Introduction
Esophageal cancer (EC) is the eighth most common
type of cancer and the sixth most common cause of cancer-associated
mortality worldwide (1,2). In 2003 a study published in New England
Journal of Medicine indicated that the worldwide overall 5-year
survival rate of EC was <15% (3).
Poor outcomes in patients with esophageal cancer are associated
with diagnosis at advanced (metastatic) stages and the propensity
for metastases (4,5). Therefore, there is a requirement to
investigate the underlying mechanisms of EC progression,
particularly metastasis, to identify potential biomarkers for
prognosis and diagnosis.
AEP, currently the only known asparaginyl
endopeptidase in the mammalian genome, is a member of the C13
family in the MEROPS database classification of peptidases, whereas
all other lysosomal cysteine proteases identified to date are
grouped in the C1 family (6,7). The strict specificity of AEP to
asparagine bonds is notable (8). AEP
has been demonstrated to contribute important functions in kidney
physiology, immunity, atherogenesis and bone metabolism (9–15). In
previous years, high AEP expression has been observed in a variety
of solid tumors and acute lymphoblastic leukemia (16–21).
Furthermore, AEP expression positively correlated with
clinicopathological and biological variables in colorectal, and
breast cancer (19,20). Although cancer cells that highly
express AEP have been revealed to exhibit enhanced migratory and
invasive capacity through the activation of
pro-matrix-metalloproteinase 2 (MMP2), and cathepsins (17,22,23), the
pathological functions and underlying mechanisms of AEP in
esophageal cancer remain elusive.
In the present study, it was demonstrated that AEP
expression was elevated in a cohort of esophageal cancer tissues.
Patients with EC with high AEP expression exhibited a significantly
poorer overall survival rate. Additionally, loss of function
experiments revealed that knockdown of AEP significantly reduced
the migration and invasion ability of EC cells through
downregulation of MMP2 and MMP3. The results of the present study
indicate that high AEP expression promotes progression and
indicates poor prognosis in patients with esophageal cancer,
indicating it a novel prognostic biomarker or potential therapeutic
target in esophageal cancer.
Materials and methods
Patients and tissue samples
The present study was approved by the Ethics
Committee of the Fourth Hospital of Hebei Medical University
(Hebei, China). Written informed consent was obtained from patients
or guardians on behalf of the minors enrolled in the study. A total
of 146 patients with histologically confirmed esophageal cancer at
the Fourth Hospital of Hebei Medical University were recruited for
this study between January 2005 and December 2013. There were 111
male patients and 35 female patients with a median age of 57 years
(range, 34–72 years). The specimens were obtained during surgical
resection and matched adjacent normal tissues were also collected.
Their diagnoses were independently re-reviewed by two pathologists,
classified by the World Health Organization criteria (24).
Cell lines
Esophageal cancer EC109 and TE-1 cell lines were
obtained from the Type Culture Collection of the Chinese Academy of
Sciences (Shanghai, China) and the two cell lines were cultured in
RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) at 37°C and 5% CO2. EC109 and TE-1
cells were used to determine baseline AEP expression and EC109
cells were selected to conduct following experiments.
Immunohistochemistry (IHC)
A total of 146 blocks of tissue microarray
containing EC tissues were constructed using a Microarrayer. Serial
4-µm sections were obtained from each block, with the first slide
being stained for hematoxylin and eosin to confirm pathologic
diagnosis, and the subsequent slides stained for further IHC.
Tissue microarray slides were routinely
deparaffinizated and rehydrated. For antigen retrieval, the slides
were heated at 98°C in a citrate buffer (pH 9.0) for a total of 20
min and cooled naturally to room temperature. Sections were
incubated in 0.3% hydrogen peroxide for 20 min to inactivate
endogenous peroxidases at room temperature. The sections were
blocked with 5% normal horse serum (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) in PBS for 30 min and then incubated with the
monoclonal primary antibody against AEP (1:100 dilution; cat. no.
AF2199; R&D Systems, Inc., Minneapolis, MN, USA), overnight at
4°C. The following day, sections were stained using a highly
sensitive streptavidin-biotin-peroxidase detection system
(MaxVision TM HRP-Polymer anti-Mouse IHC kit; cat. no. KIT-5001;
Maixin Biotechnology, Fuzhou, China) and counterstained with
hematoxylin. A negative control was also incorporated using
pre-immune IgG instead of the primary antibody. All slides were
observed and image captured using a light microscope.
Evaluation of
immunohistochemistry
Two sections per specimen were evaluated by two
pathologists independently. Immunoreactive staining was
characterized quantitatively according to the percentage of
positive cells and staining intensity without prior knowledge of
any of the clinicopathological information. The following
proportion scores were assigned as: 0, 0% of the tumor cells showed
positive staining; 1, 0–10% stained; 2, 11–50% stained; 3, 51–75%
stained and 4, 75–100% stained. The intensity of staining was rated
on a scale of 0 to 3: 0, negative; 1, weak; 2, moderate and 3,
strong. The proportion and intensity scores were combined to obtain
a total score (range 0–12). All patients were designated into
negative (score 0), low (score 1–4), moderate (score 5–8) and high
(score 9–12) groups based on AEP expression.
Western blot analysis
To analyze AEP expression in EC109 and TE-1 cell
lines, western blot assays were performed. Briefly, cells were
lysed using radioimmunoprecipitation assay buffer [50 mM Tris–HCl
(pH 7.5), 150 mM NaCl, 1% Triton X-100, 0.5% Na-deoxycholate]
containing protease inhibitors (CompleteMini; Roche Applied
Science, Penzberg, Germany). The concentration of protein was
determined using a BCA kit (Beyotime Institute of Biotechnology,
Shanghai, China). Total protein (20–30 µg per lane) of the lysates
were separated on 8–12% SDS-PAGE gels and transferred to
polyvinylidene fluoride membranes. The membranes were firstly
blocked with 5–10 ml western blocking reagent (Quickblock™; cat.
no. P0220; Beyotime Institute of Biotechnology) at room temperature
for 2 h and then incubated with primary antibodies, goat anti-human
AEP (cat. no. AF2199; R&D Systems; 1:1,000) and rabbit
anti-actin (cat. no. EP1123Y; EMD Millipore, Billerica, CA, USA;
1:10,000), overnight at 4°C. Subsequently, membranes were incubated
with a horseradish peroxidase-conjugated secondary antibody (donkey
anti-goat; cat. no. A0181; 1:10,000; or goat anti-rabbit; cat. no.
A0208; 1:10,000; Beyotime Institute of Biotechnology). The
membranes were incubated with secondary antibodies at room
temperature for 2 h. The bound antibodies were detected using an
enhanced chemiluminescence kit (Pierce; Thermo Fisher Scientific,
Inc.; cat. no. PI32209).
Lentiviral vector mediated
AEP-knockdown
Lentiviral vectors for human AEP-specific short
hairpin RNA (shRNA) carrying a green fluorescent protein (GFP)
sequence were constructed by Hanyin Co. (Shanghai, China). The
recombinant AEP knockdown lentivirus and the negative control (NC)
lentivirus (GFP-lentivirus; Hanyin Co., Shanghai, China) were
prepared, and titered to 109 TU/ml (transfection unit). The AEP
shRNA sequences were AEP-KD1, 5′-GCTCTTGGTGGATCATCAA-3′; and
AEP-KD2, 5′-GCATGTTCAATGGGAGCTTGGA-3′. After 48 h, the knockdown
efficiency was confirmed via reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) and western blotting. To obtain
the stable AEP-knockdown cell line, EC109 cells were seeded at a
density of 2×105 cells/well in 6-well dishes. The cells
were then infected with the same titer virus with 8 µg/ml polybrene
on the following day. At ~72 h post-viral infection, GFP expression
was confirmed under a fluorescence microscope, and the culture
medium was replaced with RPMI-1640 containing 4 µg/ml puromycin
(Thermo Fisher Scientific, Inc.). The cells were then cultured for
at least 14 days at 37°C. The puromycin-resistant cell clones were
isolated, amplified in medium containing 2 µg/ml puromycin for 7–9
days and transferred to a medium without puromycin.
Matrigel-Transwell assay
EC109 and AEP-knockdown EC109 cells (1,000 per well)
were then plated in the top chamber of Transwell assay inserts (EMD
Millipore) with a Matrigel-coated membrane containing 8-µm pores in
200 ml of serum-free RPMI-1640 medium. The assays were conducted in
triplicate. The inserts were then placed into the bottom chamber of
a 24-well plate containing RPMI 1640 supplemented with 10% FBS as a
chemoattractant. After 24 h, the top layer of the insert was
scraped with a sterile cotton swab to remove any remaining cells.
The invading cells on the bottom surface were stained with 0.1%
crystal violet at room temperature for 2 h, examined, counted, and
imaged using a light microscope. The number of cells in five random
fields of each chamber was counted, and an average number of cells
were calculated.
Scratch assay
EC109 and AEP-knockdown EC109 cells were then plated
into 6-well plates in 200 ml of serum-free RPMI 1640 medium at a
density of 10,000 per well. The assays were conducted in
triplicate. The inserts were then placed into the bottom chamber of
a 24-well plate containing RPMI 1640 with 10% FBS as a
chemoattractant. After 24 h, the top layer of the insert was
scraped with a sterile cotton swab to remove any remaining cells.
The invading cells on the bottom surface were stained 0.1% crystal
violet at room temperature for 2 h, examined, counted, and images
were captured using a light microscope. The number of cells in five
random fields of each chamber was counted, and an average number of
cells were calculated.
Statistical analysis
Survival was calculated starting from the date of
surgery to date of death or last follow-up. Survival curves for AEP
were plotted using the Kaplan-Meier and compared using the log-rank
test. Cox proportional hazard models were used for univariate and
multivariate analysis to test clinical features for their
associations with overall survival. In the multivariate Cox model,
variables with P<0.1 from the univariate model were included. In
addition to AEP expression, the following variables were
considered: Age, sex, grading and tumor location. Median times and
hazard ratios are presented with 95% confidence intervals (CIs).
All statistical analyses were performed using SPSS for Windows
v.17.0 (SPSS, Chicago, IL, USA). Two-tailed P<0.05 was
considered to indicate a statistically significant difference.
Results
High AEP expression in esophageal
carcinoma is associated with poor prognosis
Immunostaining was conducted to analyze the
expression and intracellular location of AEP in 146 patient samples
with esophageal carcinoma. Representative expression patterns in
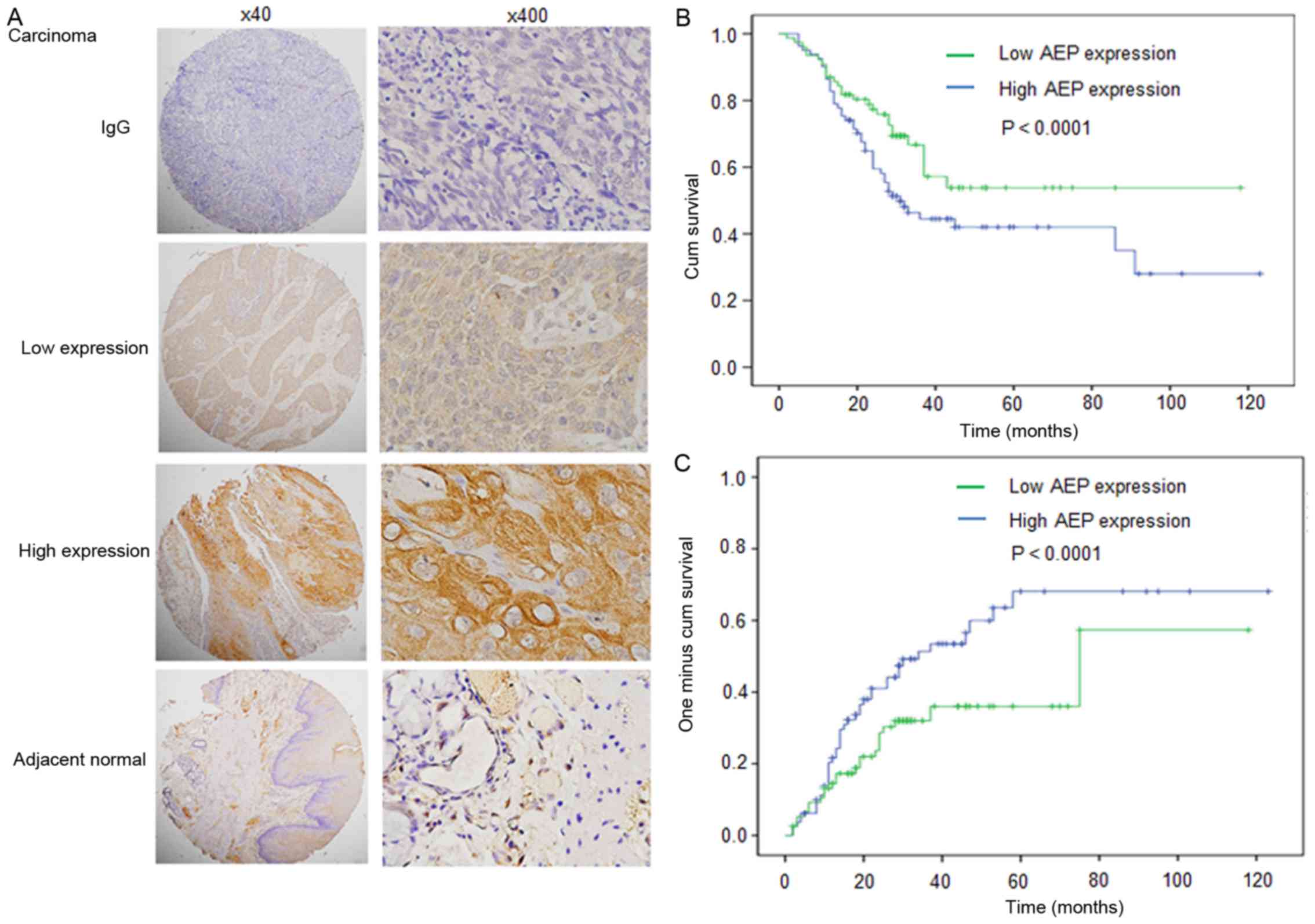
esophageal carcinoma samples are presented in Fig. 1A. Positive staining of AEP revealed
predominantly cytoplasmic localization in cancerous tissues and AEP
expression was higher in cancerous tissues compared with adjacent
normal tissues (Fig. 1A).
According to AEP expression in esophageal carcinoma
samples, all cases were distributed into two sub-groups: Low AEP
expression group (n=70) and high AEP expression group (n=76)
(Fig. 1A; Table I). Following the evaluation of
immunohistochemical staining, AEP levels in high-grade cases were
significantly increased compared with that in low-grade cases
(Table I; P=0.066).
 | Table I.Associations between tumor AEP
expression and clinicopathologic features. |
Table I.
Associations between tumor AEP
expression and clinicopathologic features.
|
| AEP |
|---|
|
|
|
|---|
| Characteristics | Low | High | P-value |
|---|
| Age, years |
|
|
|
| ≤60 | 38 | 46 | 0.146 |
|
>60 | 32 | 30 |
|
| Sex |
|
|
|
|
Female | 20 | 15 | 0.276 |
| Male | 50 | 61 |
|
| Drinking history |
|
|
|
| No | 25 | 23 | 0.137 |
| Yes | 45 | 53 |
|
| Smoking history |
|
|
|
| No | 25 | 23 | 0.300 |
| Yes | 45 | 53 |
|
| Family cancer
history |
|
|
|
| No | 49 | 58 | 0.208 |
| Yes | 21 | 18 |
|
| T Stage |
|
|
|
| I and
II | 29 | 24 | 0.144 |
| III and
IV | 41 | 52 |
|
| N Stage |
|
|
|
| N0 | 44 | 43 | 0.273 |
| N1 and
N2 | 26 | 33 |
|
| Tumor
differentiation |
|
|
|
|
I–II | 49 | 60 | 0.147 |
|
III | 21 | 16 |
|
| TNM stage |
|
|
|
| I | 49 | 43 | 0.066 |
|
II–III | 21 | 33 |
|
To evaluate the association of AEP expression with
patient prognosis, a log-rank test and Kaplan-Meier analysis were
introduced to assess the effect of AEP expression on patient
survival and relapse. The log-rank test (univariate analysis)
revealed that patients with low level of AEP expression in tumor
tissues demonstrated significantly longer overall survival compared
with patients with high AEP expression (n=146; Fig. 1B-C; Table
II; P=0.019). Factors including the drinking history, N stage
and TNM stage also affected OS. However, AEP expression did not
affect time to relapse.
 | Table II.Univariate and multivariate analyses
of factors associated with overall survival and time to
relapse. |
Table II.
Univariate and multivariate analyses
of factors associated with overall survival and time to
relapse.
|
| OS | TTR |
|---|
|
|
|
|
|---|
|
|
| Multivariate |
| Multivariate |
|---|
|
|
|
|
|
|
|---|
| Variables | Univariate
P-value | HR | 95%CI | P-value | Univariate
P-value | HR | 95%CI | P-value |
|---|
| Age, years (>60
vs. ≤60) | 0.204 |
|
| NA | 0.199 |
|
| NA |
| Sex (male vs.
female) | 0.197 |
|
| NA | 0.050 |
|
| NA |
| Drinking
history | 0.033 | 1.591 | 0.945–2.680 | 0.080 | 0.263 |
|
| NA |
| (yes vs. no) |
|
|
|
|
|
|
|
|
| Smoking
history | 0.545 |
|
| NA | 0.135 |
|
| NA |
| (yes vs. no) |
|
|
|
|
|
|
|
|
| Family cancer
history | 0.587 |
|
| NA | 0.444 |
|
| NA |
| (yes vs. no) |
|
|
|
|
|
|
|
|
| T stage | 0.059 |
|
| NA | 0.238 |
|
| NA |
| (III and IV vs. I
and II) |
|
|
|
|
|
|
|
|
| N stage (N1 and N2
vs. N0) | 0.000 | 2.000 | 0.828–4.830 | 0.123 | 0.003 | 1.997 | 0.554–7.202 | 0.290 |
| Differentiation
(III vs. I–II) | 0.661 |
|
| NA | 0.831 |
|
| NA |
| TNM stage (III–II
vs. I) | 0.000 | 1.683 | 0.694–4.080 | 0.249 | 0.002 | 2.081 | 0.584–7.409 | 0.258 |
| AEP tumor (high vs.
low) | 0.019 | 1.669 | 0.982–2.838 | 0.058 | 0.115 |
|
| NA |
Further, multivariate COX regression analysis was
also performed to explore whether AEP was an independent prognostic
factor for patient survival. As shown in Table II, AEP expression was not an
independent prognosis factor (HR, 1.669; 95% CI, 0.982–2.838;
P=0.058).
AEP expression in esophageal cancer
cell lines
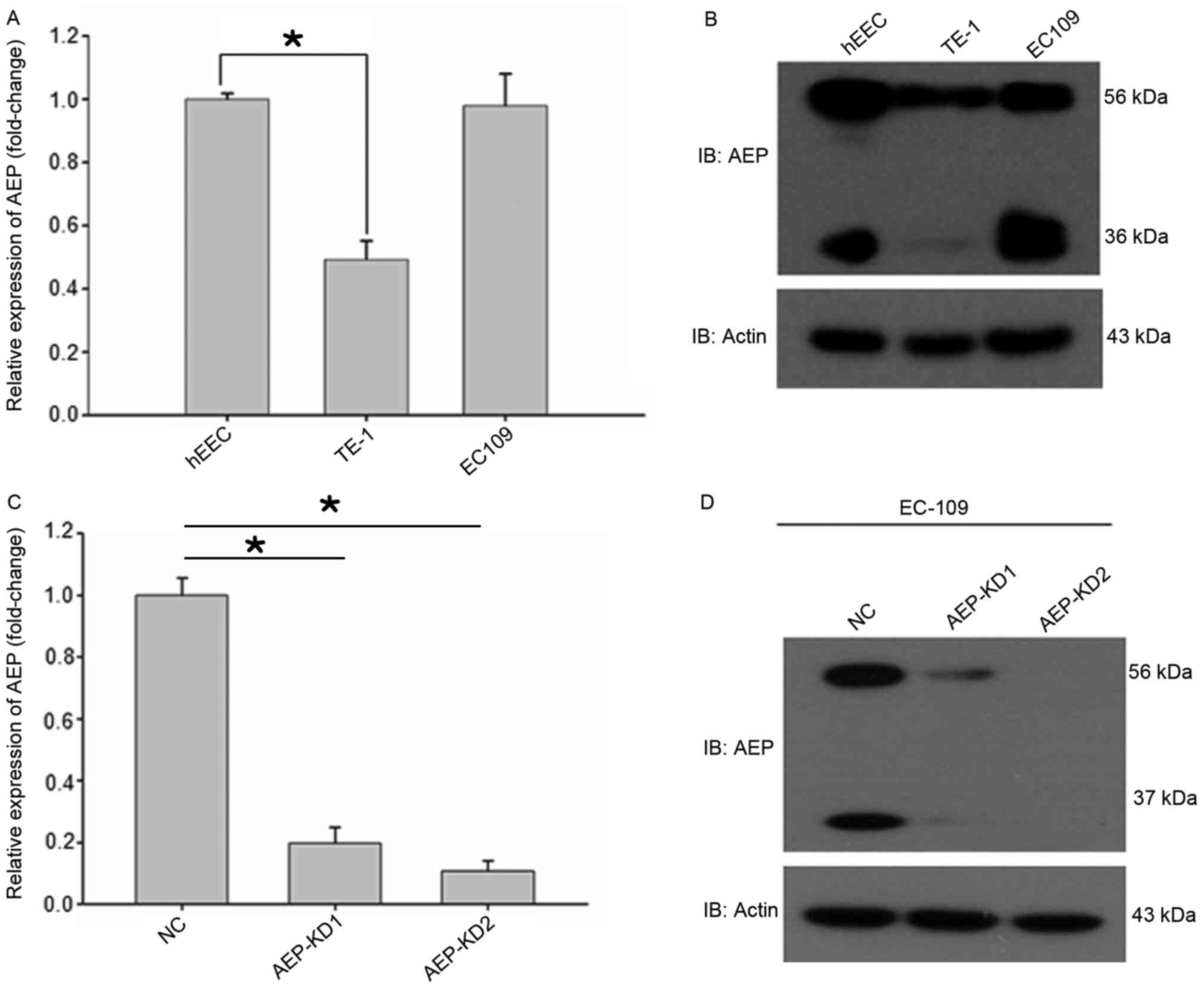
AEP is reported to be overexpressed in multiple
types of human solid tumors and acute lymphoblastic leukemia
compared with normal tissues (16–21). In
the present study, it was demonstrated that AEP mRNA and protein
levels were increased in EC109 and TE-1 esophageal cancer cell
lines. The messenger RNA level of AEP was analyzed by reverse
transcription-quantitative polymerase chain reaction using the
PrimeScript RT reagent kit and TaKaRa Premix Ex Taq kit. The
protein expression level of AEP was analyzed by western blot
analysis. AEP has two molecular mass isoforms, the inactive zymogen
(pro-AEP) of ~56 kDa and the mature enzyme (active AEP) of ~36 kDa
(Fig. 2) (5). qPCR and western blot analysis
demonstrated that AEP expression levels were highest in the hEEC
cell line, followed by the EC109, and TE-1 cells (Fig. 2A and B).
Effects of AEP on the migration and
invasion of esophageal cancer cells
To investigate the effects of AEP on esophageal
cancer metastasis, the migration and invasion ability of esophageal
cancer cells were analyzed. Control and AEP-silenced EC109 cells
were subjected to a migration assay. A target-shRNA to knockdown
endogenous AEP in esophageal cancer cell line EC109 was employed.
Two different shRNAs were designed to exclude off-target effects.
Efficient AEP knockdown was demonstrated by significantly decreased
AEP protein levels in EC109 cells with stably transfected
recombinant shRNA (Fig. 2C and D),
thus shRNAs were considered appropriate for AEP knockdown.
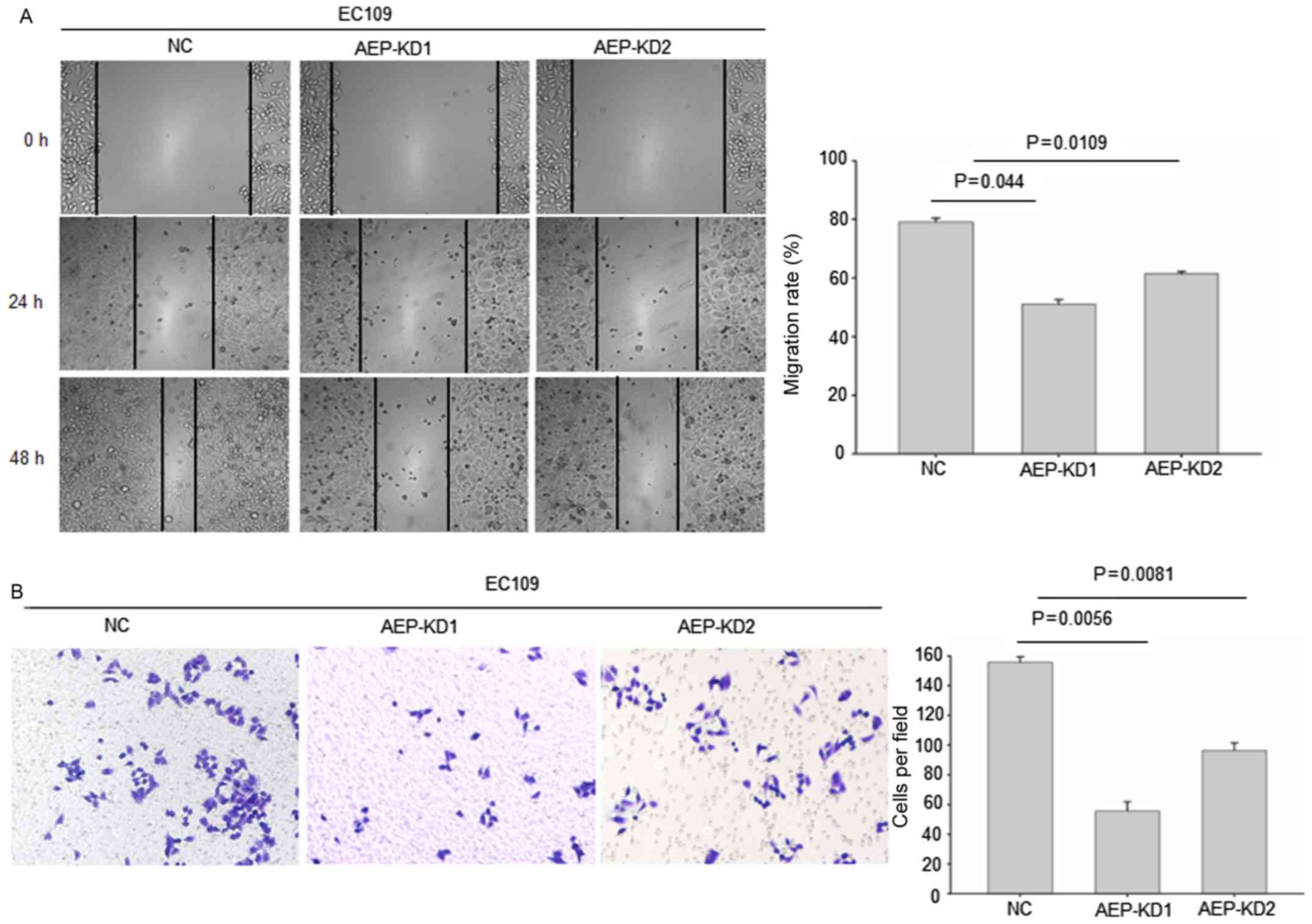
The wound healing assay data revealed that the
stable transfection of shRNA1 and shRNA2 into esophageal cancer
cells resulted in a significant inhibition of cell migration
capacity (Fig. 3A), compared with NC
shRNA. In addition, silencing of AEP significantly decreased the
invasion capacity into Matrigel as demonstrated by the Transwell
assay (Fig. 3B). The tumor cell
migration and invasion assay indicated that AEP depletion reduced
the invasion, and migration capability of EC109 cell line.
AEP knockdown inhibits EC cell
migration and metastasis through targeting MMPs
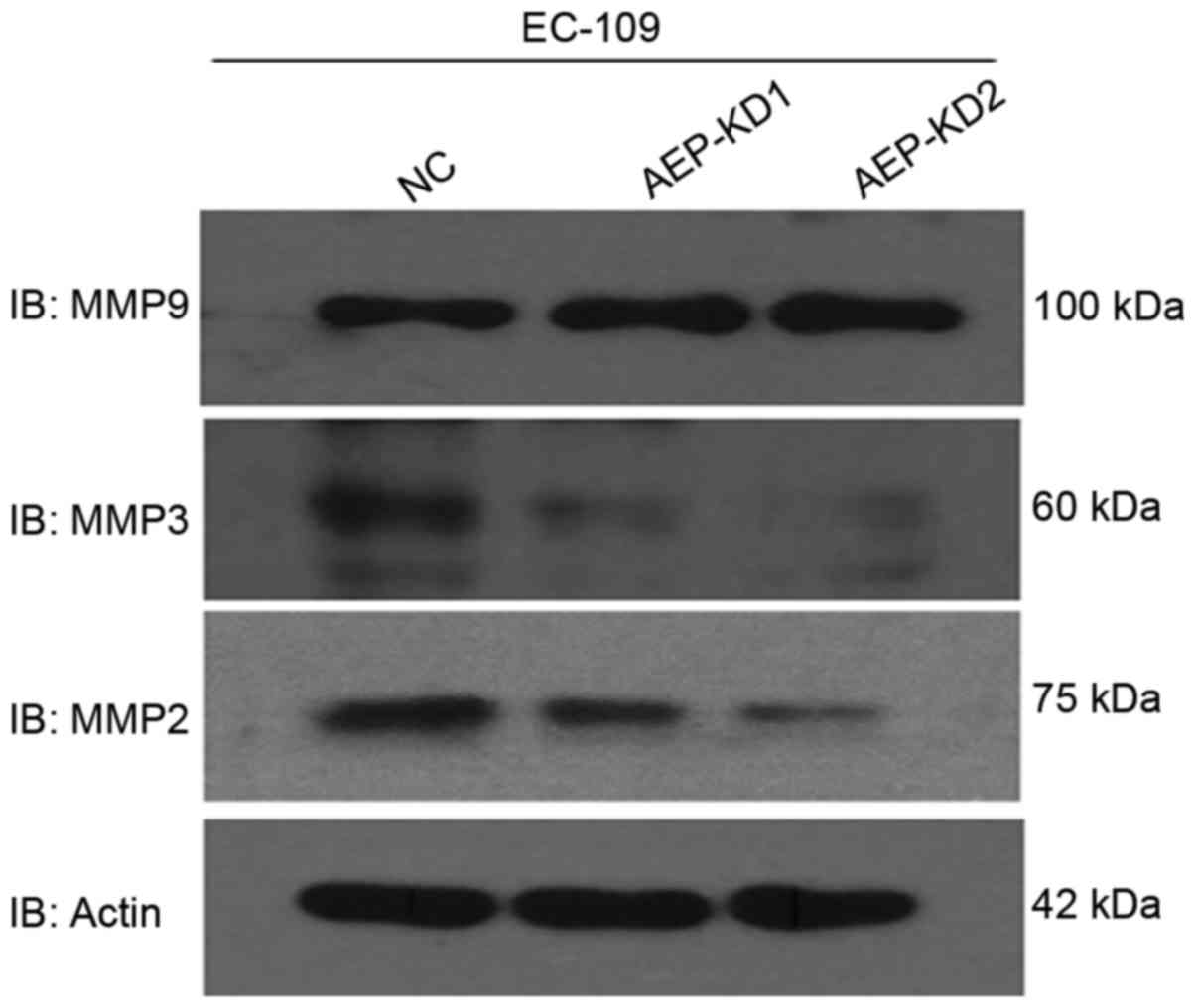
To determine how AEP influenced the invasive ability
of esophageal cancer cells, the expression of several
invasion-associated proteins following AEP knockdown, compared with
control cells was investigated by western blot analysis. Notably,
western blot analysis revealed that depletion of AEP markedly
reduced MMP2 and MMP3, but not MMP9 expression in AEP-KD-EC109
cells (Fig. 4), compared with control
cells. MMPs are known to facilitate cell invasion and metastasis by
enzymatically degrading extracellular matrix components (23). Taken together, it was confirmed that
AEP promotes metastasis through regulation of MMPs in esophageal
tumor cells.
Discussion
Despite recent advances in esophageal cancer
treatment, there has been no significant improvement in the overall
survival rate for patients with advanced esophageal cancer. Novel
strategies are necessary for early detection and to improve
treatment options in esophageal cancer.
Previous reports have indicated that AEP expression
positively correlates with clinicopathologic and biological
variables in colorectal cancer, and may be a novel oncogene
(19–22). Concordantly, it was demonstrated in
the present study that AEP was significantly overexpressed in
esophageal cancer and was associated with poor prognosis. These
observations suggest that AEP may be a potential novel diagnostic
biomarker and that AEP inhibitors or monoclonal antibodies may be
proposed as esophageal cancer therapies.
Previous studies have revealed that AEP is localized
in the front of invading cells, and forms a complex with integrins
on the surface of lamellipodia and invadopodia (17). The binding of AEP to integrins
significantly promotes its ability to activate pro-MMP2 and
cathepsin L (25). These downstream
substrates of AEP have well-established functions in metastasis,
which may partially explain the mechanism of AEP metastasis
regulation (26). Nevertheless, it is
crucial to identify unknown substrates of AEP to further clarify
its function in tumor development. The data from the present study
demonstrated that secreted AEP is critical for esophageal cancer
progression through regulation of MMPs. Degradation of the
extracellular matrix by cancer cells are important processes for
direct invasion. There are three types of enzymes that effectively
degrade extracellular matrix (ECM): MMPs, serine proteinases, and
cysteine proteinases. MMPs are known to serve important functions
in ECM remodeling during the process of tumor invasion and
metastasis. The expression of MMPs was reported to be associated
with tumor invasion and lymph node metastasis in EC (27). However, the role of AEP in esophageal
cancer progression should be further examined by animal in
vivo models.
In summary, data from the present study provides
evidence for AEP as a novel biomarker in esophagyeal cancer. In
addition, AEP may be of prognostic value and a therapeutic target
for the treatment of this disease. Targeting AEP with
Aza-Asn-epoxides and its derivatives, which are specific to AEP may
have potential therapeutic value.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–-E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vermeulen E, Zamora-Ros R, Duell EJ,
Luján-Barroso L, Boeing H, Aleksandrova K, Bueno-de-Mesquita HB,
Scalbert A, Romieu I, Fedirko V, et al: Dietary flavonoid intake
and esophageal cancer risk in the European prospective
investigation into cancer and nutrition cohort. Am J Epidemiol.
178:570–581. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pennathur A, Farkas A, Krasinskas AM,
Ferson PF, Gooding WE, Gibson MK, Schuchert MJ, Landreneau RJ and
Luketich JD: Esophagectomy for T1 esophageal cancer: Outcomes in
100 patients and implications for endoscopic therapy. Ann Thorac
Surg. 87:1048–1055. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hayashi Y, Correa AM, Hofstetter WL,
Vaporciyan AA, Mehran RJ, Rice DC, Suzuki A, Lee JH, Bhutani MS,
Welsh J, et al: Patients with high body mass index tend to have
lower stage of esophageal carcinoma at diagnosis. Dis Esophagus.
25:614–622. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zheng J, Xu C, Chu D, Zhang X, Li J, Ji G,
Hong L, Feng Q, Li X, Wu G, et al: Human leukocyte antigen G is
associated with esophageal squamous cell carcinoma progression and
poor prognosis. Immunol Lett. 161:13–19. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Harris KS, Durek T, Kaas Q, Poth AG,
Gilding EK, Conlan BF, Saska I, Daly NL, van der Weerden NL, Craik
DJ and Anderson MA: Efficient backbone cyclization of linear
peptides by a recombinant asparaginyl endopeptidase. Nat Commun.
6:101992015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao L, Hua T, Crowley C, Ru H, Ni X, Shaw
N, Jiao L, Ding W, Qu L, Hung LW, et al: Structural analysis of
asparaginyl endopeptidase reveals the activation mechanism and a
reversible intermediate maturation stage. Cell Res. 24:344–358.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Miller G, Matthews SP, Reinheckel T,
Fleming S and Watts C: Asparagine endopeptidase is required for
normal kidney physiology and homeostasis. FASEB J. 25:1606–1617.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Descamps D, Le Gars M, Balloy V, Barbier
D, Maschalidi S, Tohme M, Chignard M, Ramphal R, Manoury B and
Sallenave JM: Toll-like receptor 5 (TLR5), IL-1β secretion, and
asparagine endopeptidase are critical factors for alveolar
macrophage phagocytosis and bacterial killing. Proc Natl Acad Sci
USA. 109:pp. 1619–1624. 2012; View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sepulveda FE, Maschalidi S, Colisson R,
Heslop L, Ghirelli C, Sakka E, Lennon-Duménil AM, Amigorena S,
Cabanie L and Manoury B: Critical role for asparagine endopeptidase
in endocytic Toll-like receptor signaling in dendritic cells.
Immunity. 31:737–748. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Morita Y, Araki H, Sugimoto T, Takeuchi K,
Yamane T, Maeda T, Yamamoto Y, Nishi K, Asano M, Shirahama-Noda K,
et al: Legumain/asparaginyl endopeptidase controls extracellular
matrix remodeling through the degradation of fibronectin in mouse
renal proximal tubular cells. FEBS Lett. 581:1417–1424. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Choi SJ, Reddy SV, Devlin RD, Menaa C,
Chung H, Boyce BF and Roodman GD: Identification of human
asparaginyl endopeptidase (legumain) as an inhibitor of osteoclast
formation and bone resorption. J Biol Chem. 274:27747–27753. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xing W, Baylink D, Kesavan C, Hu Y, Kapoor
S, Chadwick RB and Mohan S: Global gene expression analysis in the
bones reveals involvement of several novel genes and pathways in
mediating an anabolic response of mechanical loading in mice. J
Cell Biochem. 96:1049–1060. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dickinson D: Cysteine peptidases of
mammals: Their biological roles and potential effects in the oral
cavity and other tissues in health and disease. Crit Rev Oral Biol
Med. 13:238–275. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gawenda J, Traub F, Lück HJ, Kreipe H and
von Wasielewski R: Legumain expression as a prognostic factor in
breast cancer patients. Breast Cancer Res Treat. 102:1–6. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Brix K, McInnes J, Al-Hashimi A, Rehders
M, Tamhane T and Haugen MH: Proteolysis mediated by cysteine
cathepsins and legumain-recent advances and cell biological
challenges. Protoplasma. 252:755–774. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Holland M, Castro FV, Alexander S, Smith
D, Liu J, Walker M, Bitton D, Mulryan K, Ashton G, Blaylock M, et
al: RAC2, AEP, and ICAM1 expression are associated with CNS disease
in a mouse model of pre-B childhood acute lymphoblastic leukemia.
Blood. 118:638–649. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen JM, Dando PM, Rawlings ND, Brown MA,
Young NE, Stevens RA, Hewitt E, Watts C and Barrett AJ: Cloning,
isolation, and characterization of mammalian legumain, an
asparaginyl endopeptidase. J Biol Chem. 272:8090–8098. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu C, Sun C, Huang H, Janda K and
Edgington T: Overexpression of legumain in tumors is significant
for invasion/metastasis and a candidate enzymatic target for
prodrug therapy. Cancer Res. 63:2957–2964. 2003.PubMed/NCBI
|
|
21
|
Murthy RV, Arbman G, Gao J, Roodman GD and
Sun XF: Legumain expression in relation to clinicopathologic and
biological variables in colorectal cancer. Clin Cancer Res.
11:2293–2299. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ohno Y, Nakashima J, Izumi M, Ohori M,
Hashimoto T and Tachibana M: Association of legumain expression
pattern with prostate cancer invasiveness and aggressiveness. World
J Urol. 31:359–364. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu JK, Forman S, Moorthy CR and Benzil
DL: Update on treatment modalities for optic nerve sheath
meningiomas. Neurosurg Focus. 14:e72003. View Article : Google Scholar
|
|
24
|
Napier KJ, Scheerer M and Misra S:
Esophageal cancer: A Review of epidemiology, pathogenesis, staging
workup and treatment modalities. World J Gastrointest Oncol.
6:112–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu X, Wang Z, Zhang G, Zhu Q, Zeng H,
Wang T, Gao F, Qi Z, Zhang J and Wang R: High TRAF6 expression is
associated with esophageal carcinoma recurrence and prompts cancer
cell invasion. Oncol Res. 25:485–493. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lin Y, Qiu Y, Xu C, Liu Q, Peng B,
Kaufmann GF, Chen X, Lan B, Wei C, Lu D, et al: Functional role of
asparaginyl endopeptidase ubiquitination by TRAF6 in tumor invasion
and metastasis. J Natl Cancer Inst. 106:dju0122014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Murray GI, Duncan ME, O'Neil P, McKay JA,
Melvin WT and Fothergill JE: Matrix metalloproteinase-1 is
associated with poor prognosis in oesophageal cancer. J Pathol.
185:256–261. 1998. View Article : Google Scholar : PubMed/NCBI
|