Introduction
Colorectal cancer is a common malignant tumor of the
digestive tract, with high morbidity and mortality rates. Its
incidence ranks fourth in malignant tumors in China, and the
mortality rate ranks fifth in tumors leading to death (1). In addition, the incidence and mortality
rates of colorectal cancer in China are on the increase, which has
deleterious effects on the lives and health of patietns (2). Previous findings have shown that the
5-year survival rate of colorectal cancer patients with metastasis
was significantly lower than that of colorectal cancer patients
without metastasis (3,4). In the diagnosis of patients with
colorectal cancer, metastasis occurs in approximately 25% of
patients, which is the main reason leading to the poor curative
effect in the clinical treatment for patients (5).
Metastasis suppressor and promoter genes play key
roles in the process of tumor metastasis. An increasing number of
scholars have been focused on the study of tumor metastasis
suppressor genes. A previous study demonstrated that the effect of
metastasis suppressor genes on tumor metastasis is more critical
than that of metastasis promoter genes, and the reduction in the
expression of metastasis suppressor genes or gene loss may induce
the invasion and metastasis of tumor cells (6). The KiSS-1 gene was initially
identified in non-metastatic melanoma cells, and is an important
tumor metastasis suppressor gene (7).
KiSS-1 genes are located at the long arm of human chromosome
1, and the gene encoding proteins can bind to the G protein-coupled
receptor 54 (GPR54), resulting in physiological effects of
inhibition of the proliferation, invasion and metastasis of tumor
cells and induction of the differentiation and apoptosis of tumor
cells (8). Further findings have
shown that the expression level of KiSS-1 genes is
downregulated in a variety of tumors, including pancreatic
(9), gastric (10), breast (11) and bladder cancer (12), and promotes tumor invasion and
metastasis.
The findings of Siegel et al indicated that
the expression level of KiSS-1 proteins in the tumor tissues of
patients with colorectal cancer was significantly lower than that
in the normal colorectal tissues. Additionally, KiSS-1 acts as a
tumor suppressor gene in the occurrence and development of
colorectal cancer (13).
The present study aimed to investigate the
expression of KiSS-1 in the tumor tissues of patients with
colorectal cancer and its effects on pathological parameters and
the prognosis of patients with colorectal cancer. Reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
immunohistochemistry were used to analyze the expression levels of
KiSS-1 in the tumor tissues and cancer-adjacent normal tissues of
patients with colorectal cancer. The effects of KiSS-1 expression
on pathological parameters and the prognosis of patients with
colorectal cancer were analyzed combined with clinical data.
Materials and methods
Data
For the clinical data, 56 patients with colorectal
cancer treated in the Department of Surgery of our hospital from
May 2009 to December 2011 were selected. The patients included 32
males and 24 females aged (55.7±9.3) years who received surgical
treatment for the first time, and had no history of chemotherapy
and radiotherapy. Tumor tissues and cancer-adjacent normal tissues
>10 cm away from the tumor were taken from the patients with
colorectal cancer during surgery. A section of the tissues was
immediately stored in liquid nitrogen, while the other section was
fixed in 10% neutral formalin solution. The patients were diagnosed
with colorectal cancer via pathological analysis. The study was
approved by the Medical ethics committee of the Dezhou People's
Hospital (Shandong, China). All the patients or their family
members signed the informed consent.
The main reagents used were TRIzol, reverse
transcription and RT-qPCR kits (Invitrogen, Carlsbad, CA, USA);
primer synthesis [Takara Biomedical Technology (Dalian) Co., Ltd.,
Dalian, China]; rabbit anti-human KiSS-1 polyclonal antibody
(18375–1-AP), general immuno histochemical test kit for
anti-rat/rabbit (KIHC-1) (Wuhan Sanying Biotechnology Co. Ltd.,
Wuhan, China), glyceraldehyde-3-phosphate dehydrogenase (GADPH)
antibodies, horseradish peroxidase (HRP)-labeled antibodies (Wuhan
Proteintech Group, Inc., Wuhan, China); and immunohistochemical
staining kit SP-9001 (Beijing Zhongshan Golden Bridge Biological
Technology Co., Ltd., Beijing, China).
Detection of the expression of KiSS-1
messenger ribonucleic acid (mRNA) in tissue specimens of patients
by RT-qPCR
Total RNA in the frozen tissues of patients was
extracted using TRIzol kits, and RNA concentration and purity were
measured using an ultraviolet spectrophotometer (Hitachi, Tokyo,
Japan). When the ratio of A260/A280 was between 1.8 and 2.0, the
samples were considered to be qualified. Reverse transcription was
conducted according to the protocol. PCR was carried out according
to the recommended method of RT-qPCR kits with the obtained
complementary deoxyribonucleic acid (cDNA) as the template. Primer
sequences are shown in Table I.
Specific reaction conditions were: pre-denaturation at 94°C for 5
min, denaturation at 95°C for 30 sec, annealing at 60°C for 30 sec,
and extension at 72°C for 30 sec and the amplification was
conducted for 30 cycles. With GAPDH as the control, the expression
level of KiSS-1 mRNA was calculated using the relative
quantification method. The formula used was: ∆Ct (target gene) = Ct
(target gene) - Ct (control gene).
 | Table I.Primer sequences of RT-qPCR. |
Table I.
Primer sequences of RT-qPCR.
| Gene name | Primer sequence |
|---|
| KiSS-1 | F:
5′-ACCTGGCTCTTCTCACCAAG-3′ |
|
| R:
5′-TAGCAGCTGGCTTCCTCTC-3′ |
| GAPDH | F:
5′-GCACCGTCAAGGCTGAGAAC-3′ |
|
| R:
5′-TGGTGAAGACGCCAGTGGA-3′ |
Detection of the expression of KiSS-1
proteins by immunohistochemistry
Tissues fixed in 10% neutral formalin solution were
taken and embedded in paraffin, and then continuously cut into 4-µm
slices. Immunohistochemical staining was performed by
streptavidin-perosidase (SP) two-step method, and the procedures
were carried out according to the protocol of the kits. Routine
dewaxing was performed, citric acid solution was used for repairing
protein, and goat serum was applied for sealing. KiSS primary
antibodies (diluted at 1:200) were added drop by drop, and the
tissues were incubated in a refrigerator at 4°C overnight. After
washing by pre-cooling phosphate-buffered saline (PBS) solution
three times, secondary antibodies (diluted at 1:1,000) were added
drop by drop. The tissues were incubated at room temperature for 30
min and washed by pre-cooling PBS three times. Diaminobenzidine
(DAB) development was then conducted, hematoxylin was used for
re-dyeing, and finally gum was used for mounting.
Five ×400 visual fields were randomly selected from
each slice. Results of KiSS-1 protein-positive staining were that
pale brown and chocolate-brown particles were precipitated in the
cytoplasm. According to the percentage of stained cells and the
staining intensity, scores were evaluated comprehensively: 0 points
for positive cells <5%, 1 point for 5–25%, 2 points for 26–50%
and 3 points for >51%; 0 points for basic colorlessness, 1 point
for light yellow, 2 points for pale brown and 3 points for
chocolate-brown. These scores were added, and ≤3 points represented
negative expression, while >3 points represented positive
expression.
Relationship of the expression of
KiSS-1 in the tumor tissues of patients with colorectal cancer with
pathological parameters and the prognosis of patients
According to the expression level of KiSS-1 in
colorectal cancer tissues, 56 patients with colorectal cancer were
divided into the KiSS-1 positive and KiSS-1 negative expression
groups. Clinicopathological parameters of patients were examined
and the association between the expression of KiSS-1 and
pathological parameters of patients was analyzed. The 56 patients
were followed up after surgery for 5 years. The survival time was
calculated from the first day after surgery, and the follow-up was
carried out once a month.
Statistical treatment
Data were processed using Statistical Product and
Service Solutions (SPSS) 17.0 software (IBM Corp., Armonk, NY,
USA). Measurement data were expressed as mean ± standard deviation,
and t-test was used for comparisons between two groups. The
χ2 test was used for intergroup comparisons of countable
data. The univariate survival analysis was performed using
Kaplan-Meier method. P≤0.05 was considered to indicate a
statistically significant difference.
Results
Detection of the expression of KiSS-1
in tumor tissues of patients with colorectal cancer by RT-qPCR
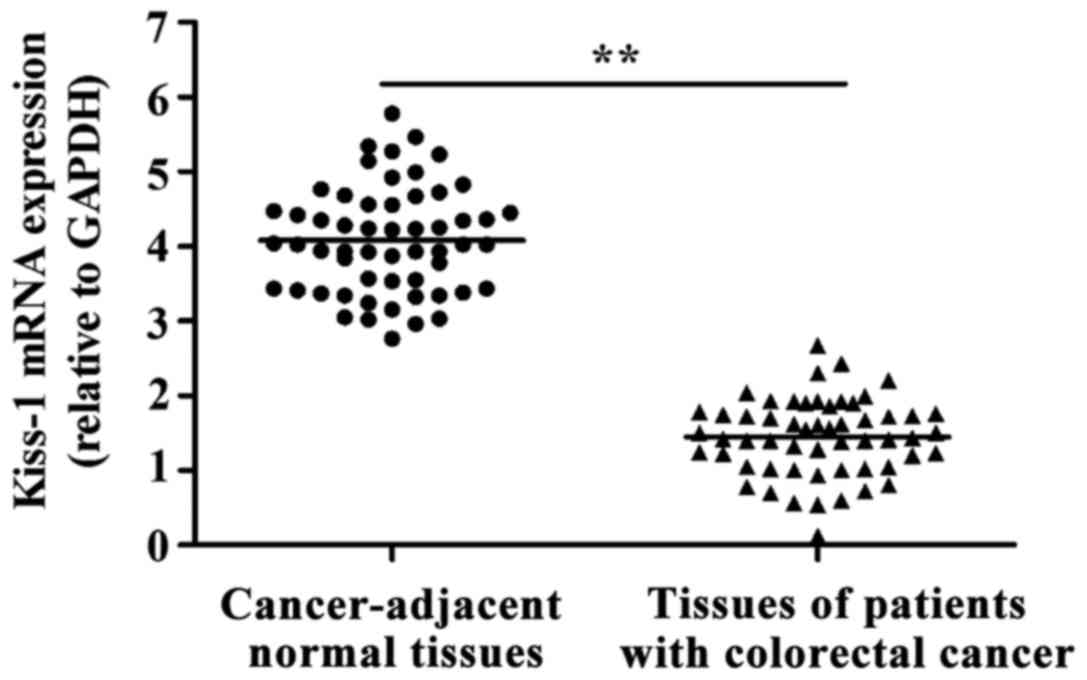
As shown in Fig. 1,
the expression of KiSS-1 mRNA in colorectal cancer tissues was
significantly decreased compared with that in cancer-adjacent
normal tissues (P<0.01).
Detection of the expression of KiSS-1
in tumor tissue specimens of patients with colorectal cancer by
immunohistochemistry
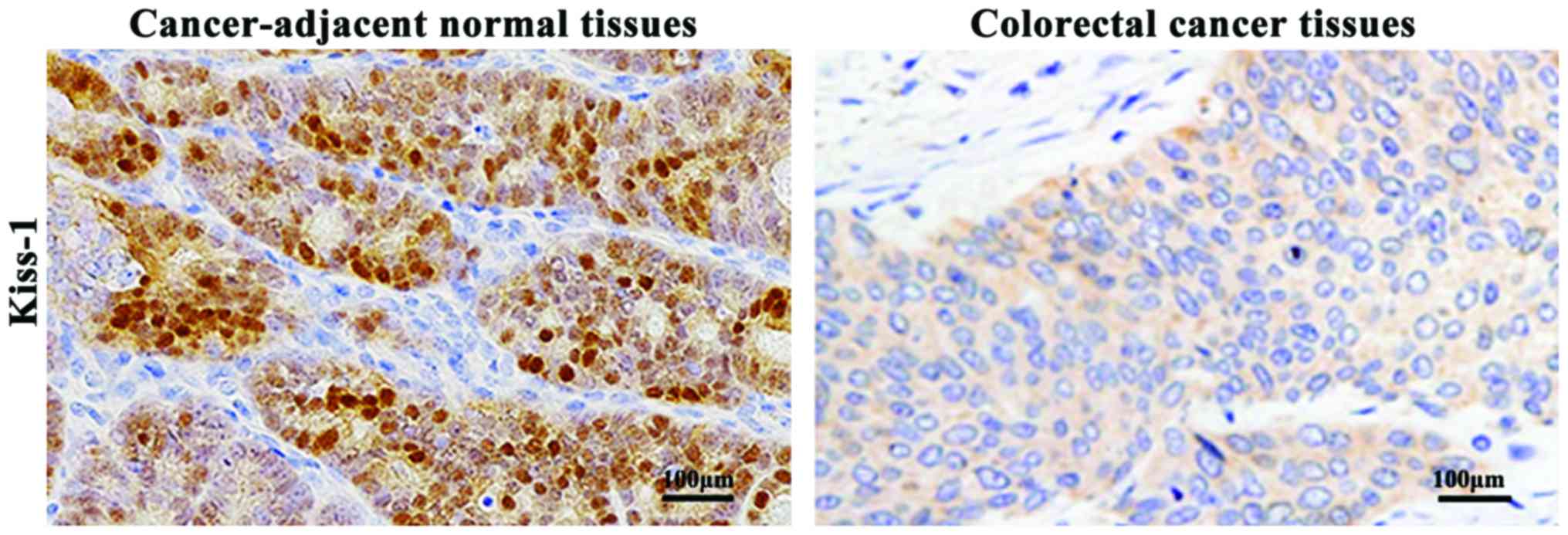
As shown in Fig. 2,
immunohistochemistry results indicated that the positiveness of
KiSS-1 proteins was manifested as light brown or dark brown
precipitates in the cytoplasm. The expression levels of KiSS-1
proteins in colorectal cancer tissues were significantly decreased
compared with that in the cancer-adjacent normal tissues
(P<0.01).
Results of the statistical analysis according to the
standard of evaluation indicated that the positive expression rate
of KiSS-1 proteins in colorectal cancer tissues was 26.79% (15/56),
and the negative expression rate was 73.21% (41/56). The positive
expression rate of KiSS-1 proteins in the cancer-adjacent normal
tissues was 80.36% (45/56), and the negative expression rate was
19.64% (11/56).
Association between the expression of
KiSS-1 in tumor tissues of patients with colorectal cancer and
clinicopathological parameters
According to the results of immunohistochemistry,
patients with colorectal cancer were divided into the KiSS-1
positive and KiSS-1 negative expression groups, and the association
between the expression of KiSS-1 in the tumor tissues of patients
with colorectal cancer and clinicopathological parameters was
examined. The results showed that the negative expression of KiSS-1
in tumor tissues of patients was correlated with tumor
differentiation degree, invasion and metastasis as well as clinical
staging (Table II).
 | Table II.Association of the abnormal expression
of KiSS-1 and clinicopathological parameters of patients with
colorectal cancer. |
Table II.
Association of the abnormal expression
of KiSS-1 and clinicopathological parameters of patients with
colorectal cancer.
|
|
| KiSS-1 |
|---|
|
|
|
|
|---|
| Clinicopathological
parameters | Case | Negative expression
(case, %) | χ2 | P-value |
|---|
| Sex |
|
| 0.12 | >0.05 |
| Male | 32 | 24 (75.00) |
|
|
|
Female | 24 | 17 (70.83) |
|
|
| Age (years) |
|
| 0.05 | >0.05 |
| ≥50 | 35 | 26 (74.290 |
|
|
|
<50 | 21 | 15 (71.43) |
|
|
| Tumor size (cm) |
|
| 0.51 | >0.05 |
| ≥5 | 33 | 23 (69.70) |
|
|
|
<5 | 23 | 18 (78.26) |
|
|
| Differentiation
degree |
|
| 14.24 | <0.01 |
| Low | 22 | 10 (45.45) |
|
|
|
Moderate/high | 34 | 31 (91.18) |
|
|
| Invasion and
metastasis |
|
| 12.63 | <0.05 |
| Yes | 36 | 32 (88.89) |
|
|
| No | 20 | 9
(45.00) |
|
|
| Clinical staging |
|
| 10.36 | <0.01 |
| I–II | 25 | 13 (52.00) |
|
|
|
III–IV | 31 | 28 (90.32) |
|
|
Analysis of the survival condition and
prognosis of patients
The 5-year follow up was statistically analyzed, and
showed that of the 56 patients with colorectal cancer, 31 survived
and 25 succumbed to the disease. The 5-year overall survival rate
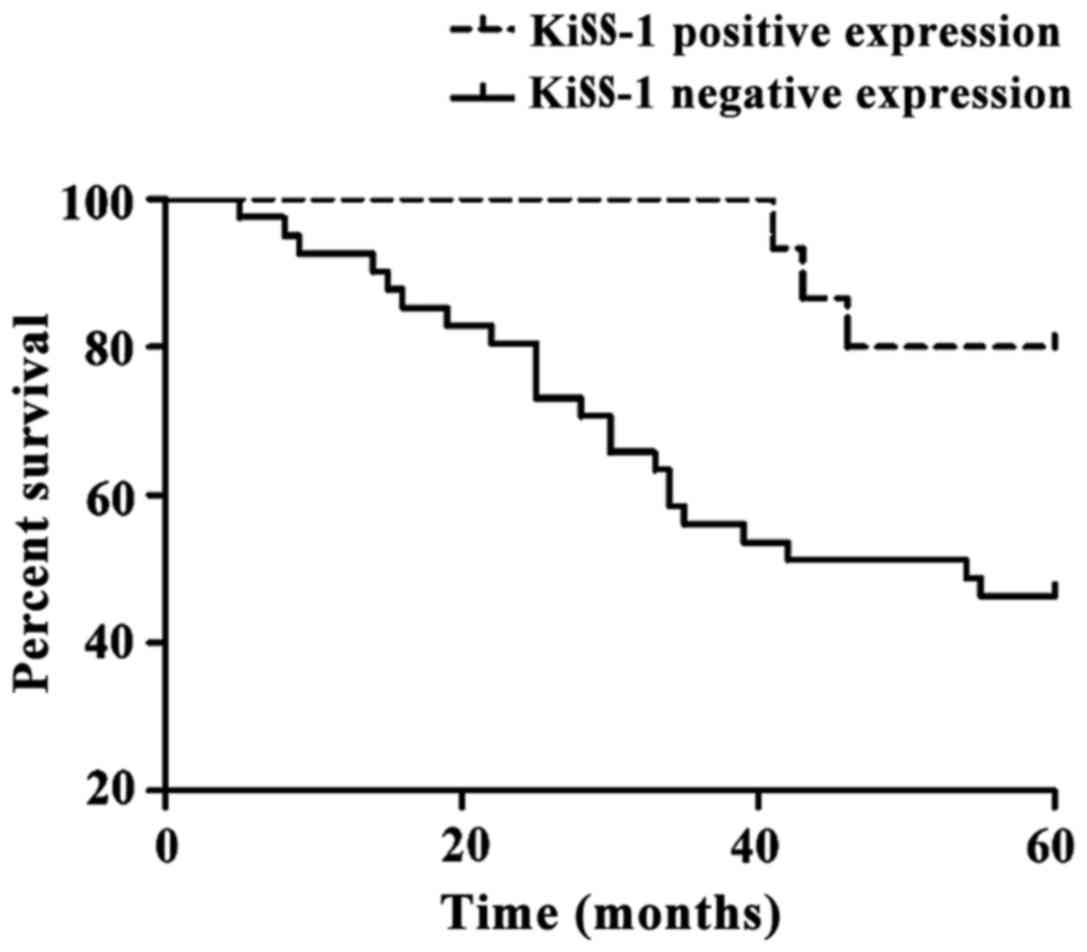
was 55.36% (31/56). Results of the Kaplan-Meier analysis of
survival curves for colorectal cancer patients are shown in
Fig. 3. Patients with negatively
expressed KiSS-1 had worse prognosis. Results of the univariate
survival analysis are shown in Table
III. KiSS-1 significantly affected the overall survival rate of
colorectal cancer, which was an independent factor affecting the
survival of patients with colorectal cancer (P<0.05).
 | Table III.Univariate survival analysis of the
association between the expression of KiSS-1 and the overall
survival rate of patients with colorectal cancer. |
Table III.
Univariate survival analysis of the
association between the expression of KiSS-1 and the overall
survival rate of patients with colorectal cancer.
| Group | Case | 5-year survival
case | 5-year survival rate
(%) | Wald (log-rank) | P-value |
|---|
| KiSS-1 |
|
|
| 5.36 | <0.05 |
| Positive
expression | 15 | 12 | 73.33 |
|
|
| Negative
expression | 41 | 19 | 48.78 |
|
|
Discussion
Colorectal cancer is a common malignant tumor of the
digestive tract with a high metastatic rate, whose incidence ranks
second in female tumors and third in male tumors worldwide
(1).
In recent years, the KiSS-1 gene has become a
hot spot in tumor research. Previous findings have shown that
KiSS-1 proteins can inhibit tumor cell proliferation and invasion
and induce tumor cell apoptosis (14). The coding product of KiSS-1
genes is polypeptide with the same length as that of 54 amino
acids, which can bind specifically to GPR54 or AXOR12. After
binding to GPR54, polypeptide induces phosphatidylinositol
4,5-bisphosphate (PIP2) hydrolysis to elevate the intracellular
calcium concentration, thus activating intracellular signaling
pathways to play roles in inhibiting cell proliferation and
differentiation (15,16). Stafford et al showed that
proteins encoded by KiSS-1 bind to GPR54 to activate phospholipase
C, producing the second messenger inositol trisphosphate (IP3) and
diglycerides in cells to inhibit cell proliferation and promote
cell apoptosis (8). Other findings
have shown that KiSS-1 can suppress the invasion and metastasis of
tumor cells, and its mechanism is mainly to downregulate the
expression of matrix metallopeptidase-9 (MMP-9) in order to carry
out its roles (17).
The present study aimed to study the expression of
KiSS-1 in tissues of colorectal cancer patients and its effects on
pathological parameters and the prognosis of patients with
colorectal cancer. In the present study, RT-qPCR was used to detect
the expression of KiSS-1 mRNA in tumor tissues and cancer-adjacent
normal tissues of patients with colorectal cancer. The results
showed that the transcription level of KiSS-1 mRNA in the tissues
of patients with colorectal cancer was significantly lower than
that in the cancer-adjacent normal tissues. The results of
immunohistochemistry also confirmed that the expression level of
KiSS-1 proteins in tissues of patients with colorectal cancer was
significantly decreased compared to that in cancer-adjacent normal
tissues. In order to determine the association between its
expression and the pathological parameters of patients, statistical
analysis was conducted, which indicated that the negative
expression of KiSS-1 in the tumor tissues of patients was
correlated with the degree of tumor differentiation, invasion and
metastasis as well as clinical staging. The univariate Kaplan-Meier
survival analysis was used to assess the effect of KiSS-1
expression on the 5-year overall survival rate of patients. The
results showed that KiSS-1 significantly affected the overall
survival of colorectal cancer, and patients with lowly expressed
KiSS-1 have lower 5-year survival rate and worse prognosis.
Dhar et al found the presence of lowly
expressed KiSS-1 genes in tissues of patients with gastric
cancer, and KiSS-1 is closely associated with the metastasis of
gastric cancer (18). Studies of
Masui et al suggested that the expression of KiSS-1
genes in pancreatic cancer tissues was significantly lower than
that in normal tissues, and the low expression of KiSS-1
genes may be associated with the metastasis and staging of
pancreatic cancer (19). Results of
the aforementioned studies are consistent with those of the current
study. In the present study, the expression of KiSS-1 was confirmed
to be downregulated in the tumor tissues of patients with
colorectal cancer. It was also shown that the expression of KiSS-1
was correlated with the degree of tumor differentiation, invasion
and metastasis, clinical staging and prognosis.
In summary, KiSS-1 is lowly expressed in tumor
tissues of colorectal cancer patients, which is closely associated
with the occurrence and development of colorectal cancer, the
degree of differentiation, invasion and metastasis, as well as
clinical staging. KiSS-1 also affects the 5-year survival rate of
patients. Patients with lowly expressed KiSS-1 have a lower 5-year
survival rate. Consequently, the expression of KiSS-1 of tumor
tissues of patients can be used as a reference factor for the
prognosis estimation for colorectal cancer patients.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Zhang S, Zhao P, Zeng H
and Zou X: Report of cancer incidence and mortality in China, 2010.
Ann Transl Med. 2:612014.PubMed/NCBI
|
|
3
|
Slipicevic A, Holm R, Emilsen E, Ree
Rosnes AK, Welch DR, Mælandsmo GM and Flørenes VA: Cytoplasmic
BRMS1 expression in malignant melanoma is associated with increased
disease-free survival. BMC Cancer. 12:73–79. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xie J, Xiang DB, Wang H, Zhao C, Chen J,
Xiong F, Li TY and Wang XL: Inhibition of Tcf-4 induces apoptosis
and enhances chemosensitivity of colon cancer cells. PLoS One.
7:e456172012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Elferink MA, de Jong KP, Klaase JM,
Siemerink EJ and de Wilt JH: Metachronous metastases from
colorectal cancer: A population-based study in North-East
Netherlands. Int J Colorectal Dis. 30:205–212. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Samant RS, Seraj MJ, Saunders MM, Sakamaki
TS, Shevde LA, Harms JF, Leonard TO, Goldberg SF, Budgeon L, Meehan
WJ, et al: Analysis of mechanisms underlying BRMS1 suppression of
metastasis. Clin Exp Metastasis. 18:683–693. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee JH, Miele ME, Hicks DJ, Phillips KK,
Trent JM, Weissman BE and Welch DR: KiSS-1, a novel human malignant
melanoma metastasis-suppressor gene. J Natl Cancer Inst.
88:1731–1737. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stafford LJ, Xia C, Ma W, Cai Y and Liu M:
Identification and characterization of mouse metastasis-suppressor
KiSS1 and its G-protein-coupled receptor. Cancer Res. 62:5399–5404.
2002.PubMed/NCBI
|
|
9
|
Masui T, Doi R, Mori T, Toyoda E, Koizumi
M, Kami K, Ito D, Peiper SC, Broach JR, Oishi S, et al: Metastin
and its variant forms suppress migration of pancreatic cancer
cells. Biochem Biophys Res Commun. 315:85–92. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guan-Zhen Y, Ying C, Can-Rong N, Guo-Dong
W, Jian-Xin Q and Jie-Jun W: Reduced protein expression of
metastasis-related genes (nm23, KISS1, KAI1 and p53) in lymph node
and liver metastases of gastric cancer. Int J Exp Pathol.
88:175–183. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cho SG, Li D, Stafford LJ, Luo J,
Rodriguez-Villanueva M, Wang Y and Liu M: KiSS1 suppresses
TNFalpha-induced breast cancer cell invasion via an inhibition of
RhoA-mediated NF-kappaB activation. J Cell Biochem. 107:1139–1149.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cebrian V, Fierro M, Orenes-Piñero E, Grau
L, Moya P, Ecke T, Alvarez M, Gil M, Algaba F, Bellmunt J, et al:
KISS1 methylation and expression as tumor stratification biomarkers
and clinical outcome prognosticators for bladder cancer patients.
Am J Pathol. 179:540–546. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mandai M, Konishi I, Kuroda H, Nanbu K,
Matsushita K, Yura Y, Hamid AA and Mori T: Expression of abnormal
transcripts of the FHIT (fragile histidine triad) gene in ovarian
carcinoma. Eur J Cancer. 34:745–749. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kotani M, Detheux M, Vandenbogaerde A,
Communi D, Vanderwinden JM, Le Poul E, Brézillon S, Tyldesley R,
Suarez-Huerta N, Vandeput F, et al: The metastasis suppressor gene
KiSS-1 encodes kisspeptins, the natural ligands of the orphan G
protein-coupled receptor GPR54. J Biol Chem. 276:34631–34636. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Muir AI, Chamberlain L, Elshourbagy NA,
Michalovich D, Moore DJ, Calamari A, Szekeres PG, Sarau HM,
Chambers JK, Murdock P, et al: AXOR12, a novel human G
protein-coupled receptor, activated by the peptide KiSS-1. J Biol
Chem. 276:28969–28975. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ohtaki T, Shintani Y, Honda S, Matsumoto
H, Hori A, Kanehashi K, Terao Y, Kumano S, Takatsu Y, Masuda Y, et
al: Metastasis suppressor gene KiSS-1 encodes peptide ligand of a
G-protein-coupled receptor. Nature. 411:613–617. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dhar DK, Naora H, Kubota H, Maruyama R,
Yoshimura H, Tonomoto Y, Tachibana M, Ono T, Otani H and Nagasue N:
Downregulation of KiSS-1 expression is responsible for tumor
invasion and worse prognosis in gastric carcinoma. Int J Cancer.
111:868–872. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Masui T, Doi R, Mori T, Toyoda E, Koizumi
M, Kami K, Ito D, Peiper SC, Broach JR, Oishi S, et al: Mtastin and
its variant forms suppress migration of pancreatic cancer cells.
Biochem Biophys Res Commun. 315:85–92. 2004. View Article : Google Scholar : PubMed/NCBI
|