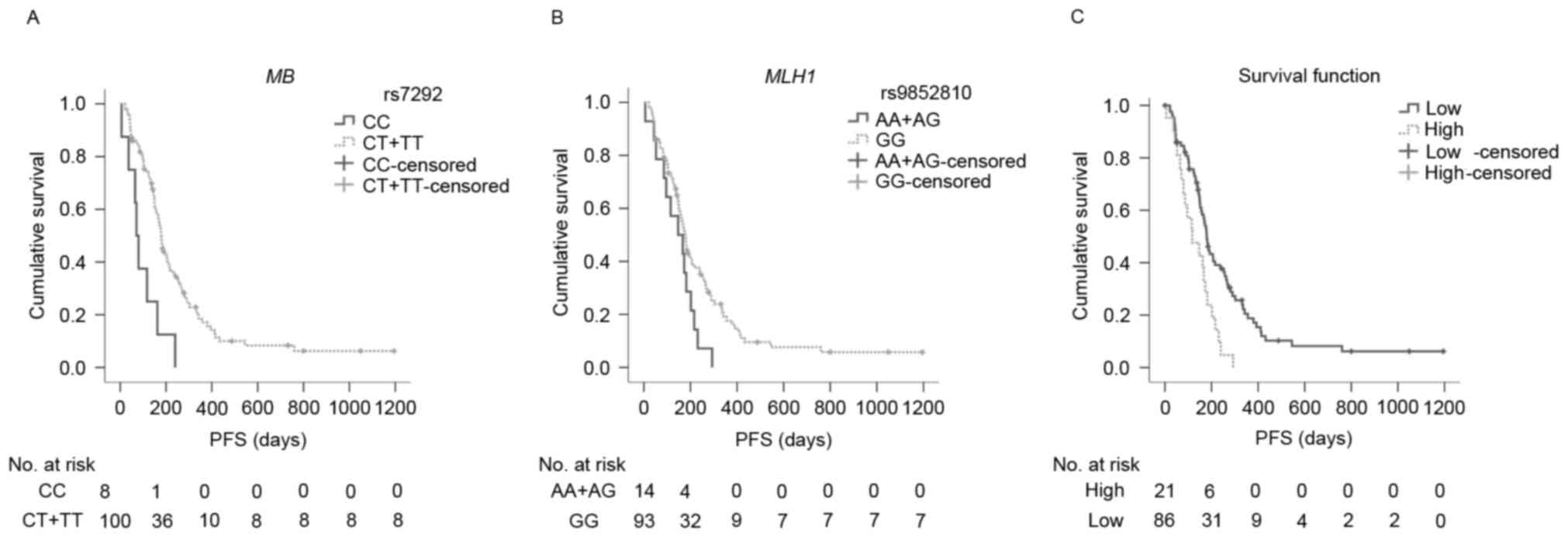 | Figure 2.PFS curves for MB rs7292, MLH1
rs9852810, and the prognostic index among subgroups. (A) PFS curves
of rs7292 among subgroups. Median PFS time for CC carriers (solid
line) and CT/TT carriers (dotted line) were 71.0 days (95% CI:
50.2–91.8) and 179.0 days (95% CI: 162.7–191.1), respectively
(P=0.001); (B) PFS curves of rs9852810 among subgroups. Median
overall survival time for AA/AG carriers (solid line), GG carriers
(dotted line) were 147.0 days (95% CI: 51.7–242.3) and 178.0 days
(95% CI: 155.8–200.2), respectively (P=0.040); (C) PFS curves of
prognostic index among subgroups. The median PFS time for low-risk
(0 risk, solid line), and high-risk (1 or 2 risks, dotted line)
were 180.0 days (95% CI: 161.5–198.5) and 117.0 days (95% CI:
40.7–193.3) respectively (P=0.002). The high-risk group had a
2.2-fold increased risk of progression compared with the low-risk
group (HR=2.223, 95% CI=1.335–3.704; P=0.002). PFS,
progression-free survival; CI, confidence interval; HR, hazard
ratio; MB, myoglobin; MLH1, MutL homolog 1. |
 | Table III.PFS and OS analysis with the
Kaplan-Meier method and log-rank test. |
Table III.
PFS and OS analysis with the
Kaplan-Meier method and log-rank test.
| Clinical
characteristic | Clinical
factors | Patients (n) | Median PFS
(days) | 95% CI |
P-valuea | Median OS
(days) | 95% CI |
P-valuea |
|---|
| MB rs7292 | CC | 8 | 71.0 | 50.2–91.8 | 0.001 | 244.0 | 132.4–355.6 | 0.245 |
|
| CT+TT | 100 | 179.0 | 162.7–191.1 |
| 465.0 | 302.8–627.2 |
|
| ABCG2
rs2231142 | AA | 13 | 182.0 | 159.2–204.7 | 0.705 | 297.0 | 173.2–420.8 | 0.278 |
|
| AC+CC | 94 | 173.0 | 152.3–193.7 |
| 465.0 | 292.0–638.0 |
|
| MLH1 rs1800734 | AA | 35 | 159.0 | 112.0–206.0 | 0.897 | 367.0 | 50.3–683.7 | 0.590 |
|
| AG+GG | 73 | 179.0 | 158.5–199.5 |
| 465.0 | 309.3–620.7 |
|
| MLH1 rs9852810 | AA+AG | 14 | 147.0 | 51.7–242.3 | 0.040 | 241.0 | 100.0–382.1 | 0.287 |
|
| GG | 93 | 178.0 | 155.8–200.2 |
| 465.0 | 207.7–622.3 |
|
| PARP-1
rs1136410 | CC | 17 | 202.0 | 158.0–246.0 | 0.528 | 1378.0 | b | 0.156 |
|
| CT+TT | 90 | 169.0 | 148.1–189.9 |
| 403.0 | 252.4–533.6 |
|
 | Table IV.Multivariate analysis of prognostic
factors of PFS following EOF treatment. |
Table IV.
Multivariate analysis of prognostic
factors of PFS following EOF treatment.
|
| PFS |
|---|
|
|
|
|---|
| Clinical
characteristic | Hazard ratio | 95% CI |
P-valuea |
|---|
| Pathological
grade |
|
|
|
| Low and
undifferentiated | 1 |
| 0.005 |
|
Moderate and high | 0.252 | 0.097–0.657 |
|
|
Unclassified | 0.633 | 0.382–1.046 |
|
| No. of lesions |
|
|
|
| 1 | 1 |
| 0.020 |
| 2 | 0.159 | 0.030–0.854 |
|
| ≥3 | 0.952 | 0.300–3.025 |
|
| Synchronous
metastasis |
|
|
|
|
Presence | 1 |
| 0.628 |
|
Absence | 1.184 | 0.597–2.348 |
|
| MB rs7292 |
|
|
|
| CC | 1 |
| <0.001 |
|
CT+TT | 0.135 | 0.057–0.321 |
|
| MLH1 rs9852810 |
|
|
|
|
AA+AG | 1 |
| 0.024 |
| GG | 0.494 | 0.267–0.913 |
|
PFS analysis based on the prognostic
index
According to the favorable SNPs for PFS (MB rs7292
CT/TT genotype and MLH1 rs9852810 GG genotype), patients were
classified into a low-risk group (involving one or two of the two
SNPs) and a high-risk group (involving neither of the two SNPs).
Patients from the low-risk group revealed a significantly longer
PFS than patients from the high-risk group (median PFS: 180.0 and
117.0 days, respectively; P=0.002; Fig.
2C). In comparison with the low-risk group, the high-risk group
demonstrated a 2.2-fold increased risk of disease progression
(HR=2.223, 95% CI=1.335–3.704; P=0.002).
Haplotype analysis
Haplotype analysis was performed in SNPs that were
in strong LD. The analysis demonstrated that the GA haplotype of
MLH1 rs1800734/rs9852810 was associated with unfavorable PFS
(HR=1.84, 95% CI=1.07–3.16; P=0.027), while no haplotype of MLH1
rs1800734/rs9852810 was associated with OS (Table V). The haplotypes of MB had no
significant influence on survival (Table
VI).
 | Table V.Analyses of associations between
haplotypes of MLH1 and PFS/OS. |
Table V.
Analyses of associations between
haplotypes of MLH1 and PFS/OS.
|
|
| PFS | OS |
|---|
|
|
|
|
|
|---|
| Haplotypes | Haplotype
frequencies (%) | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| AGa | 51.00 |
|
|
|
|
|
|
| GAa | 6.73 | 1.84 | 1.07–3.16 | 0.027 | 1.50 | 0.75–3.00 | 0.253 |
| GGa | 42.31 | 0.95 | 0.70–1.27 | 0.712 | 0.87 | 0.58–1.30 | 0.490 |
 | Table VI.Analyses of associations between
haplotypes of MB and PFS/OS. |
Table VI.
Analyses of associations between
haplotypes of MB and PFS/OS.
|
|
| PFS | OS |
|---|
|
|
|
|
|
|---|
| Haplotypes | Haplotype
frequencies (%) | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| TAa | 75.00 |
|
|
|
|
|
|
| CGa | 25.00 | 1.09 | 0.79–1.51 | 0.60 | 1.20 | 0.78–1.84 | 0.41 |
Discussion
Hypoxia is common in numerous types of solid tumor,
including GC, and is considered to be an important factor
associated with aggressive phenotypes and treatment resistance of
tumors (6,32). Several studies have investigated the
influence of hypoxia on the chemosensitivity, metastasis, and
prognosis of GC. Liu et al (4)
demonstrated that GC cells developed resistance to 5-FU,
vincristine, cisplatin, etoposide, and adriamycin under conditions
of hypoxia. Matsuoka et al (5)
demonstrated that hypoxia may stimulate the EMT process of GC
cells. In addition, Osinsky et al (6) revealed that low levels of oxygenation in
tumor tissues were associated with poor survival in patients with
GC.
The present study explored the relationship between
polymorphisms of genes associated with tumor hypoxia and the
clinical outcome of patients with MGC treated with EOF, and to the
best of our knowledge, revealed for the first time that the CT/TT
genotype of MB rs7292 and the GG genotype of MLH1 rs9852810 were
independent favorable predictive factors of PFS for patients with
MGC treated with the EOF regimen. Since PFS more directly reflects
the therapeutic effect of first-line chemotherapy, MB rs7292 and
MLH1 rs9852810 may serve as biomarkers that predict the short-term
efficacy of the first-line EOF regimen in patients with MGC.
The MB rs7292 CT/TT genotype was demonstrated to be
an independent favorable predictor of PFS for patients with MGC
treated with EOF chemotherapy, while MB rs7292 CT/TT carriers
appeared to possess a tendency toward an improved OS compared with
CC carriers (for CT/TT and CC genotypes, median OS: 465.0 and 244.0
days, respectively; P=0.245). Although the difference was not
statistically significant, considering the markedly separative
tendency of OS curves and the limited sample size of the present
study, there may still be a link between the CT/TT genotype and
survival. With respect to the prognostic function of MB rs7293,
since MB rs7292 and rs7293 were in complete LD (D'=1.000,
r2=1.000), the results of the analysis of the rs7293 GG
genotype were identical to those of the rs7292 CC genotype (data
not shown).
The function of MB in GC has not previously been
reported, and the function of MB in tumors varies and remains
controversial. For example, one previous study demonstrated that
71% of 1,027 breast cancer cases demonstrated MB expression, with a
significant correlation with hypoxia markers (HIF-2α, CAIX) and an
improved prognosis (9). Furthermore,
another previous study revealed that MB was induced in breast
cancer cells by prolonged hypoxia, partially by HIF-1/2-dependent
transactivation, and additionally revealed that MB may damage
mitochondria in hypoxic cancer cells, indicating the
tumor-suppressive potential of MB in breast cancer (10). However, the function of MB in
non-small-cell lung cancer is controversial. For instance, Galluzzo
et al (33) demonstrated that
overexpression of MB in non-small-cell lung cancer cells suppressed
the hypoxia response, inhibited tumor growth, reduced vessel
density, promoted tumor differentiation and suppressed tumor
metastasis. On the other hand, Oleksiewicz et al (13) reported that patients with
non-small-cell lung cancer with low MB levels had a longer survival
duration than patients with high MB expression. Hypoxia may also
induce the expression of MB in prostate cancer (11) and renal cell carcinoma (12). However, to date, no studies have
reported the polymorphisms of MB in malignant tumors, and the
function of MB in gastric cancer has not been previously
demonstrated. Further research is required to elucidate whether the
change of rs7292 may affect the expression of MB, and whether and
how MB may influence the efficacy of the EOF treatment in patients
with GC.
The link between MLH1 rs1800734 and rs9852810 and GC
was also reported in the present study. In addition to the
prolonged PFS of patients with MGC carrying the GG genotype of MLH1
rs9852810, MLH1 rs9852810 GG carriers were revealed to possess an
improved OS compared with A carriers (for GG and AA/AG genotypes,
median OS: 465.0 and 241.0 days, respectively; P=0.287). The
results demonstrating that OS difference did not reach statistical
significance may be attributed to the relatively small sample size
and the therapy following first-line chemotherapy. Additionally,
haplotype analysis revealed that the GA haplotype of MLH1
rs1800734/rs9852810 was associated with poorer PFS compared with
the common haplotype AG; the reason may be that the GG genotype of
rs9852810 had a significantly favorable PFS.
As a key DNA MMR gene, the expression of MLH1 was
demonstrated to be reduced under hypoxia as a result of
transcriptional, translational, or methylation modification
(18,34,35). In
addition, several studies have revealed that methylation of the
MLH1 gene promoter was associated with the stages of carcinogenesis
and progression as well as oxaliplatin resistance in GC (36–38).
With respect to the influence of the polymorphisms
of MLH1 on cancer, Langeberg et al (39) reported that the rs9852810 A allele was
associated with a higher risk of prostate cancer incidence and
recurrence, as well as being associated with more aggressive types
of prostate cancer. Furthermore, rs1800734 was located in the CpG
island of MLH1, and the A allele of rs1800734 was significantly
associated with MLH1 methylation and high-degree microsatellite
instability in colorectal cancer (40–42). In
GC, Zhu et al (43)
demonstrated that the G allele of MLH1 rs1800734 was associated
with a decreased risk of GC. However, the present study
demonstrated no significant association between rs1800734 and the
clinical outcome of patients with MGC who received EOF
chemotherapy, a result that may be explained by the controversy
surrounding the influence of microsatellite instability on GC
(44,45). Since the influence of the two
polymorphisms on the survival and treatment response of patients
with MGC has, to the best of our knowledge, never previously been
reported, and additionally whether the two SNPs may alter the
function of MLH1 and subsequently influence GC progression is
unknown, additional work is required to validate the predictive
value of those polymorphisms for GC.
When the patients were classified into a low-risk
group and a high-risk group according to the number of favorable
SNPs for PFS (MB rs7292 CC genotype and MLH1 rs9852810 A allele)
they possessed, the high-risk group was revealed to possess a
significantly shorter PFS; thus, this type of classification may
assist in a more comprehensive identification of patients with poor
prognosis.
To conclude, to the best of our knowledge, the
present study revealed for the first time, that in Chinese patients
with MGC treated with the EOF regimen as first-line chemotherapy,
the CT/TT genotype of MB rs7292 and the GG genotype of MLH1
rs9852810 were independent favorable predictive factors of PFS, and
that these may serve as biomarkers to predict the short-term
efficacy of the first-line EOF regimen in patients with MGC. Since
all the SNPs of the four hypoxia-associated genes investigated in
the present study have seldom been reported in GC, the results of
the present study will require confirmation with larger cohort
studies and further functional investigations.
Acknowledgements
The present study was supported by the Natural
Science Foundation of Shanghai, Shanghai, China (grant no.
13ZR1408200) and the Foundation of the Shanghai Municipal Science
and Technology Commission, Shanghai, China (grant no.
134119a8600).
References
|
1
|
Bang YJ, Van Cutsem E, Feyereislova A,
Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T,
et al: Trastuzumab in combination with chemotherapy versus
chemotherapy alone for treatment of HER2-positive advanced gastric
or gastro-oesophageal junction cancer (ToGA): A phase 3,
open-label, randomised controlled trial. Lancet. 376:687–697. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Van Cutsem E, Moiseyenko VM, Tjulandin S,
Majlis A, Constenla M, Boni C, Rodrigues A, Fodor M, Chao Y, Voznyi
E, et al: Phase III study of docetaxel and cisplatin plus
fluorouracil compared with cisplatin and fluorouracil as first-line
therapy for advanced gastric cancer: A report of the V325 study
group. J Clin Oncol. 24:4991–4997. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Balkwill FR, Capasso M and Hagemann T: The
tumor microenvironment at a glance. J Cell Sci. 125:5591–5596.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang SY, Song BQ, Dai SL, Yang KX, Jin Z
and Shi KW: Effects of hypoxia-inducible factor-1α silencing on
drug resistance of human pancreatic cancer cell line Patu8988/5-Fu.
Hepatogastroenterology. 61:2395–2401. 2014.PubMed/NCBI
|
|
5
|
Zhao M, Zhang Y, Zhang H, Wang S, Zhang M,
Chen X, Wang H, Zeng G, Chen X, Liu G and Zhou C: Hypoxia-induced
cell stemness leads to drug resistance and poor prognosis in lung
adenocarcinoma. Lung Cancer. 87:98–106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hockel M and Vaupel P: Tumor hypoxia:
Definitions and current clinical, biologic, and molecular aspects.
J Natl Cancer Inst. 93:266–276. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Röhwer N, Dame C, Haugstetter A,
Wiedenmann B, Detjen K, Schmitt CA and Cramer T: Hypoxia-inducible
factor 1alpha determines gastric cancer chemosensitivity via
modulation of p53 and NF-kappaB. PLoS One. 5:e120382010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fraser J, de Mello LV, Ward D, Rees HH,
Williams DR, Fang Y, Brass A, Gracey AY and Cossins AR:
Hypoxia-inducible myoglobin expression in nonmuscle tissues. Proc
Natl Acad Sci USA. 103:pp. 2977–2981. 2006; View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kristiansen G, Rose M, Geisler C,
Fritzsche FR, Gerhardt J, Lüke C, Ladhoff AM, Knüchel R, Dietel M,
Moch H, et al: Endogenous myoglobin in human breast cancer is a
hallmark of luminal cancer phenotype. Br J Cancer. 102:1736–1745.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kristiansen G, Hu J, Wichmann D, Stiehl
DP, Rose M, Gerhardt J, Bohnert A, Ten Haaf A, Moch H, Raleigh J,
et al: Endogenous myoglobin in breast cancer is hypoxia-inducible
by alternative transcription and functions to impair mitochondrial
activity: A role in tumor suppression? J Biol Chem.
286:43417–43428. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Meller S, Bicker A, Montani M, Ikenberg K,
Rostamzadeh B, Sailer V, Wild P, Dietrich D, Uhl B, Sulser T, et
al: Myoglobin expression in prostate cancer is correlated to
androgen receptor expression and markers of tumor hypoxia. Virchows
Arch. 465:419–427. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Behnes CL, Bedke J, Schneider S, Küffer S,
Strauss A, Bremmer F, Ströbel P and Radzun HJ: Myoglobin expression
in renal cell carcinoma is regulated by hypoxia. Exp Mol Pathol.
95:307–312. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Oleksiewicz U, Daskoulidou N, Liloglou T,
Tasopoulou K, Bryan J, Gosney JR, Field JK and Xinarianos G:
Neuroglobin and myoglobin in non-small cell lung cancer:
Expression, regulation and prognosis. Lung Cancer. 74:411–418.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Krishnamurthy P and Schuetz JD: The ABC
transporter Abcg2/Bcrp: Role in hypoxia mediated survival.
Biometals. 18:349–358. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Krishnamurthy P, Ross DD, Nakanishi T,
Bailey-Dell K, Zhou S, Mercer KE, Sarkadi B, Sorrentino BP and
Schuetz JD: The stem cell marker Bcrp/ABCG2 enhances hypoxic cell
survival through interactions with heme. J Biol Chem.
279:24218–24225. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mihaylova VT, Bindra RS, Yuan J, Campisi
D, Narayanan L, Jensen R, Giordano F, Johnson RS, Rockwell S and
Glazer PM: Decreased expression of the DNA mismatch repair gene
Mlh1 under hypoxic stress in mammalian cells. Mol Cell Biol.
23:3265–3273. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rodríguez-Jiménez FJ, Moreno-Manzano V,
Lucas-Dominguez R and Sánchez-Puelles JM: Hypoxia causes
downregulation of mismatch repair system and genomic instability in
stem cells. Stem Cells. 26:2052–2062. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nakamura H, Tanimoto K, Hiyama K, Yunokawa
M, Kawamoto T, Kato Y, Yoshiga K, Poellinger L, Hiyama E and
Nishiyama M: Human mismatch repair gene, MLH1, is transcriptionally
repressed by the hypoxia-inducible transcription factors, DEC1 and
DEC2. Oncogene. 27:4200–4209. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Y, Yang Y, Lu Y, Herman JG, Brock MV,
Zhao P and Guo M: Predictive value of CHFR and MLH1 methylation in
human gastric cancer. Gastric Cancer. 18:280–287. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Meyers M, Wagner MW, Hwang HS, Kinsella TJ
and Boothman DA: Role of the hMLH1 DNA mismatch repair protein in
fluoropyrimidine-mediated cell death and cell cycle responses.
Cancer Res. 61:5193–5201. 2001.PubMed/NCBI
|
|
21
|
Tentori L, Leonetti C, Muzi A, Dorio AS,
Porru M, Dolci S, Campolo F, Vernole P, Lacal PM, Praz F and
Graziani G: Influence of MLH1 on colon cancer sensitivity to
poly(ADP-ribose) polymerase inhibitor combined with irinotecan. Int
J Oncol. 43:210–218. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vyas S and Chang P: New PARP targets for
cancer therapy. Nat Rev Cancer. 14:502–509. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Klymenko T, Brandenburg M, Morrow C, Dive
C and Makin G: The novel Bcl-2 inhibitor ABT-737 is more effective
in hypoxia and is able to reverse hypoxia-induced drug resistance
in neuroblastoma cells. Mol Cancer Ther. 10:2373–2383. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim JW, Ho WJ and Wu BM: The role of the
3D environment in hypoxia-induced drug and apoptosis resistance.
Anticancer Res. 31:3237–3245. 2011.PubMed/NCBI
|
|
25
|
Erdélyi K, Pacher P, Virág L and Szabó C:
Role of poly(ADP-ribosyl) ation in a ‘two-hit’ model of hypoxia and
oxidative stress in human A549 epithelial cells in vitro. Int J Mol
Med. 32:339–346. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Therasse P, Arbuck SG, Eisenhauer EA,
Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van
Oosterom AT, Christian MC and Gwyther SG: New guidelines to
evaluate the response to treatment in solid tumors. European
Organization for research and treatment of cancer, national cancer
institute of the United States, national cancer institute of
Canada. J Natl Cancer Inst. 92:205–216. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Geng R, Chen Z, Zhao X, Qiu L, Liu X, Liu
R, Guo W, He G, Li J and Zhu X: Oxidative stress-related genetic
polymorphisms are associated with the prognosis of metastatic
gastric cancer patients treated with epirubicin, oxaliplatin and
5-fluorouracil combination chemotherapy. PLoS One. 9:e1160272014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shi YY and He L: SHEsis, a powerful
software platform for analyses of linkage disequilibrium, haplotype
construction, and genetic association at polymorphism loci. Cell
Res. 15:97–98. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tregouet DA and Garelle V: A new JAVA
interface implementation of THESIAS: Testing haplotype effects in
association studies. Bioinformatics. 23:1038–1039. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu X, Chen Z, Zhao X, Huang M, Wang C,
Peng W, Yin J, Li J, He G, Li X and Zhu X: Effects of IGF2BP2,
KCNQ1 and GCKR polymorphisms on clinical outcome in metastatic
gastric cancer treated with EOF regimen. Pharmacogenomics.
16:959–970. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Oken MM, Creech RH, Tormey DC, Horton J,
Davis TE, McFadden ET and Carbone PP: Toxicity and response
criteria of the Eastern Cooperative Oncology Group. Am J Clin
Oncol. 5:649–656. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Teicher BA: Hypoxia and drug resistance.
Cancer Metastasis Rev. 13:139–168. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Galluzzo M, Pennacchietti S, Rosano S,
Comoglio PM and Michieli P: Prevention of hypoxia by myoglobin
expression in human tumor cells promotes differentiation and
inhibits metastasis. J Clin Invest. 119:865–875. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lu Y, Wajapeyee N, Turker MS and Glazer
PM: Silencing of the DNA mismatch repair gene MLH1 induced by
hypoxic stress in a pathway dependent on the histone demethylase
LSD1. Cell Rep. 8:501–513. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Min L, Chen Q, He S, Liu S and Ma Y:
Hypoxia-induced increases in A549/CDDP cell drug resistance are
reversed by RNA interference of HIF-1α expression. Mol Med Rep.
5:228–232. 2012.PubMed/NCBI
|
|
36
|
Teng Y, Dai D, Shen W and Liu H: Effect of
methylation of hMLH1 gene promotor on stage tumorigenesis and
progression of human gastric cancer. Zhonghua Wei Chang Wai Ke Za
Zhi. 18:166–170. 2015.(In Chinese). PubMed/NCBI
|
|
37
|
Guo H, Yan W, Yang Y and Guo M: Promoter
region methylation of DNA damage repair genes in human gastric
cancer. Zhonghua Yi Xue Za Zhi. 94:2193–2196. 2014.(In Chinese).
PubMed/NCBI
|
|
38
|
Milane L, Ganesh S, Shah S, Duan ZF and
Amiji M: Multi-modal strategies for overcoming tumor drug
resistance: Hypoxia, the Warburg effect, stem cells, and
multifunctional nanotechnology. J Control Release. 155:237–247.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Langeberg WJ, Kwon EM, Koopmeiners JS,
Ostrander EA and Stanford JL: Population-based study of the
association of variants in mismatch repair genes with prostate
cancer risk and outcomes. Cancer Epidemiol Biomarkers Prev.
19:258–264. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen H, Taylor NP, Sotamaa KM, Mutch DG,
Powell MA, Schmidt AP, Feng S, Hampel HL, de la Chapelle A and
Goodfellow PJ: Evidence for heritable predisposition to epigenetic
silencing of MLH1. Int J Cancer. 120:1684–1688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Samowitz WS, Curtin K, Wolff RK, Albertsen
H, Sweeney C, Caan BJ, Ulrich CM, Potter JD and Slattery ML: The
MLH1-93 G>A promoter polymorphism and genetic and epigenetic
alterations in colon cancer. Genes Chromosomes Cancer. 47:835–844.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Raptis S, Mrkonjic M, Green RC, Pethe VV,
Monga N, Chan YM, Daftary D, Dicks E, Younghusband BH, Parfrey PS,
et al: MLH1-93G>A promoter polymorphism and the risk of
microsatellite-unstable colorectal cancer. J Natl Cancer Inst.
99:463–474. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Rho JK, Choi YJ, Lee JK, Ryoo BY, Na II,
Yang SH, Kim CH, Yoo YD and Lee JC: Gefitinib circumvents
hypoxia-induced drug resistance by the modulation of HIF-1alpha.
Oncol Rep. 21:801–807. 2009.PubMed/NCBI
|
|
44
|
Liu L, Sun L, Zhang H, Li Z, Ning X, Shi
Y, Guo C, Han S, Wu K and Fan D: Hypoxia-mediated up-regulation of
MGr1-Ag/37LRP in gastric cancers occurs via
hypoxia-inducible-factor 1-dependent mechanism and contributes to
drug resistance. Int J Cancer. 124:1707–1715. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Willger SD, Puttikamonkul S, Kim KH,
Burritt JB, Grahl N, Metzler LJ, Barbuch R, Bard M, Lawrence CB and
Cramer RA Jr: A sterol-regulatory element binding protein is
required for cell polarity, hypoxia adaptation, azole drug
resistance, and virulence in Aspergillus fumigatus. PLoS Pathog.
4:e10002002008. View Article : Google Scholar : PubMed/NCBI
|