Introduction
Renal cell carcinoma (RCC) accounts for ~2-3% of all
malignancies worldwide (1). Despite
an increasing proportion of patients with early stage tumors at
diagnosis and the development of novel treatment strategies, a
quarter still present with locally advanced or metastatic disease,
and eventually, one third of patients submitted to surgical
nephrectomy develop recurrence, or metastasis (2). The clinical outcomes of RCC vary widely,
emphasizing the need for prompt and accurate prognostic
stratification. To date, the best prognostic system for overall
survival (OS) is the tumor node metastasis (TNM) staging system in
RCC, although it is insufficient to significantly improve the
management of patients (3,4). Important prognostic factors of RCC
include tumor size, histological subtype, nuclear grade, local
extent of the tumor and evidence of metastatic disease at
presentation (5). Therefore,
identification of novel reliable predictive and independent
prognostic factors is critical for improving therapeutic modalities
and for prolonging the survival of patients with RCC.
Sex-determining region Y (SRY)-box protein (SOX)
genes arise from the founding member Sry, the mammalian
testis-determining factor. Currently, 20 SOX proteins are known in
mammals and characterized by a conserved high mobility group
DNA-binding domain (6,7). Members of the SOX family are
developmental regulators with functions in sex determination,
chondrogenesis, hematopoiesis, neural crest development and
neurogenesis (8). Numerous findings
support the involvement of different SOX genes in cancer
development. The expression of SOX genes is reported to be
associated with prognosis in various cancer types, including lung
(9,10), brain (11), hepatocellular (12), gastric (13), prostate (14) and cervical squamous cell carcinoma
(15). However, the potential role of
SOX family members in RCC, and its biological functions on the
initiation, progression and outcome of the disease remain unclear.
Therefore, the present study aimed to analyze the prognostic value
of SOX genes in patients with RCC using public database and
validate these findings in a patient cohort.
Materials and methods
Patients and samples
The Cancer Genome Atlas (TCGA) database makes
available gene expression level and clinical data on RCC from the
website of Cancer Genomics Browser of University of California
Santa Cruz (https://genome-cancer.ucsc.edu/). Only patients with
fully characterized tumors, intact OS, disease-free survival (DFS),
and RNAseq information were included as described previously
(16). Patients receiving
pretreatment were excluded. Clinicopathological characteristics,
including age, gender, tumor size, TNM stage, tumor grade, stage,
laterality, OS and DFS were collected. A total of 16 patients whose
samples were confirmed with non-clear cell RCC were excluded
(17). Finally, 505 patients
confirmed with primary clear cell RCC with detailed SOX expression
data were included in the current study.
Network analysis of genes that were associated with
OS was performed using the tools from cBioPortal (http://www.cbioportal.org/public-portal/cgds_r.jsp).
Genes were considered in the same network if they were in the same
complex or interacted with each other with >12% of changes.
Information from 20 members of the SOX family and the associated
genes obtained from the TCGA RNAseq database are listed in Table IV.
 | Table IV.Gene IDs of SOXs family members and
related genes. |
Table IV.
Gene IDs of SOXs family members and
related genes.
| Official gene
symbol | Full name | UniGene |
|---|
| SOX1 | SRY (sex
determining region Y)-box 1 | Hs.202526 |
| SOX2 | SRY (sex
determining region Y)-box 2 | Hs.518438 |
| SOX3 | SRY (sex
determining region Y)-box 3 | Hs.157429 |
| SOX4 | SRY (sex
determining region Y)-box 4 | Hs.643910 |
| SOX5 | SRY (sex
determining region Y)-box 5 | Hs.657542 |
| SOX6 | SRY (sex
determining region Y)-box 6 | Hs.368226 |
| SOX7 | SRY (sex
determining region Y)-box 7 | Hs.709543 |
| SOX8 | SRY (sex
determining region Y)-box 8 | Hs.243678 |
| SOX9 | SRY (sex
determining region Y)-box 9 | Hs.647409 |
| SOX10 | SRY (sex
determining region Y)-box 10 | Hs.376984 |
| SOX11 | SRY (sex
determining region Y)-box 11 | Hs.432638 |
| SOX12 | SRY (sex
determining region Y)-box 12 | Hs.43627 |
| SOX13 | SRY (sex
determining region Y)-box 13 | Hs.201671 |
| SOX14 | SRY (sex
determining region Y)-box 14 | Hs.248184 |
| SOX15 | SRY (sex
determining region Y)-box 15 | Hs.95582 |
| SOX17 | SRY (sex
determining region Y)-box 17 | Hs.98367 |
| SOX18 | SRY (sex
determining region Y)-box 18 | Hs.8619 |
| SOX21 | SRY (sex
determining region Y)-box 21 | Hs.187577 |
| SOX30 | SRY (sex
determining region Y)-box 30 | Hs.529462 |
| POU5F1 | POU class 5
homeobox 1 | Hs.249184 |
| POU2F1 | POU class 2
homeobox 1 | Hs.283402 |
| NR5A1 | Nuclear receptor
subfamily 5 group A member 1 | Hs.495108 |
In order to validate the prognostic value of these
genes, a validation cohort from Fudan University Shanghai Cancer
Center (FUSCC; Shanghai, China) was established, including 192
patients with histologically confirmed clear cell RCC between
February 2009 and June 2012 who underwent radical nephrectomy or
nephron sparing nephrectomy. Patient characteristics parallel to
TCGA data, including age, gender, tumor size, TNM stage, tumor
grade, stage and tumor position were obtained from clinical
records. Patients with missing data on the aforementioned variables
were excluded.
Trained research nurses followed up the cohort by
telephone once every 3–6 months, and recorded the events of
clinical interest, including tumor recurrence, progression and
metastasis. All tissue samples were collected during surgeries and
stored at −70°C in the tissue bank of FUSCC.
The present study was approved by the institutional
review board of FUSCC and written informed consent was obtained
from all patients.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
For the validation cohort, 192 frozen tissue samples
(100 mg) were harvested and ground into a fine powder. Total RNA
was isolated using TRIzol® reagent (cat. no. 15596-026;
Invitrogen, Thermo Fisher Scientific, Inc.). A PrimeScript RT
reagent kit (cat. no. K1622, Thermo Fisher Scientific, Inc.) was
used to synthesize first strand cDNA from total RNA according to
the manufacturer's protocol. Next, SYBR-Green RT-qPCR assays were
performed using an ABI 7900HT system (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The thermocycling conditions were as
follows: 50°C for 5 min, 95°C for 2 min followed by 40 cycles of
95°C for 3 sec and 60°C for 30 sec. The expression level of RNA was
normalized to the level of β-actin with 2−∆∆Cq method
(18). The primers for RT-qPCR
analysis were synthesized by Sangon Biotech Co., Ltd. (Shanghai,
China) and the sequences were as follows: SOX2 forward,
5′-TGGACAGTTACGCGCACAT-3′ and reverse, 5′-CGAGTAGGACATGCTGTAGGT-3′;
SOX12 forward, 5′-AAGAGGCCGATGAACGCATT-3′ and reverse,
5′-TAGTCCGGGTAATCCGCCAT-3′; SOX15 forward,
5′-GCGACTACCCCGACTACAAG-3′ and reverse, 5′-TTGCAGTGGGAAGAGCCATA-3′;
β-actin forward, 5′-AGCGAGCATCCCCCAAAGTT-3′ and reverse,
5′-GGGCACGAAGGCTCATCATT-3′.
Statistical analysis
Survival endpoints were mortality due to any cause
for OS and recurrence at any site for DFS. DFS and OS rates were
calculated based on the Kaplan-Meier method, and the curves were
compared with log-rank tests. Variables of SOX gene expression with
P<0.10 in univariate and multivariate Cox's proportional hazard
models in the TCGA cohort were selected for further study. The
final Cox's Proportional Hazards model, including clinical data and
genes in the network associated with OS was performed in TCGA, and
validation cohorts. Categorical data were analyzed using Fisher's
exact χ2 test. Continuous data were analyzed using a
Student's t-test. All the statistical analyses were performed using
SPSS software version 20.0 (IBM Corp., Armonk, NY, USA). Two-tailed
P<0.05 was considered to indicate a statistically significant
difference.
Results
Clinical characteristics in TCGA and
validation cohorts
In the TCGA cohort, the median age of the 505
patients with clear cell RCC was 60.6 years old, ranging between 26
and 90 years old. A total of 325 (64.4%) were male patients and 180
(35.6%) were female patients. Tumor size, TNM stage, tumor grade,
stage and laterality are presented in Table I. The median OS of this cohort was
79.5 months and 163 patients succumbed during the follow up. In the
validation cohort, the median age of the 192 patients with clear
cell RCC was 55.5 years old, ranging between 25 and 86 years old. A
total of 135 (70.3%) were male patients and 57 (29.7%) were female
patients. Tumor size, TNM stage, tumor grade, stage and tumor
position are presented in Table I.
The median OS of this cohort was 50.2 months and 42 patients
succumbed during follow up.
 | Table I.Clinical characteristics of patients
with clear cell RCC. |
Table I.
Clinical characteristics of patients
with clear cell RCC.
|
| TCGA cohort | N=505 | Validation
cohort | N=192 |
|
|---|
|
|
|
|
|
|---|
| Variable | N | % | N | % | P-value |
|---|
| Age, median
(range) | 60.6c | (26–90) | 55.5d | (25–86) | <0.01a |
| Sex |
|
|
|
| 0.16b |
| Male | 325 | 64.4 | 135 | 70.3 |
|
|
Female | 180 | 35.6 | 57 | 29.7 |
|
| Grade |
|
|
|
| 0.80b |
|
1&2 | 232 | 46.0 | 87 | 45.3 |
|
|
3&4 | 269 | 53.3 | 105 | 54.7 |
|
| Gx | 1 | 0.2 | 0 | 0 |
|
| pT |
|
|
|
| <0.01b |
| T1 | 257 | 50.9 | 137 | 71.4 |
|
| T2 | 63 | 12.5 | 26 | 13.5 |
|
| T3 | 175 | 34.7 | 24 | 12.5 |
|
| T4 | 10 | 2.0 | 5 | 2.6 |
|
| N |
|
|
|
|
<0.01b |
| N0 | 226 | 44.8 | 183 | 95.3 |
|
| N1 | 17 | 3.4 | 2 | 1 |
|
| Nx | 262 | 51.9 | 7 | 3.6 |
|
| M |
|
|
|
|
<0.01b |
| M0 | 402 | 79.6 | 184 | 95.8 |
|
| M1 | 78 | 15.4 | 7 | 3.6 |
|
| Mx | 23 | 4.6 | 1 | 0.5 |
|
| AJCC 7th edition
stage |
|
|
|
|
<0.01b |
| I | 252 | 49.9 | 136 | 70.8 |
|
| II | 51 | 10.1 | 24 | 12.5 |
|
|
III | 122 | 24.2 | 24 | 12.5 |
|
| IV | 80 | 15.8 | 8 | 4.2 |
|
| Position |
|
|
|
|
<0.01b |
|
Left | 237 | 46.9 | 87 | 45.4 |
|
|
Right | 266 | 52.7 | 98 | 51 |
|
|
Bilateral | 2 | 0.4 | 7 | 3.6 |
|
SOX gene expression and the clinical
outcomes in the TCGA, and validation cohorts
In the univariate Cox's proportion hazard ratio
analysis, age, T stage, metastasis, tumor stage, tumor grade,
hemoglobin level, white blood cell count and platelet count,
expression of SOX1, SOX2, SOX6, SOX7, SOX11, SOX12, SOX13, SOX15,
SOX17, and SOX30 were significantly associated with prognosis
regarding OS in patients with clear cell RCC in the TCGA cohorts
(Table II). Subsequently, the
variables with significance in the univariate analysis were used
for further multivariate analysis. After adjustment for all the
potential prognostic factors, analysis indicated that age [hazard
ratio (HR), 1.032; 95% confidence interval (CI), 1.018–1.047;
P<0.001], tumor stage (HR, 1.995; 95% CI, 1.411–2.820;
P<0.001), tumor grade (HR, 1.300; 95% CI, 1.020–1.657; P=0.034),
SOX2 (HR, 1.130; 95% CI, 1.002–1.275, P=0.046), SOX12 (HR, 1.379;
95% CI, 1.060–1.793; P=0.017) and SOX15 (HR, 1.245; 95% CI,
1.063–1.459; P=0.007) were independent predictors of OS (Table II). Finally, the expression levels of
SOX2, SOX12 and SOX15 were selected for further study.
 | Table II.Univariate and multivariate survival
analysis of SOXs family and related genes for patients with clear
cell RCC in the TCGA cohort. |
Table II.
Univariate and multivariate survival
analysis of SOXs family and related genes for patients with clear
cell RCC in the TCGA cohort.
|
| Univariate | Multivariate | Multivariate |
|---|
|
|
|
|
|
|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age | 1.028
(1.015–1.041) | <0.001 | 1.032
(1.018–1.047) | <0.001 | 1.033
(1.018–1.049) | <0.001 |
| Gendera | 0.950
(0.693–1.302) | 0.752 | 0.947
(0.666–1.346) | 0.760 | 0.968
(0.676–1.386) | 0.859 |
| T | 1.992
(1.685–2.355) | <0.001 | 0.764
(0.531–1.097) | 0.145 | 0.716
(0.489–1.049) | 0.087 |
| Na | 0.992
(0.562–1.752) | 0.978 | 0.845
(0.714–1.002) | 0.052 | 0.860
(0.723–1.023) | 0.089 |
| Ma | 2.459
(1.921–3.149) | <0.001 | 1.237
(0.780–1.962) | 0.367 | 1.178
(0.721–1.924) | 0.513 |
| Stage | 1.954
(1.707–2.236) | <0.001 | 1.995
(1.411–2.820) | <0.001 | 2.083
(1.432–3.031) | <0.001 |
| Grade | 2.398
(1.941–2.965) | <0.001 | 1.300
(1.020–1.657) | 0.034 | 1.349
(1.031–1.765) | 0.029 |
|
Positiona | 0.695
(0.512–0.944) | 0.019 | 0.740
(0.532–1.028) | 0.073 | 0.732
(0.521–1.030) | 0.073 |
| SOX1 | 1.207
(1.106–1.317) | <0.001 | 0.950
(0.861–1.049) | 0.311 | 0.961
(0.867–1.065) | 0.450 |
| SOX2 | 1.221
(1.119–1.334) | <0.001 | 1.130
(1.002–1.275) | 0.046 | 1.123
(1.001–1.272) | 0.047 |
| SOX3 | 1.087
(0.909–1.300) | 0.361 | – | – | – | – |
| SOX4 | 1.065
(0.864–1.312) | 0.557 | – | – | – | – |
| SOX5 | 0.969
(0.840–1.117) | 0.665 | – | – | – | – |
| SOX6 | 0.773
(0.698–0.855) | <0.001 | 0.987
(0.872–1.118) | 0.841 | 0.986
(0.864–1.126) | 0.837 |
| SOX7 | 0.875
(0.753–1.017) | 0.082 | – | – | – | – |
| SOX8 | 1.041
(0.914–1.186) | 0.553 | – | – | – | – |
| SOX9 | 0.942
(0.836–1.062) | 0.330 | – | – | – | – |
| SOX10 | 1.030
(0.909–1.168) | 0.642 | – | – | – | – |
| SOX11 | 1.126
(1.032–1.228) | 0.008 | 1.094
(0.981–1.219) | 0.107 | 1.089
(0.974–1.218) | 0.134 |
| SOX12 | 2.041
(1.626–2.563) | <0.001 | 1.379
(1.060–1.793) | 0.017 | 1.360
(1.026–1.803) | 0.033 |
| SOX13 | 0.636
(0.530–0.762) | <0.001 | 0.860
(0.676–1.094) | 0.219 | 0.867
(0.666–1.128) | 0.289 |
| SOX14 | 0.866
(0.226–3.313) | 0.833 | – | – | – | – |
| SOX15 | 1.290
(1.131–1.470) | <0.001 | 1.245
(1.063–1.459) | 0.007 | 1.269
(1.072–1.501) | 0.006 |
| SOX17 | 0.790
(0.675–0.924) | 0.003 | 0.945
(0.765–1.168) | 0.602 | 0.962
(0.773–1.197) | 0.729 |
| SOX18 | 0.847
(0.859–1.133) | 0.986 | – | – | – | – |
| SOX21 | 0.946
(0.803–1.114) | 0.506 | – | – | – | – |
| SOX30 | 1.216
(1.0371.425) | 0.016 | 1.132
(0.950–1.348) | 0.167 | 1.116
(0.930–1.338) | 0.237 |
| POU5F1 |
|
|
|
| 0.983
(0.883–1.095) | 0.754 |
| POU2F1 |
|
|
|
| 0.947
(0.727–1.233) | 0.685 |
| NR5A1 |
|
|
|
| 1.026
(0.894–1.178) | 0.716 |
To further evaluate the prognostic value of SOX2,
SOX12 and SOX15, the continuous variables were dichotomized by the
median cut-off value (0.47 for SOX2; 9.22 for SOX12; 3.14 for
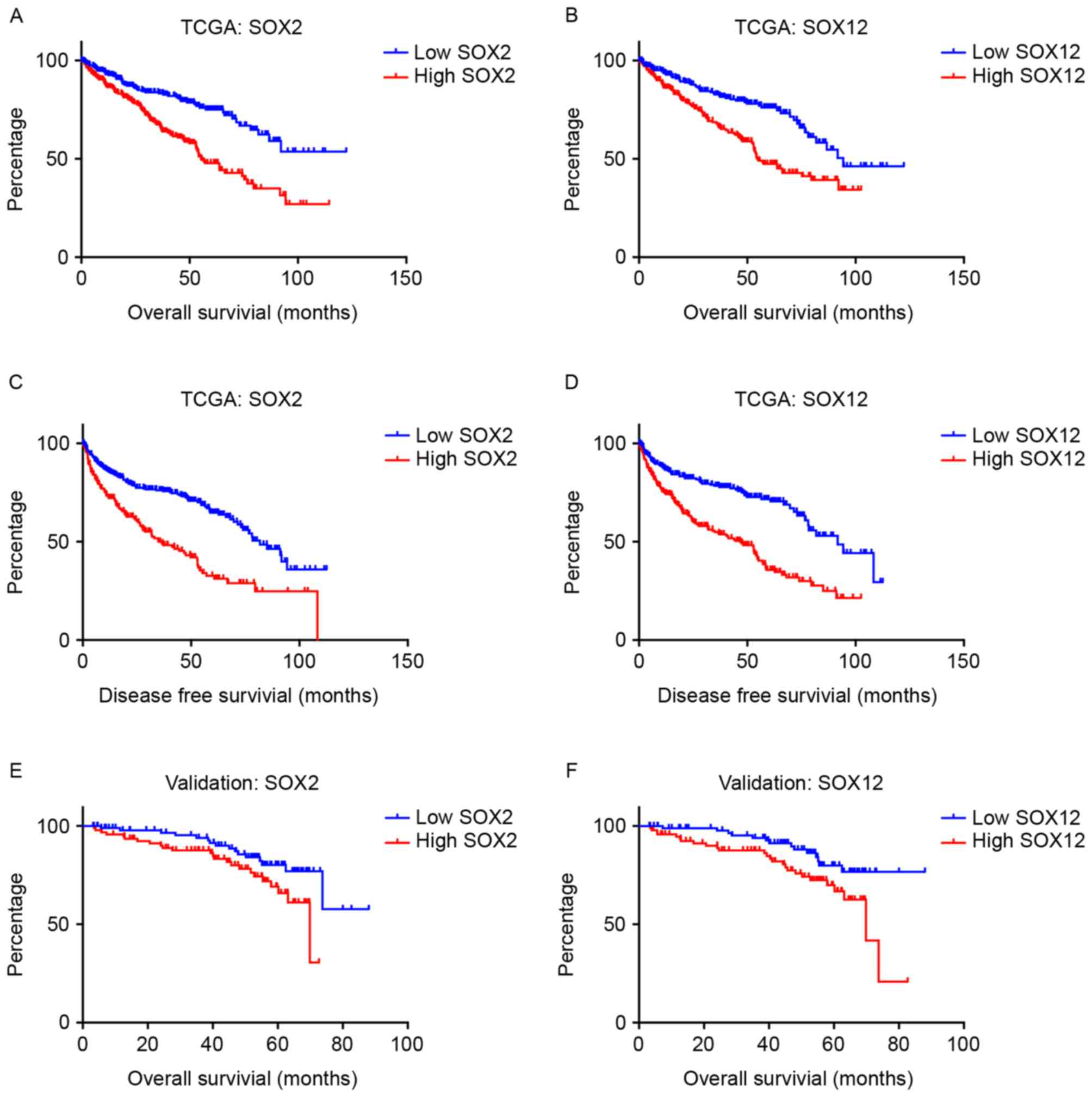
SOX15). Kaplan-Meier curves demonstrated that high expression of
SOX2, SOX12 and SOX15 was associated with poor prognosis for OS,
and DFS (Fig. 1).
The interaction network was established based on
three situations, stated as ‘react with’, ‘in same component’ and
‘state change’. The cut-point of state change was set as 12%
(16). As a result, POU class 5
homeobox 1 (POU5F1), POU2F1 and nuclear receptor subfamily 5 group
A member 1 (NR5A1) were identified to be associated with SOX2, and
added to the multivariate Cox's proportional hazard model.
Multivariate analysis revealed that the expression of SOX2 (HR,
1.123; 95% CI, 1.001–1.272; P=0.047), SOX12 (HR, 1.360; 95% CI,
1.026–1.803; P=0.033) and SOX15 (HR, 1.269; 95% CI, 1.072–1.501;
P=0.006) remained significantly associated with the OS of patients
with clear cell RCC in the cohort (Table
II). However, when the results were validated in the FUSCC
cohort, only SOX2 and SOX 12 were significant predictors of OS
(Fig. 1).
Multivariate logistic regression analysis was
performed to determine clinical factors that may affect the
expression of SOX2 and SOX12. Tumor stage [odds ratio (OR), 1.257;
95% CI, 1.053–1.501; P=0.011] and tumor grade (OR, 1.436, 95% CI,
1.086–1.898; P=0.011) were significantly associated with SOX12
expression, while only tumor stage (OR, 1.954; 95% CI, 1.069–1.518;
P=0.007) was significantly associated with SOX2 expression
(Table III).
 | Table III.Multivariate logistic regression
analysis of factors that might affect the expression of SOX2 and
SOX12 in the TCGA Cohort with clear cell RCC. |
Table III.
Multivariate logistic regression
analysis of factors that might affect the expression of SOX2 and
SOX12 in the TCGA Cohort with clear cell RCC.
|
| SOX2 | SOX12 |
|---|
|
|
|
|
|---|
| Variable | OR (95% CI) | P-value | OR (95% CI) | P-value |
|---|
| Age | 1.000
(0.985–1.015) | 0.967 | 1.013
(0.998–1.029) | 0.085 |
| Gendera | 1.165
(0.779–1.770) | 0.427 | 0.968
(0.499–1.080) | 0.117 |
| Stage | 1.954
(1.069–1.518) | 0.007 | 1.257
(1.053–1.501) | 0.011 |
| Grade | 2.398
(0.849–1.464) | 0.435 | 1.436
(1.086–1.898) | 0.011 |
|
Positiona | 0.749
(0.745–1.506) | 0.749 | 0.732
(0.521–1.030) | 0.835 |
Discussion
To the best of our knowledge, for the first time,
the present study has demonstrated that expression of SOX family
genes was associated with OS in patients with clear cell RCC.
Multivariate analysis revealed that SOX2, SOX12 and SOX15 may serve
an important role in the prognosis of patients with clear cell RCC.
When validated in the cohort from FUSCC, SOX2 and SOX12 remained
independent prognostic factors.
SOX family members participate in numerous important
biological processes, particularly in cell differentiation during
embryonic development. It has been considered that the origins of
cancer may be associated with the aberrant reactivation of
embryonic development or stem cell programs within normal tissues.
Conversely, numerous oncogenes and tumor suppressor genes were also
identified to be essential in embryogenesis (19). SOX2, known as a major stemness marker,
is high expressed in cancer stem cells, which have the ability to
renew itself and generate the diversity of cell types. It is
considered that SOX2 confers a less differentiated phenotype and
its high level of expression may promote a potential for metastasis
(20). SOX12 may act as oncogenes,
tumor suppressor genes or both depending on the cancer types. In
squamous esophageal, colorectal and small cell lung cancer, SOX2 is
associated with poor prognosis, and is activated through DNA
amplification (21–24). However, SOX2 acts as tumor suppressor
gene in gastric cancer and non-small cell lung cancers (25–27), which
further emphasizes the context-specific nature of SOX involvement
in carcinogenesis.
SOX12 is a member of group C of SOX transcription
factors. Another two members named SOX4 and SOX11 were reported to
serve key roles in cardiac, neuronal, and other major developmental
processes, as well as be involved in cancer development, but the
roles of SOX12 remain unknown (7,28). A
recent study revealed that SOX12 had significant prognostic value
in human hepatic cell carcinoma. Overexpression of SOX12 was
significantly correlated with loss of tumor encapsulation,
microvascular invasion and a higher tumor-nodule-metastasis stage.
Furthermore, SOX12 expression was an independent and significant
risk factor for recurrence, and reduced survival time following
curative resection (29). The
underlying mechanism may be that SOX12 expression was positively
associated with Twist1, fibroblast growth factor binding protein 1
and forkhead box Q1 expression levels, which serve a central role
in tumor invasion, and metastasis (29). However, in colon cancer, a genome-wide
screen identified SOX12 as a metastatic suppressor affecting
Wnt/Tcf signaling (30). Although
evidence has suggested that the SOX family serves an important role
in human cancer, little is known regarding its involvement in RCC.
The results of the present study indicated an association between
the outcome of patients with clear cell RCC, and the expression of
SOX2 and SOX12, but the underlying mechanism remains unknown.
The role of SOX15 in human cancer is also relatively
understudied compared with other SOX family members. Overexpression
of SOX15 was associated with worse clinical outcome in the TCGA
cohort, but the result could not be validated in the FUSCC cohort.
Furthermore, a previous study demonstrated that SOX15
overexpression inhibited the proliferation of human testicular
embryonic carcinoma cells (31).
Another study indicated SOX15 remarkably suppressed tumor formation
via suppression of the Wnt/β-catenin signaling pathway in
pancreatic ductal adenocarcinoma (32).
The strength of the current study is that the
clinical information was obtained from two large populations with a
long-time follow-up. However, the limitation is the heterogeneous
patient characteristics between the two cohorts. Secondly, the
prognosis of clear cell RCC is affected by numerous factors in
addition to tumor stage and tumor grade, including surgical
performance, and response to adjuvant therapy. Thus, expression of
SOX genes as prognostic markers in clinical routine should be
further validated in a multicenter prospective study.
Taken together, SOX2 and SOX12 were identified as
independent prognostic factors for OS, and DFS of clear cell RCC.
This study indicated that SOX family genes may serve an important
role in clear cell RCC. This novel method of identifying
prognosis-associated genes may by applied in different types of
cancer.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ficarra V, Galfano A, Novara G, Iafrate M,
Brunelli M, Secco S, Cavalleri S, Martignoni G and Artibani W: Risk
stratification and prognostication of renal cell carcinoma. World J
Urol. 26:115–125. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim SP, Alt AL, Weight CJ, Costello BA,
Cheville JC, Lohse C, Allmer C and Leibovich BC: Independent
validation of the 2010 American Joint Committee on Cancer TNM
classification for renal cell carcinoma: Results from a large,
single institution cohort. J Urol. 185:2035–2039. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sorbellini M, Kattan MW, Snyder ME, Reuter
V, Motzer R, Goetzl M, McKiernan J and Russo P: A postoperative
prognostic nomogram predicting recurrence for patients with
conventional clear cell renal cell carcinoma. J Urol. 173:48–51.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bowles J, Schepers G and Koopman P:
Phylogeny of the SOX family of developmental transcription factors
based on sequence and structural indicators. Dev Biol. 227:239–255.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Castillo SD and Sanchez-Cespedes M: The
SOX family of genes in cancer development: Biological relevance and
opportunities for therapy. Expert Opin Ther Targets. 16:903–919.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kamachi Y and Kondoh H: Sox proteins:
Regulators of cell fate specification and differentiation.
Development. 140:4129–4144. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Castillo SD, Matheu A, Mariani N,
Carretero J, Lopez-Rios F, Lovell-Badge R and Sanchez-Cespedes M:
Novel transcriptional targets of the SRY-HMG box transcription
factor SOX4 link its expression to the development of small cell
lung cancer. Cancer Res. 72:176–186. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chapman CJ, Thorpe AJ, Murray A,
Parsy-Kowalska CB, Allen J, Stafford KM, Chauhan AS, Kite TA,
Maddison P and Robertson JF: Immunobiomarkers in small cell lung
cancer: Potential early cancer signals. Clin Cancer Res.
17:1474–1480. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
De la Rocha AM, Sampron N, Alonso MM and
Matheu A: Role of SOX family of transcription factors in central
nervous system tumors. Am J Cancer Res. 4:312–324. 2014.PubMed/NCBI
|
|
12
|
Liao YL, Sun YM, Chau GY, Chau YP, Lai TC,
Wang JL, Horng JT, Hsiao M and Tsou AP: Identification of SOX4
target genes using phylogenetic footprinting-based prediction from
expression microarrays suggests that overexpression of SOX4
potentiates metastasis in hepatocellular carcinoma. Oncogene.
27:5578–5589. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sashikawa Kimura M, Mutoh H and Sugano K:
SOX9 is expressed in normal stomach, intestinal metaplasia, and
gastric carcinoma in humans. J Gastroenterol. 46:1292–1299. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Matheu A, Collado M, Wise C, Manterola L,
Cekaite L, Tye AJ, Canamero M, Bujanda L, Schedl A, Cheah KS, et
al: Oncogenicity of the developmental transcription factor Sox9.
Cancer Res. 72:1301–1315. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Malki S, Bibeau F, Notarnicola C, Roques
S, Berta P, Poulat F and Boizet-Bonhoure B: Expression and
biological role of the prostaglandin D synthase/SOX9 pathway in
human ovarian cancer cells. Cancer Lett. 255:182–193. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lu X, Wan F, Zhang H, Shi G and Ye D:
ITGA2B and ITGA8 are predictive of prognosis in clear cell renal
cell carcinoma patients. Tumour Biol. 37:253–262. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Buttner F, Winter S, Rausch S, Reustle A,
Kruck S, Junker K, Stenzl A, Agaimy A, Hartmann A, Bedke J, et al:
Survival prediction of clear cell renal cell carcinoma based on
gene expression similarity to the proximal tubule of the nephron.
Eur Urol. 68:1016–1020. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Garraway LA and Sellers WR: Lineage
dependency and lineage-survival oncogenes in human cancer. Nat Rev
Cancer. 6:593–602. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lengerke C, Fehm T, Kurth R, Neubauer H,
Scheble V, Müller F, Schneider F, Petersen K, Wallwiener D, Kanz L,
et al: Expression of the embryonic stem cell marker SOX2 in
early-stage breast carcinoma. BMC Cancer. 11:422011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Neumann J, Bahr F, Horst D, Kriegl L,
Engel J, Luque RM, Gerhard M, Kirchner T and Jung A: SOX2
expression correlates with lymph-node metastases and distant spread
in right-sided colon cancer. BMC Cancer. 11:5182011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lundberg IV, Löfgren Burström A, Edin S,
Eklöf V, Öberg Å, Stenling R, Palmqvist R and Wikberg ML: SOX2
expression is regulated by BRAF and contributes to poor patient
prognosis in colorectal cancer. PLoS One. 9:e1019572014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang F, Gao Y, Geng J, Qu D, Han Q, Qi J
and Chen G: Elevated expression of SOX2 and FGFR1 in correlation
with poor prognosis in patients with small cell lung cancer. Int J
Clin Exp Pathol. 6:2846–2854. 2013.PubMed/NCBI
|
|
24
|
Forghanifard MM, Ardalan Khales S,
Javdani-Mallak A, Rad A, Farshchian M and Abbaszadegan MR: Stemness
state regulators SALL4 and SOX2 are involved in progression and
invasiveness of esophageal squamous cell carcinoma. Med Oncol.
31:9222014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang S, Tie J, Wang R, Hu F, Gao L, Wang
W, Wang L, Li Z, Hu S, Tang S, et al: SOX2, a predictor of survival
in gastric cancer, inhibits cell proliferation and metastasis by
regulating PTEN. Cancer Lett. 358:210–219. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Otsubo T, Akiyama Y, Hashimoto Y, Shimada
S, Goto K and Yuasa Y: MicroRNA-126 inhibits SOX2 expression and
contributes to gastric carcinogenesis. PLoS One. 6:e166172011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen Y, Huang Y, Huang Y, Chen J, Wang S
and Zhou J: The prognostic value of SOX2 expression in non-small
cell lung cancer: A meta-analysis. PLoS One. 8:e711402013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Thu KL, Becker-Santos DD, Radulovich N,
Pikor LA, Lam WL and Tsao MS: SOX15 and other SOX family members
are important mediators of tumorigenesis in multiple cancer types.
Oncoscience. 1:326–335. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang W, Chen Z, Shang X, Tian D, Wang D,
Wu K, Fan D and Xia L: Sox12, a direct target of FoxQ1, promotes
hepatocellular carcinoma metastasis through up-regulating Twist1
and FGFBP1. Hepatology. 61:1920–1933. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Duquet A, Melotti A, Mishra S, Malerba M,
Seth C, Conod A and Ruiz i Altaba A: A novel genome-wide in vivo
screen for metastatic suppressors in human colon cancer identifies
the positive WNT-TCF pathway modulators TMED3 and SOX12. EMBO Mol
Med. 6:882–901. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yan HT, Shinka T, Sato Y, Yang XJ, Chen G,
Sakamoto K, Kinoshita K, Aburatani H and Nakahori Y: Overexpression
of SOX15 inhibits proliferation of NT2/D1 cells derived from a
testicular embryonal cell carcinoma. Mol Cells. 24:323–328.
2007.PubMed/NCBI
|
|
32
|
Thu KL, Radulovich N, Becker-Santos DD,
Pikor LA, Pusic A, Lockwood WW, Lam WL and Tsao MS: SOX15 is a
candidate tumor suppressor in pancreatic cancer with a potential
role in Wnt/β-catenin signaling. Oncogene. 33:279–288. 2014.
View Article : Google Scholar : PubMed/NCBI
|