Introduction
Colorectal cancer (CRC) is the third most common
cancer in men (746,000 cases, 10.0% of the total) and the second
most common in women (614,000 cases, 9.2% of the total) worldwide
(1,2).
Despite a good outcome of treatment for early CRC, the clinical
outcome for advanced CRC is unsatisfactory (1,2).
Therefore, elucidating the molecular mechanisms underlying CRC
progression is critical for developing novel therapeutic strategies
to improve the prognosis of patients with advanced CRC.
Increasing evidence indicates that tumors are
hierarchically organized, and only a small fraction of cells
possess tumorigenic growth potential; these are termed cancer stem
cells (CSCs) (3). The existence of
CSCs was demonstrated in CRC, and cluster of differentiation
(CD)133 was identified as the marker for colorectal CSCs (4,5).
CD133+ tumor cells behave like stem-like cells, and
thereby produce more differentiated tumor cells in CRC (4,5). Despite
numerous previous studies on these undifferentiated cancer cells,
the signature profile and regulation of colorectal CSCs remains
largely unknown.
Sirtuins are NAD+-dependent deacetylases
that have been implicated in a diverse range of biological
processes and various diseases, including cancer (6). SIRT6, one of the sirtuins, is primarily
localized in the nucleus of the cells (6). It deacetylates histones H3 at either
lysine 9 (H3K9) or lysine 56 (H3K56) and represses its target gene
expression (7,8). Through its deacetylation activity, SIRT6
regulates a variety of cellular processes including chromosome
stability, inflammation, cell metabolism, apoptosis and senescence
(7,8).
Previous studies have indicated that SIRT6 serves
important roles in the development of cancer: SIRT6 expression is
downregulated in a variety of types of cancer, including hepatic
and breast cancer, and head and neck squamous cell carcinoma
(9–11). Downregulation of SIRT6 is positively
correlated with poor prognosis and low tumor-free survival rates in
CRC and pancreatic ductal adenocarcinoma (12,13).
Deletion of SIRT6 in mice increases the number, size and
aggressiveness of tumors (12,13). SIRT6
was also demonstrated to regulate cancer cell metabolism and
promote cancer cell apoptosis (12,14). These
studies indicated that SIRT6 serves as a general tumor suppressor
in human cancer. However, the exact function of SIRT6 in colorectal
CSCs remains unknown.
Cell division cycle 25A (CDC25A) is a member of the
CDC25 phosphatases family that regulates cell cycle progression by
inhibiting phosphorylation of the cyclin-dependent kinases (CDKs)
subunit, thereby activating cyclin-CDK complexes (15). Elevated expression of CDC25A has been
observed in various types of human cancer, and is often associated
with high grade tumors and poor prognosis (16). Overexpression of CDC25A leads to
genomic instability and tumorigenesis, while downregulation of
CDC25A inhibits cellular transformation and tumorigenesis (15–17).
Concurrently, overexpression of CDC25A in cancer tissues is
accompanied by disorders in the cell cycle (15–17),
suggesting that CDC25A is involved in the development and
progression of tumors by regulating the cell cycle.
The present study investigated the role of SIRT6 in
colorectal CSCs and the underlying mechanism. The results revealed
a suppressive role of SIRT6 in colorectal CSCs growth and provided
a potential target for CRC treatment.
Materials and methods
Tissue samples
The CRC tissue specimens were obtained from 10 male
and 2 female patients at the Linyi People's Hospital (Linyi, China)
following surgical resection between September 2016 and December
2016. The age of the patients ranged between 38 and 83 years. The
types of CRC were all adenocarcinoma. The Ethics Committee of the
Linyi People's Hospital approved the present study and written
informed consent was obtained from all patients.
Cell culture
CRC SW480 cells were obtained from the American Type
Culture Collection (Manassas, VA, USA) and maintained in Dulbecco's
modified Eagle's medium (DMEM; Life Technologies; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (Life Technologies; Thermo Fisher Scientific, Inc.).
Cells were cultured at 37°C in an atmosphere containing 5%
CO2 and 100% humidity.
Flow cytometry analysis
Allophycocyanin (APC)-conjugated CD133 antibodies
(AC133 epitope; Miltenyi Biotec GmbH, Bergisch Gladbach, Germany)
were used for colorectal CSCs identification. Briefly, SW480 cells
were digested using trypsin (Thermo Fisher Scientific, Inc.). CRC
tissue was digested using collagenase (1.5 mg/ml, Life
Technologies; Thermo Fisher Scientific, Inc.) and hyaluronidase (20
mg/ml, Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) in PBS for 1
h. Cells were centrifuged at 700 × g for 10 min at room
temperature. Single-cell suspensions were then incubated with
APC-CD133 antibodies on ice for 1 h in the dark. The cells were
then isolated using FACSARIA (BD Biosciences, San Jose, CA, USA)
flow cytometer. CD133-positive cells were considered to be
colorectal CSCs. The populations of CSCs and non-CSCs from SW480
cell lines were then re-analyzed on a flow cytometer (Beckman
Coulter, Brea, CA, USA). Data were analyzed using FlowJo software
(version 7.6.1; FlowJo LLC, Ashland, OR, USA).
Transfection
SW480 CSCs were transiently transfected with
pcDNA3.1 vector (Thermo Fisher Scientific, Inc.) or plasmid
expressing SIRT6 gene (SIRT6 gene was constructed to pcDNA3.1)
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol. Cells were
subjected to the following experiments 24 h after transfection.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from CSC cells was extracted using
TRIzol® Reagent (Life Technologies; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol. Reverse
transcription was performed at 37°C for 1 h and then inactivated at
70°C for 15 min according to the manufacture's protocol (Life
Technologies; Thermo Fisher Scientific, Inc.). qPCR assays were
performed using the SYBR GREEN PCR Mix (Takara Bio Inc., Otsu,
Japan) following the manufacturer's protocol. Briefly, PCR mixture
were heated at 95°C for 30 sec and then subjected to 40 cycles. For
each cycle, the PCR mixture were first heated at 95°C for 5 sec and
then amplified at 60°C for 30 sec. The expression levels of SIRT6
and CDC25A were normalized to the expression level of β-actin
(ACTB). Primers 5′-CAAGTGTAAGACGCAGTACG-3′ (forward) and
5′-GATGGTGTCCCTCAGCTCTC-3′ (reverse) were used for SIRT6
amplification. Primers 5′-TGACATCTTTCAGCTCATCG-3′ (forward) and
5′-CAGACAAAGTGGCTGTCACAG-3′ (reverse) were used for CDC25A
amplification. Primers 5′-TTGCGTTACACCCTTTCTTG-3′ (forward) and
5′-CACCTTCACCGTTCCAGTTT-3′ (reverse) were used for ACTB
amplification. Data were analyzed using the 2−ΔΔCq
method (18).
Apoptosis assay
Colorectal CSCs were transfected with the empty
vector or SIRT6-expressing plasmids for 48 h. Cells were then
harvested and stained with Annexin V-fluorescein isothiocyanate
(FITC; BioVision, Inc., Milpitas, CA, USA) for 5 min in the dark at
room temperature. Cells were then analyzed by flow cytometry as
aforementioned.
Cell proliferation assay
SW480 CSC cells were transfected with the empty
vector or SIRT6-expressing plasmids. Following 24 h transfection,
cells were seeded in 96-well plates at the density of
5×103 cells/well. Cell number was detected using CCK-8
kit at 1, 2, 3, 4, and 5 days. Briefly, CCK-8 reagent was added to
each well and the cells were incubated for 2 h at 37°C. The optical
density OD at 450 nm was measured by using a microplate reader
(BioTek Instruments, Winooski, VT, USA).
Cell cycle analysis
For the cell cycle analysis, SW480 CSCs transfected
with the empty vector or SIRT6-expressing plasmids were fixed with
100% ice-cold ethanol for 10 min, stained with propidium iodide (5
µM; BioVision, Inc.) for 5 min at room temperature and examined
with a flow cytometer as aforementioned.
Colony formation assay
Cells were seeded in 6-well plates at the density of
1,000 cells/well and maintained in DMEM containing 10% FBS. After 2
weeks, the cells were fixed in 4% paraformaldehyde for 10 min at
room temperature, and stained with crystal violet (Sigma-Aldrich;
Merck KGaA) for 10 min at room temperature. The colony number was
counted in 10 random fields of view using a wide-field light
microscope at ×5 magnification (Eclipse TS100; Nikon Corporation,
Tokyo, Japan).
Protein extraction and
immunoblotting
The whole cell lysate was extracted using lysis
buffer (100 mM Tris, pH 7.8, 1% Triton X-100, 150 mM NaCl, 0.5 mM
EDTA), and cleared by centrifugation at 12,000 × g for 10 min at
4°C. The protein concentration was determined using the BCA (Thermo
Fisher Scientific, Inc.) according to the manufacture's protocol. A
total of 50 µg of proteins were subjected to SDS-PAGE (12% gel)
electrophoresis and transferred to a nitrocellulose membrane (Life
Technologies; Thermo Fisher Scientific, Inc.). The membrane was
blocked with 3% milk in TBST buffer (20 mM Tris-HCl, pH 8.0, 150 mM
NaCl, 0.05% Tween-20) for 1 h at room temperature. The membrane was
then incubated with antibodies targeting SIRT6 (1:1,000; rabbit;
12486; Cell Signaling Technology, Inc., Danvers, MA, USA), CDC25A
(1:1,000; rabbit; 3652; Cell Signaling Technology) or β-actin
(1:2,000; rabbit; 4970; Cell Signaling Technology) at 4°C
overnight. Subsequent to 3 washes with Tris-buffered saline
Tween-20 (TBST buffer), the membrane was then incubated with
FITC-conjugated secondary antibodies (goat anti-rabbit; 926–32211;
1:5,000; LI-COR Biosciences, Lincoln, NE, USA) at room temperature
for 1 h and detected with Odyssey Imaging System (version 2.1;
LI-COR Biosciences).
Microarray analysis
Total RNA from control or SIRT6-expressing CSC cells
was extracted using TRIzol reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. RNA was subjected to
Agilent microarray analysis according to the manufacturer's
protocol (KangCheng, Shanghai, China).
Chromatin immunoprecipitation
(ChIP)
ChIP assays were performed using the ChIP assay kit
(17–371; Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol. Briefly, SW480 CSCs were cross-linked with
1% formaldehyde for 10 min at 37°C. The samples were sonicated on
ice in 30 sec intervals 6 times at the frequency of 20–60 kHz using
Bioruptor Pico (Diagenode s.a., Seraing, Belgium) to obtain ~1,000
bp fragments. The lysates were cleared by centrifugation at 12,000
× g for 10 min at 4°C. The supernatant was diluted with ChIP
dilution buffer and pre-cleared with salmon sperm DNA/protein
A-agarose (80 µl/ChIP) at 4°C for 1 h. The lysates were incubated
with 5 µg anti-SIRT6, anti-acetylated histone H3 (Lysine9; 9649;
Cell Signaling Technology), anti-acetylated histone H3 (Lysine56;
ab21623; Abcam, Cambridge, MA, USA) or control IgGs overnight at
4°C. The immunocomplexes were then collected with protein
A-agarose. The protein A-agarose beads were washed once with low
salt buffer, once with high salt buffer, once with LiCl buffer and
twice with Tris-EDTA buffer (Thermo Fisher Scientific, Inc.). The
immunocomplexes were eluted and de-crosslinked at 65°C overnight.
Following RNase digestion at 37°C for 30 min (Thermo Fisher
Scientific, Inc.) and proteinase K digestion at 45°C for 1 h
(Thermo Fisher Scientific, Inc.), the immunoprecipitated DNA was
extracted and amplified by qPCR as aforementioned. Primers
5′-TGTAGGTCGGCTTGGTTTTC-3′ (forward) and
5′-ATTAAATCCAAACAAACGTGG-3′ (reverse) were used for CDC25A promoter
amplification.
Statistical analysis
Statistical analysis was performed using SPSS
(version 16.0; SPSS, Inc., Chicago, IL, USA). Data were expressed
as the mean ± standard deviation and evaluated with a double-sided
Student's t-test. Statistical significance among >3 groups was
determined using Turkey's post-hoc analysis. P<0.05 was
considered to indicate a statistically significant difference.
Correlation analysis of SIRT6 and CDC25A expression in CSCs was
performed using Pearson analysis.
Results
SIRT6 expression is reduced in
colorectal CSCs
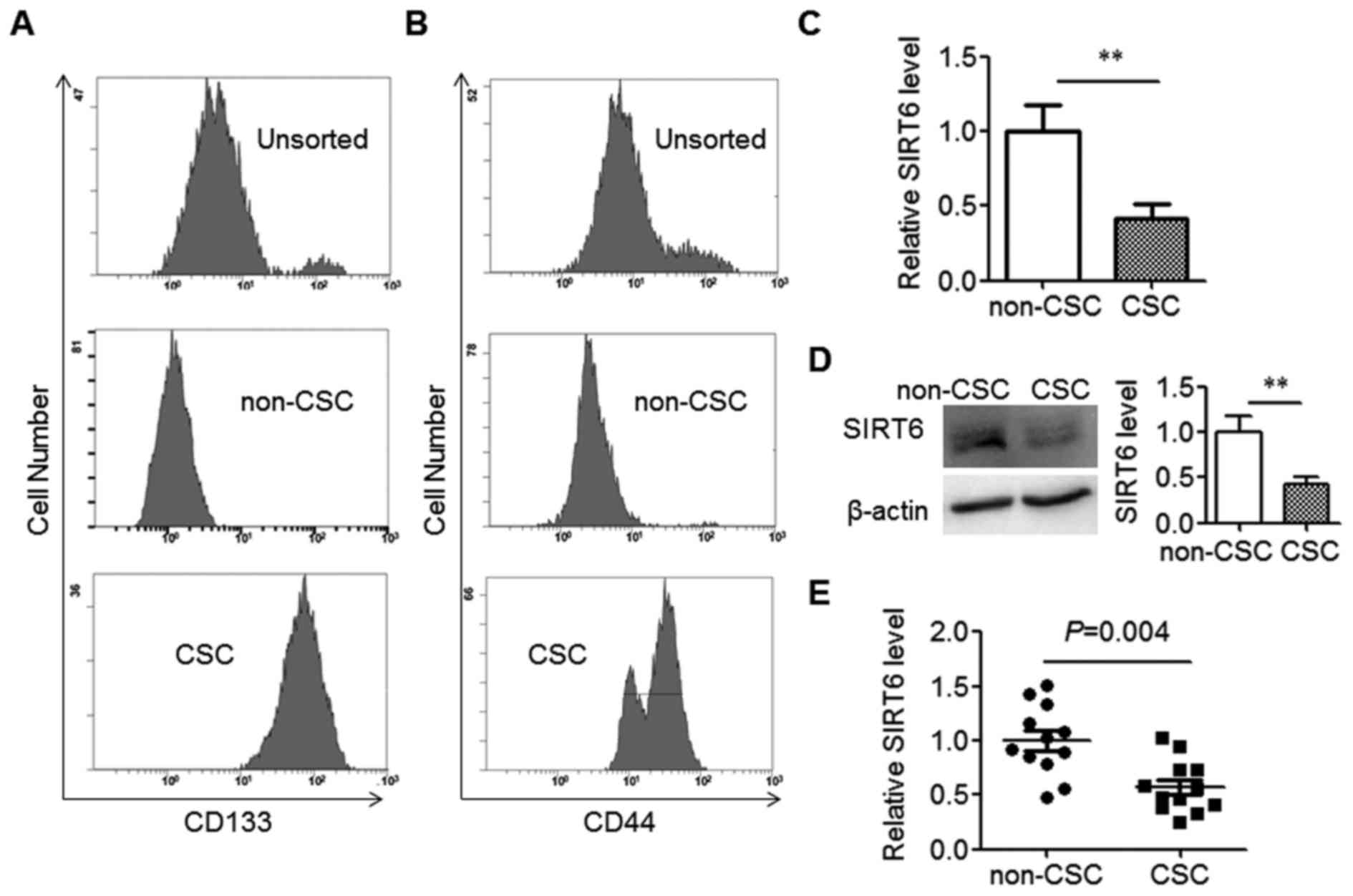
To explore the role of SIRT6 in CRC stem cells, the
CSCs population was separated from the SW480 cell line using the
marker CD133. As expected, the isolated SW480 CSCs expressed high
levels of CD133, while CD133 expression in the non-CSCs population
was low (Fig. 1A). CD44, an
additional CSCs marker, was also highly expressed in isolated SW480
CSCs as compared with non-CSCs (Fig.
1B). Next, the expression levels of SIRT6 in CSCs and non-CSCs
were detected. The data indicated that SW480 CSCs expressed
significantly decreased levels of SIRT6 compared with their
corresponding non-CSCs population (Fig.
1C and D). Immunoblotting data also demonstrated that SIRT6 had
several bands, which represented multiple isoforms (Fig. 1D).
To validate the reduced expression of SIRT6 in CSCs
isolated from SW480 cells, the CSCs and non-CSCs were collected
from the tumor tissues of patients with CRC. The data indicated
that the mRNA level of SIRT6 in colorectal CSCs was significantly
decreased compared with their corresponding non-CSCs (Fig. 1E). Taken together, the data above
demonstrated decreased SIRT6 expression in colorectal CSCs.
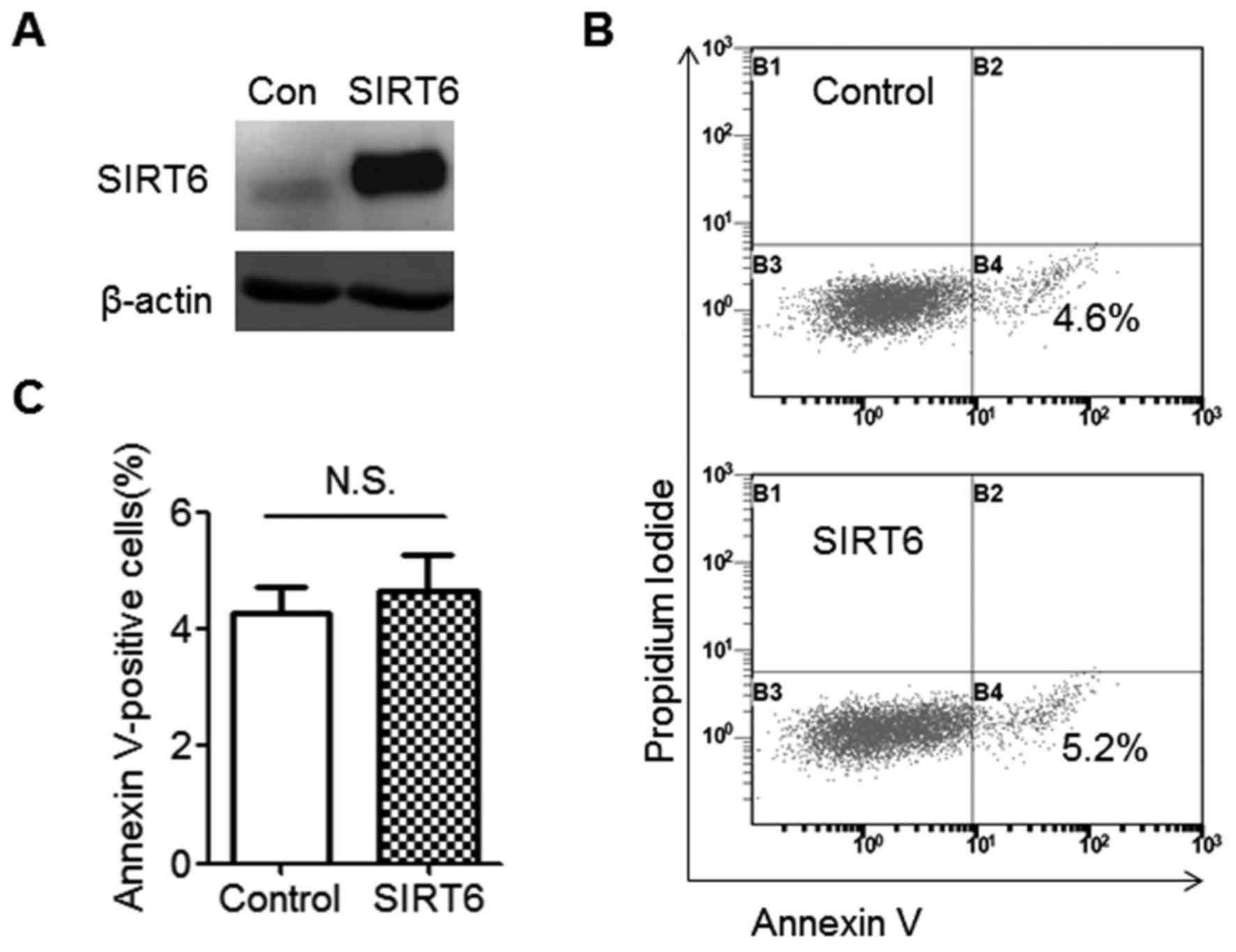
SIRT6 does not affect cell
apoptosis
SIRT6 was downregulated in colorectal CSCs; the
effect of SIRT6 on colorectal CSC behavior was therefore
investigated. As SIRT6 was previously demonstrated to induce cancer
cell apoptosis (14), SIRT6-regulated
apoptosis was first explored. The immunoblotting data suggested
that SIRT6 protein was almost undetectable in SW480 CSCs;
therefore, SIRT6 overexpression was then successfully induced
(Fig. 2A). This was also represented
at the mRNA level. SIRT6 mRNA level in the SIRT6 overexpression
group increased 200-fold compared with the control group (data not
shown). There was no difference in Annexin V staining observed
between the control and SIRT6-expressing groups, indicating that
SIRT6 does not induce cell apoptosis in SW480 CSCs (Fig. 2B and C).
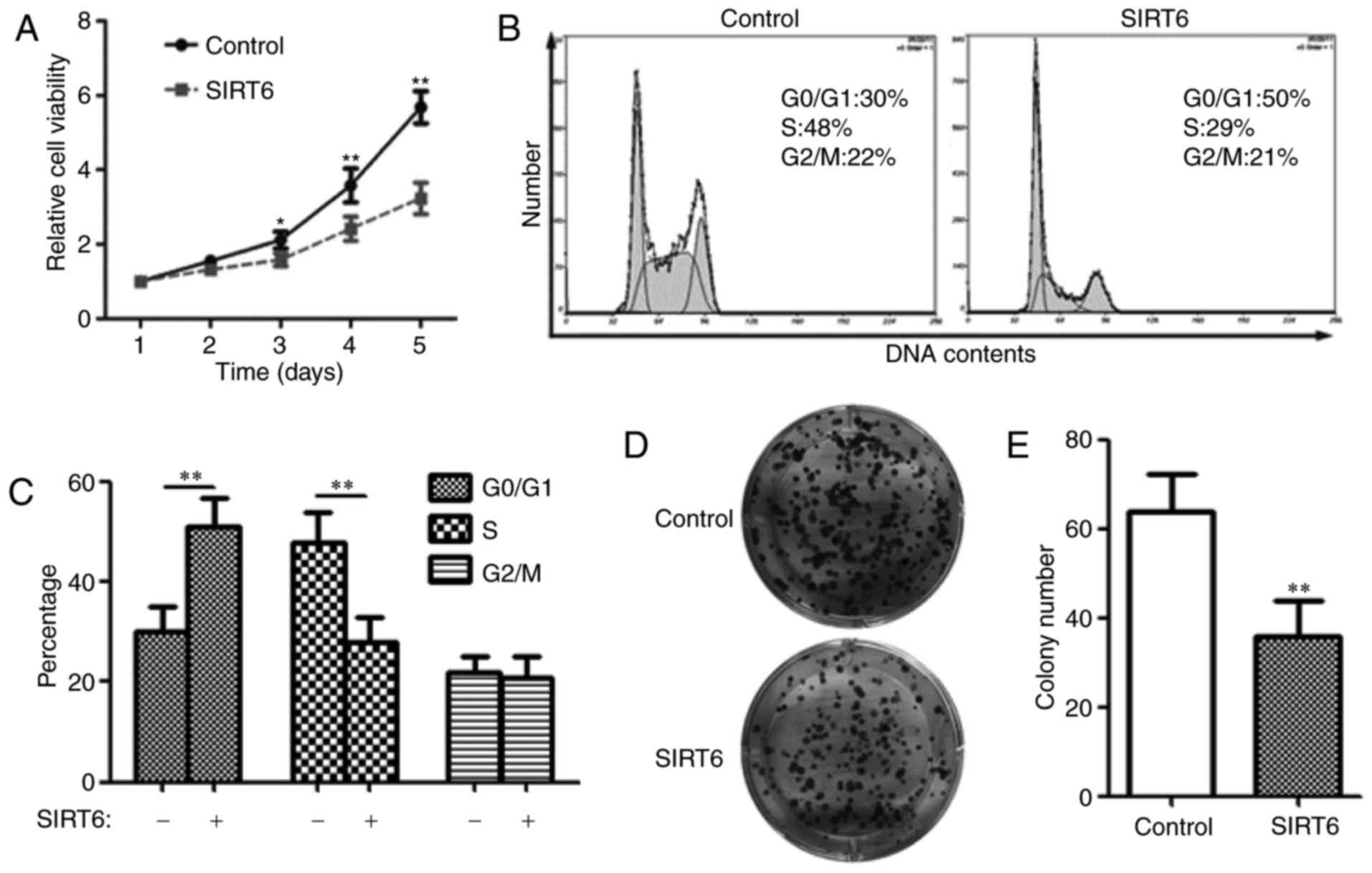
SIRT6 inhibits SW480 cell
proliferation
Next, the effect of SIRT6 on SW480 CSCs
proliferation was detected. The CCK-8 assay demonstrated that
compared with the control group, the proliferation rate was
significantly decreased in SIRT6-expressing cells (Fig. 3A), indicating that SIRT6 inhibits
SW480 CSCs proliferation. Cell proliferation is tightly controlled
by the cell cycle (19). Therefore,
the cell cycle distribution was examined using flow cytometry. The
data suggested that SIRT6 expression in SW480 CSCs increased the
number of cells in G0/G1 phase (50 vs. 30%), while decreased the
number of cells in S phase (29 vs. 48%; Fig. 3B and C), indicating that SIRT6 induces
G0/G1 arrest in colorectal CSCs.
The colony formation assay indicated that the
expression of SIRT6 in SW480 CSCs significantly decreased the
colony number (Fig. 3D and E; 36±8
vs. 64±8), additionally supporting the growth inhibitory effect of
SIRT6 on colorectal CSCs.
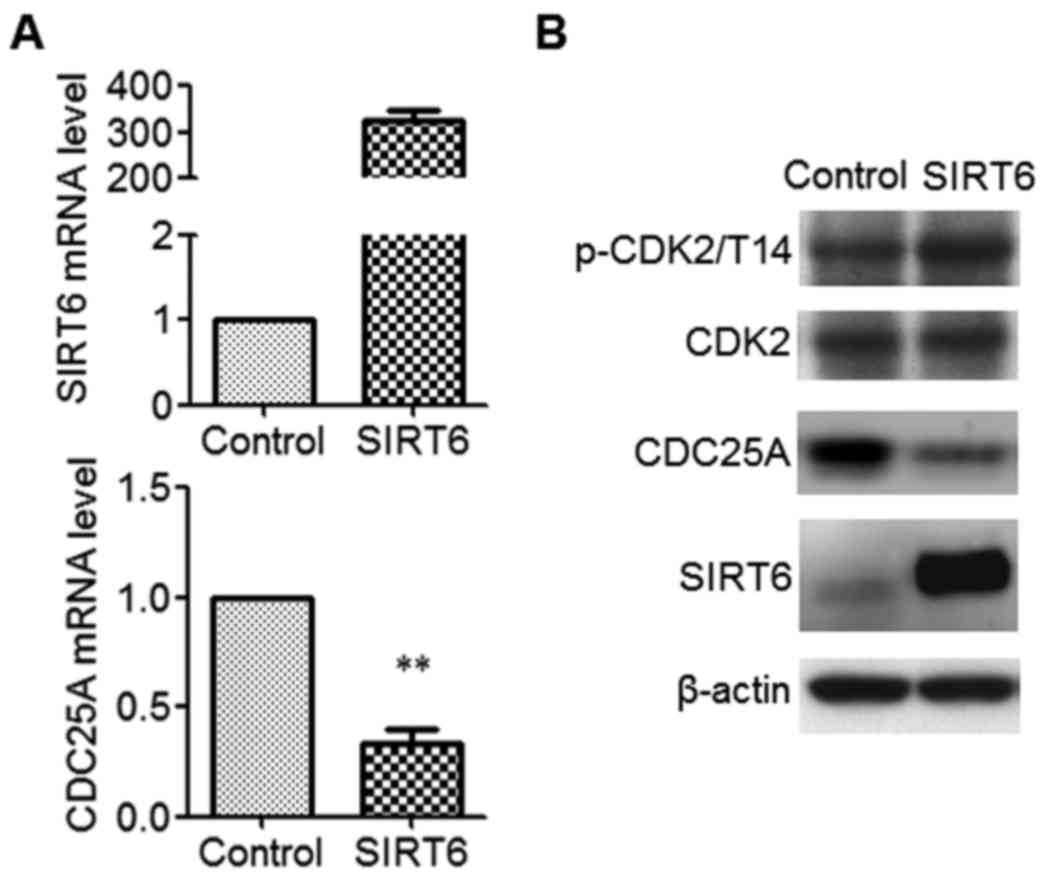
SIRT6 represses CDC25A expression in
SW480 cells
SIRT6 represses its target gene expression (7,8). To
identify the SIRT6 target genes in SW480 CSCs, SIRT6 was expressed
and microarray analysis was performed. Using a 2-fold difference
(upregulation and downregulation) as the cutoff, 120 downregulated
genes were identified following SIRT6 overexpression (data not
shown). CDC25A was among the top 10 significantly downregulated
genes, and is closely associated with cell cycle and tumorigenesis
(16). Therefore, CDC25A was selected
for additional analysis. To confirm the effect of SIRT6 on CDC25A
expression, independent overexpression experiments were performed.
The data indicated that the mRNA level (Fig. 4A) and the protein level (Fig. 4B) of CDC25A was significantly
decreased in SIRT6-expressing cells (P<0.01). Furthermore,
expression of SIRT6 increased the phosphorylation level of
cyclin-dependent kinase 2 (CDK2) at Thr14, a target of CDC25A,
without affecting the protein level of CDK2 (Fig. 4B). These data indicated that SIRT6
inhibits the expression of CDC25A in SW480 CSCs.
SIRT6 decreases the H3K9 acetylation
level at the promoter of CDC25A
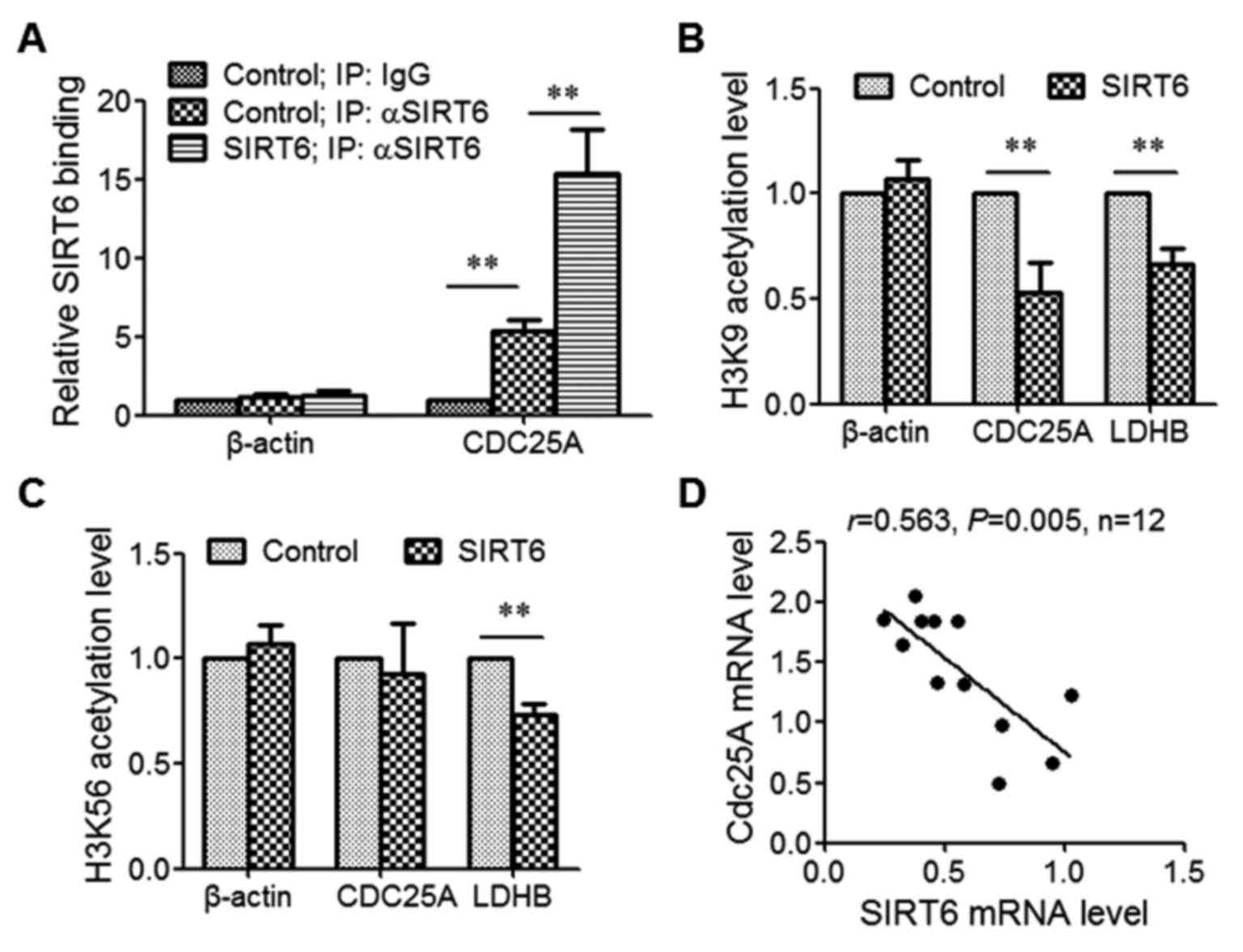
To assess whether SIRT6 regulates CDC25A expression
by directly binding to its promoter, ChIP was performed using SIRT6
antibodies. The results revealed that the level of precipitated
CDC25A promoter was increased in the SIRT6 antibody group compared
with the IgG group, demonstrating that SIRT6 binds to CDC25A
promoter. Overexpression of SIRT6 significantly increased the
binding level of SIRT6 with the CDC25A promoter (Fig. 5A).
SIRT6 deacetylates histones H3 at either lysine 9
(H3K9) or lysine 56 (H3K56) (7,8). To
determine which lysine SIRT6 deacetylated at the CDC25A promoter
region, SIRT6 was overexpressed and ChIP was performed using H3K9
acetylation or H3K56 acetylation antibodies. The data indicated
that SIRT6 expression decreased H3K9 acetylation and H3K56
acetylation at the promoter region of the Lactate dehydrogenase B
gene, which was demonstrated to be a SIRT6 target gene (12,13).
However, SIRT6 expression only decreased the level of H3K9
acetylation, not H3K56 acetylation, at the CDC25A promoter region
(Fig. 5B and C).
Finally, the expression levels of SIRT6 and CDC25A
in CRC tissues were detected. The results suggested that SIRT6
expression was negatively correlated with CDC25A expression in CRC
tissue (r=0.563, P=0.005; Fig.
5D).
Discussion
The tumor-suppressing role of SIRT6 in cancer
progression has been identified previously: Firstly, SIRT6
expression is downregulated in various types of cancer (9–11);
secondly, SIRT6 inhibits tumorigenesis and tumor development in
mouse models (12,13). However, the oncogenic role of SIRT6
has also been described: Sociali et al identified that the
inhibition of SIRT6 sensitizes pancreatic cancer cells to
chemotherapeutics (20). In addition,
SIRT6 was also demonstrated to prevent hepatocellular carcinoma
cell apoptosis by suppressing B cell lymphoma 2-associated X
protein expression (21). Therefore,
the precise role of SIRT6 may be dependent on the various types of
cancer or the specific stages and requires further investigation.
The data of the present study indicated that SIRT6 expression was
reduced in colorectal CSCs. Overexpression of SIRT6 inhibited
colorectal CSCs proliferation and induced G0/G1 cell cycle arrest
in vitro. All these data supported a suppressive role of
SIRT6 in CRC.
SIRT6 may be regulated at transcriptional or
post-transcriptional levels (22).
Ubiquitin-mediated degradation is an important aspect for SIRT6
expression regulation in human cancer (22). Thirumurthi et al demonstrated
that SIRT6 was phosphorylated at Ser (338) by the kinase AKT1,
which mediated the ubiquitination of SIRT6 by Mouse double minute 2
homolog, targeting SIRT6 for protease-dependent degradation in
breast cancer cell lines (22).
Destabilization of SIRT6 thereby promotes tumorigenesis and
trastuzumab resistance in breast cancer. The data of the present
study suggested that the mRNA level of SIRT6 was decreased in
colorectal CSCs, indicating an inhibition of SIRT6 transcription in
colorectal CSCs. Exploring potential repressors of SIRT6
transcription in colorectal CSCs may provide insight on its
expression regulation, and may identify potential targets for CRC
therapy.
Due to the critical role of CDC25A in cell cycle
regulation and cancer development, the protein level of CDC25A
requires strict regulation (15–17). This
may be achieved through multiple mechanisms, including regulating
CDC25A expression at transcriptional, translational or
post-translational levels (23).
CDC25A protein degradation is regulated by a ubiquitin-proteasome
system, and stabilization of CDC25A is one of the major causes for
the upregulation of this protein in cancer (24). Pereg et al (24) revealed that ubiquitin hydrolase
Ubiquitin-specific peptidase 17-like family member 2 (Dub3) reduces
CDC25A degradation by removing its ubiquitin chains, and by
stabilizing CDC25A, Dub3 promotes oncogenic transformation in
breast cancer. Activation of CDC25A transcription also induces the
upregulation of this protein in cancer: Human papillomavirus type
16 E7 oncoprotein was demonstrated to upregulate CDC25A
transcription by disruption of the E2F transcription factor
1-Retinoblastoma protein inhibitory complex (25). Tumor suppressors, conversely, repress
CDC25A transcription. For example, tumor protein 53 was suggested
to activate Activating transcription factor 3, resulting in its
direct binding to the CDC25A promoter and inhibition of CDC25A
transcription (26). The data of the
present study demonstrated that overexpression of SIRT6 in
colorectal CSCs resulted in significantly decreased expression of
CDC25A. The ChIP experiments demonstrated that SIRT6 binds to
CDC25A promoter and reduces H3K9 level, indicating that CDC25A
expression is regulated through an epigenetic mechanism by SIRT6 in
colorectal CSCs. Additional studies on SIRT6 regulation of CDC25A
expression in the development of CRC will increase our
understanding of the regulation of CDC25A expression in cancer.
To conclude, the present study identified that SIRT6
expression is reduced in colorectal CSCs. Furthermore, it was
demonstrated that SIRT6 inhibits cell proliferation and CDC25A
expression in colorectal CSCs. These results indicate a suppressive
role of SIRT6 in the development of CRC, and implicate its
potential application in CRC therapy.
Acknowledgements
The authors would like to thank Professor Xiangwei
Gao from Zhejiang University for providing the plasmid expressing
SIRT6 and for the critical reading of the manuscript.
Funding
The present study was supported by the Natural
Science Foundation of Zhejiang Province (grant no. LY16H040008) and
the Medical Foundation of Zhejiang Province (grant no.
2015KYB405).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WL and JL designed the experiments. WL, MW, HD, XS
and TZ performed the experiments and analysis.
Ethics approval and consent to
participate
The Ethics Committee of the Linyi People's Hospital
approved the present study and written informed consent was
obtained from all patients.
Consent for publication
Written informed consent was obtained from all
patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
O'Brien CA, Pollett A, Gallinger S and
Dick JE: A human colon cancer cell capable of initiating tumour
growth in immunodeficient mice. Nature. 445:106–110. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ricci-Vitiani L, Lombardi DG, Pilozzi E,
Biffoni M, Todaro M, Peschle C and De Maria R: Identification and
expansion of human colon-cancer-initiating cells. Nature.
445:111–115. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Michishita E, Park JY, Burneskis JM,
Barrett JC and Horikawa I: Evolutionarily conserved and
nonconserved cellular localizations and functions of human SIRT
proteins. Mol Biol Cell. 16:4623–4635. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhong L, D'Urso A, Toiber D, Sebastian C,
Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD,
Nir T, et al: The histone deacetylase Sirt6 regulates glucose
homeostasis via Hif1alpha. Cell. 140:280–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Toiber D, Erdel F, Bouazoune K, Silberman
DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor
B, Giacosa S, et al: SIRT6 recruits SNF2H to DNA break sites,
preventing genomic instability through chromatin remodeling. Mol
Cell. 51:454–468. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Min L, Ji Y, Bakiri L, Qiu Z, Cen J, Chen
X, Chen L, Scheuch H, Zheng H, Qin L, et al: Liver cancer
initiation is controlled by AP-1 through SIRT6-dependent inhibition
of survivin. Nat Cell Biol. 14:1203–1211. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Khongkow M, Olmos Y, Gong C, Gomes AR,
Monteiro LJ, Yagüe E, Cavaco TB, Khongkow P, Man EP, Laohasinnarong
S, et al: SIRT6 modulates paclitaxel and epirubicin resistance and
survival in breast cancer. Carcinogenesis. 34:1476–1486. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lai CC, Lin PM, Lin SF, Hsu CH, Lin HC, Hu
ML, Hsu CM and Yang MY: Altered expression of SIRT gene family in
head and neck squamous cell carcinoma. Tumour Biol. 34:1847–1854.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sebastián C, Zwaans BM, Silberman DM,
Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber
D, et al: The histone deacetylase SIRT6 is a tumor suppressor that
controls cancer metabolism. Cell. 151:1185–1199. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kugel S, Sebastian C, Fitamant J, Ross KN,
Saha SK, Jain E, Gladden A, Arora KS, Kato Y, Rivera MN, et al:
SIRT6 suppresses pancreatic cancer through control of Lin28b. Cell.
165:1401–1415. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Van Meter M, Mao Z, Gorbunova V and
Seluanov A: SIRT6 overexpression induces massive apoptosis in
cancer cells but not in normal cells. Cell Cycle. 10:3153–3158.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Blomberg I and Hoffmann I: Ectopic
expression of Cdc25A accelerates the G(1)/S transition and leads to
premature activation of cyclin E- and cyclin A-dependent kinases.
Mol Cell Biol. 19:6183–6194. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Boutros R, Lobjois V and Ducommun B: CDC25
phosphatases in cancer cells: Key players? Good targets? Nat Rev
Cancer. 7:495–507. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Galaktionov K, Lee AK, Eckstein J, Draetta
G, Meckler J, Loda M and Beach D: CDC25 phosphatases as potential
human oncogenes. Science. 269:1575–1577. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Otto T and Sicinski P: Cell cycle proteins
as promising targets in cancer therapy. Nat Rev Cancer. 17:93–115.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sociali G, Galeno L, Parenti MD, Grozio A,
Bauer I, Passalacqua M, Boero S, Donadini A, Millo E, Bellotti M,
et al: Quinazolinedione SIRT6 inhibitors sensitize cancer cells to
chemotherapeutics. Eur J Med Chem. 102:530–539. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ran LK, Chen Y, Zhang ZZ, Tao NN, Ren JH,
Zhou L, Tang H, Chen X, Chen K, Li WY, et al: SIRT6 overexpression
potentiates apoptosis evasion in hepatocellular carcinoma via
BCL2-associated X protein-dependent apoptotic pathway. Clin Cancer
Res. 22:3372–3382. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Thirumurthi U, Shen J, Xia W, LaBaff AM,
Wei Y, Li CW, Chang WC, Chen CH, Lin HK, Yu D and Hung MC:
MDM2-mediated degradation of SIRT6 phosphorylated by AKT1 promotes
tumorigenesis and trastuzumab resistance in breast cancer. Sci
Signal. 7:ra712014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dozier C, Mazzolini L, Cenac C, Froment C,
Burlet-Schiltz O, Besson A and Manenti S: CyclinD-CDK4/6 complexes
phosphorylate CDC25A and regulate its stability. Oncogene.
36:3781–3788. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pereg Y, Liu BY, O'Rourke KM, Sagolla M,
Dey A, Komuves L, French DM and Dixit VM: Ubiquitin hydrolase Dub3
promotes oncogenic transformation by stabilizing Cdc25A. Nat Cell
Biol. 12:400–406. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Katich SC, Zerfass-Thome K and Hoffmann I:
Regulation of the Cdc25A gene by the human papillomavirus Type 16
E7 oncogene. Oncogene. 20:543–550. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Demidova AR, Aau MY, Zhuang L and Yu Q:
Dual regulation of Cdc25A by Chk1 and p53-ATF3 in DNA replication
checkpoint control. J Biol Chem. 284:4132–4139. 2009. View Article : Google Scholar : PubMed/NCBI
|