Introduction
Lung cancer has a high incidence of tumor recurrence
and metastasis, and is the most common cause of cancer-associated
morality worldwide (1). The overall
5-year survival rate among patients with lung cancer is <15%,
and a high rate of metastasis is the primary cause of lung
cancer-associated mortality (2). A
previous study confirmed that differentially expressed in
adenocarcinoma of the lung (DAL-1), a protein that belongs to the
membrane-associated cytoskeleton protein 4.1 family, is an
efficient suppressor of epithelial-mesenchymal transition (EMT) in
lung cancer (3). EMT is a pivotal
event in lung cancer metastasis progression (4–6). However,
the regulators of DAL-1 in EMT remain unknown.
Recently, a novel batch of endogenous small
non-coding regulatory RNAs (microRNA's; miRNA's) have received
attention in the development of cancer, miRNA's bind complementary
sequences in target mRNAs, resulting in selective degradation or
selective inhibition of their translation (7). The present study investigated the
possible miRNAs of DAL-1 using bioinformatic assays, which included
miR-26b, miR-26a, miR-96 and miR-223. Among them, the regulation of
DAL-1 by miR-223 has been demonstrated to serve a role in gastric
cancer (8). The present study
revealed that the gene silencing of DAL-1 induced annexin A1
(ANXA1) protein expression levels to increase in H460 cells. ANXA1
belongs to a family of calcium/phospholipid-binding and actin
regulatory proteins (9).
Dysregulation of ANXA1 has been reported in numerous types of
neoplasm, suggesting that this protein may serve important roles in
tumor development and progression (10). The present study aimed to profile the
expression and function of miR-26a in the development of lung
cancer and to verify whether miR-26a is the regulator of DAL-1.
Materials and methods
Candidate miRNAs of DAL-1 predicted
using bioinformatics
microRNA.org (http://www.microrna.org/microrna/home.do) (11), Target Scan (http://www.targetscan.org/mamm_31/) (12) and PicTar (https://www.mdc-berlin.de/10440258/en/research/research_teams/systems_biology_of_gene_regulatory_elements/projects/pictar)
(13) softwares were used in order to
determine candidate miRNAs regulating DAL-1 expression in lung
cancer cells.
Tissue samples and cell lines
A total of 9 non-small cell lung cancer (NSCLC)
tissues and matched normal tissues were obtained from the
Department of Thoracic Surgery, The First Affiliated Hospital of
Guangzhou Medical University (Guangzhou, China). The aforementioned
lung cancer cell lines: PAa, A549, H460, 95D, H1299, H520, PLAM,
H446 and the normal lung bronchus epithelial immortalized cell line
16HBE sourced from the Central Laboratory of Guangzhou Medical
University, were cultured in RPMI-1640 basic medium (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% fetal bovine serum (FBS; Gibco, Thermo Fisher Scientific, Inc.)
at 37°C with 5% CO2.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted by TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The RNA yield
and the ratio of absorbance at 260 to 280 nm (260/280 ratio) was
measured using the Nano Drop 2000 Spectrophotometer (Nano Drop
Technologies; Thermo Fisher Scientific, Inc., Wilmington, DE, USA).
1 µg RNA was reverse transcribed in a final volume of 20 µl [4 µl
5X Prime Script Buffer (for quantitative, 1 µl Prime Script RT
Enzyme Mix I, 1 µl Oligo dT Primer (50 µM), 1 µl Random 6 mers (100
µM) and RNase Free dH2O up to 20 µl)] using the Prime
Script® RT reagent kit (cat no. DRR037A; Takara Bio,
Inc., Otsu, Japan) with random hexamers according to the
manufacturer's protocol. The reverse transcription reaction
condition was as follows: 37°C for 15 min, 85°C for 5 sec and 4°C
for 2 min. RT-qPCR was performed using ABI 7500 Real-Time PCR
Systems (Applied Biosystems; Thermo Fisher Scientific, Inc.) and
SYBR Green® Premix Ex Taq™ II kit (cat no.
DRR081A; Takara Bio, Inc.). GAPDH was used as an internal control.
The specific PCR prime sequences of the genes designed by DNAClub
software (Xiongfong Chen, Cornell University, Ithaca NY, USA) were
as follows: DAL-1, forward 5′-acctgtgatgtagagaaacgctcc-3′ and
reverse 5′-agtgccaagcaccacttcga-3′. All primers were identified
using the Gen Bank database (14)
with the National Centre for Biotechnology Information blastn
program (15) to ensure sequence
specificity. The thermocycling conditions were as follows: 95°C for
30 sec, followed by 40 cycles at 95°C for 5 sec and 60°C for 34
sec. Specificity of amplification products was confirmed by melting
curve analysis. miRNA were extracted using a mirVana miRNA
Isolation kit (Ambion; Thermo Fisher Scientific, Inc.). Specific RT
primers and Taqman probe (Applied Biosystems; Thermo Fisher
Scientific, Inc.) were used for the quantitative detection
(16) of miR-26a (cat no. 4427012)
and reference gene U6 (cat no. 4426961).
Plasmids
The miR-26a mimic/inhibitor and the control were
obtained from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The
sequence for miR-26a used is as follows:
5′-UUCAAGUAAUCCAGGAUAGGCU-3′. Takara LA Taq® (cat no.
RR02MQ; Takara Bio, Inc.) was used to amplify the 3′UTR of DAL-1.
The PCR thermocycler conditions were as follows: Pre-denaturation
at 94°C for 5 min, followed by 30 cycles at 98°C for 10 s and 68°C
for 100 s and posterior extension 72°C for 10 min. The 3′UTR of
DAL-1 was amplified using the following primers: Sense
5′-CCGCTCGAGCCAGAGGAATAACTTAGCTTGCACATG-3′ and antisense
5′-ATAAGAATGCGGCCGCCAGTGCATTTGCCATTTTTATTTCG-3′. The PCR product
was inserted into psiCHCK2 within XhoI and NotI
restriction sites (Promega Corporation, Madison, WI, USA). The
mutation experiment was performed using a KOD-Plus-mutagenesis kit
(Toyobo Co., Ltd., Osaka, Japan).
Cells culture and transfection
A549, H460 lung cancer cell lines and the human
embryonic kidney cell line HEK 293T (supplied by the Central
Laboratory of Guangzhou Medical University, Guangzhou, China), were
propagated in RPMI-1640 basic medium supplemented with 10% FBS. The
cells were maintained at 37°C in a humidified atmosphere containing
5% CO2. Transfection was performed using Lipofectamine
3000 Reagent® (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's instructions. A final
concentration of 50 nM miR-26a mimic or 150 nM miR-26a inhibitor of
their respective negative controls (NCs) were used for each
transfection in proliferation, migration, invasion and wound
scratch healing experiments. A further 2.5 µg DAL-1 short hairpin
RNA (shRNA) was used for transfection in a two-dimensional gel
electrophoresis assay. The mock group in this study is the cell
that only transfected with Lipofectamine® 3000.
Western blot analysis
Total protein samples were extracted from primary
tissues or cells using a lysis buffer (Beyotime Institute of
Biotechnology, Haimen, China) supplemented with phenylmethyl
sulfonyfluoride (PMSF). Protein quantification was performed using
a BCA protein content detection kit (cat no. KGP902; Nanjing Key
Gen Biotech Co., Ltd., Nanjing, China). The protein samples were
mixed with 6X SDS-PAGE loading buffer (cat no. P0015F; Beyotime
Institute of Biotechnology) and denatured, and separated using 10%
SDS-PAGE (30 µg protein was loaded per lane) prior to being
transferred to nitrocellulose membranes. The membranes were blocked
with 5% non-fat milk for 1 h at room temperature, prior to being
incubated with an anti-DAL-1 antibody (cat no. ab154071; Abcam,
Cambridge, UK; 1:800 dilution), an anti-ANXA1 antibody (cat no.
32934S; Cell Signaling Technology, Inc., Danvers, MA, USA; 1:1,000
dilution) or an anti-GAPDH antibody (cat no. 2118; Cell Signaling
Technology, Inc.; 1:1,000 dilution) overnight at 4°C. Protein
signals were quantified following incubation with an anti-rabbit
immunoglobulin (Ig)G, horseradish peroxidase (HRP)-linked secondary
antibody (cat no. 7074P2; Cell Signaling Technology, Inc.; 1:1,000
dilution) or anti-mouse IgG, HRP-linked secondary antibody (cat no.
7076P2; Cell Signaling Technoogy, Inc.; 1:1,000 dilution) for 1 h
at room temperature, conjugated with peroxidase using the Super
Signal® West Pico Chemiluminescent Substrate (cat no.
PH203837; Thermo Fisher Scientific, Inc.). The images of the
proteins were obtained and analyzed by the Fusion Solo 6s
Multifunctional Gel Imaging System (Vilber Lourmat, Marne
Le-Vallée, France).
Cell proliferation assay
Cell proliferation was indirectly assayed using the
Cell Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology,
Haimen, China), which stains living cells and this was carried out
according to the manufacturer's instructions. A total of 5,000
cells per well were seeded in 96-well plates and transfected with
the mimic (with a final concentration of 50 nM), inhibitor (with a
final concentration of 150 nM) or their respective NC the following
day, and incubated for 24 h at 37°C. The CCK-8 solution was used to
determine viability once every 24 h following transfection, for 7
days. The absorbance of each well was measured with a microplate
reader set at 450 nm. All experiments were performed in
triplicate.
Cell migration and invasion
assays
Cell migration and invasion was measured using a
Transwell chamber assay (EMD Millipore, Billerica, MA, USA) with
and without Matrigel (BD Biosciences, San Jose, CA, USA). For the
invasion assay, 1×105 transfected cells were plated into
the upper chambers coated with Matrigel. In the assays, cells were
cultured in RPMI-1640 in the upper chambers, and 500 µl 10%
FBS-RPMI-1640 was added to the lower chamber. Following 24 h of
incubation at 37°C, migrated cells were fixed with absolute
methanol for 30 min at room temperature. Non-migrated cells on the
upper chamber were removed using cotton swabs. Cells on the bottom
surface of the membrane were subsequently stained with 0.5% crystal
violet for 20 min. For each group, three cell images were randomly
obtained using an inverted microscope, using a magnification of
×100.
In vitro wound healing assay
A cross-hair pattern was marked on the outside
surface of the undersides of the 6-well culture plates. Cells were
plated in triplicate so as to reach 100% confluence after 1 day.
Subsequently an ~2 cm-long and ~0.5 mm-wide wound was made by
dragging a plastic 10 µl pipette-tip along a straight edge with
moderate and consistent pressure, to scratch the cell monolayer.
Following this, the culture medium was replaced immediately. Cells
were imaged immediately and one day subsequent to wounding, at ×50
magnification at identical locations along the scratches.
Dual-luciferase reporter assay
The target sequence of DAL-1 3′UTR was cloned into
the dual-luciferase reporter vector, check-2 (Promega Corporation,
Madison, WI, USA) HEK-293T cells were plated into 48-well plates
(~2×104 cell per well) with ~50–60% confluency, 24 h
prior to transfection. A mixture of 50 nM miR-26a mimic and 200 ng
CHEK-DAL-1 reporter vector, was transfected into cells using the
Lipofectamine® 3000 reagent. Following 48 h, luciferase
activity levels were determined using a dual-luciferase reporter
assay system (Promega Corporation) and normalized by dividing
firefly luciferase by Renilla luciferase, according to the
manufacturer's instructions. Each transfection was performed in
triplicate.
Two-dimensional gel electrophoresis
assay
This experiment was performed as previously detailed
(17). A mock group was used where
cells were transfected with Lipofectamine® 3000 reagent
alone. An NC group was used where cells were transfected with
Lipofectamine® 3000 reagent and shRNA-NC. Finally an
untreated group was used where cells were untreated.
Statistical analysis
All experiments were repeated three times. All data
are presented as the mean ± standard deviation, and were analyzed
using SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA). One-way
analysis of variance with a Least Significant Difference t-test was
used to determine the statistical significance of the differences
of miR-26a expression among nine lung cancer cell lines and a 16HBE
cell line. A two-tailed Student's t-test was used to determine the
statistical significance of the differences. P<0.05 was
considered to indicate a statistically significant difference.
Results
Candidate miRNAs of DAL-1 predicted by
bioinformatics
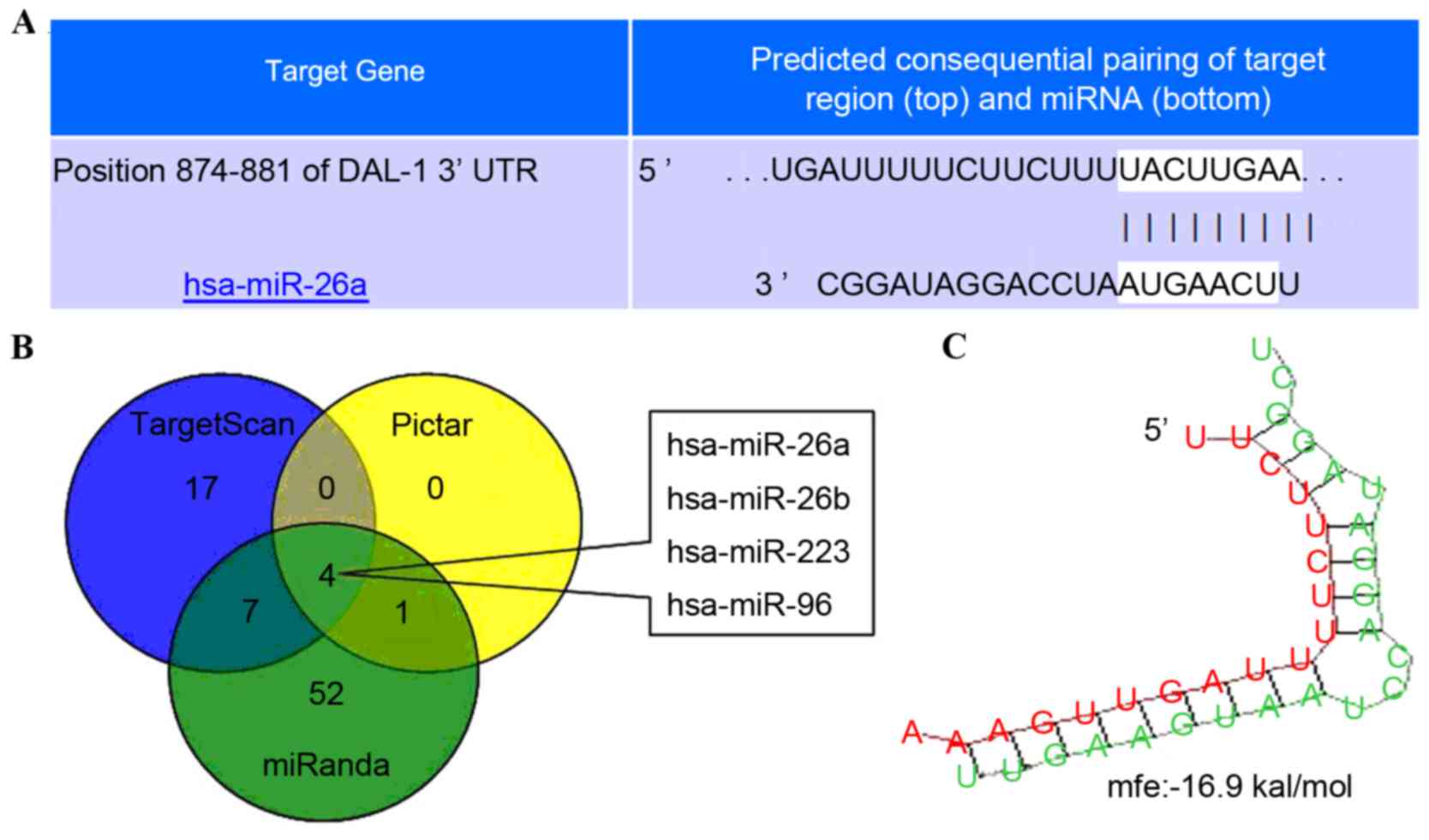
microRNA.org, Target Scan and Pic Tar
softwares were used to determine the candidate miRNA regulating
DAL-1 expression in lung cancer cells. The predicted results
revealed that miR-26a, miR-26b, miR-223 and miR-96 are able to
regulate DAL-1 by combining with DAL-1 mRNA 3′UTR (Fig. 1A-C).
Expression of miR-26a in lung cancer
tissues and cell lines
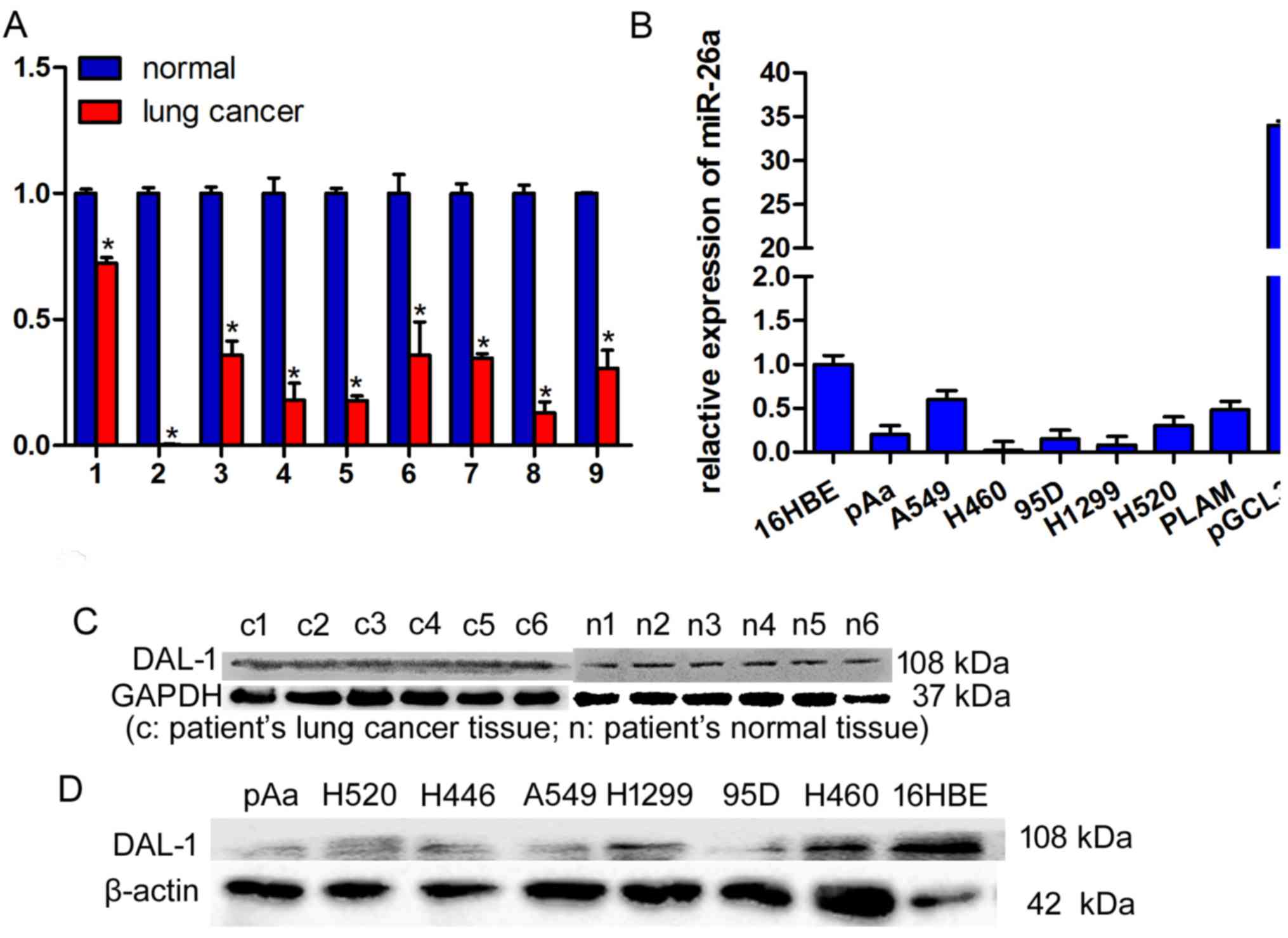
The expression levels of miR-26a in nine lung cancer
tissues, nine matched adjacent normal lung tissues and various lung
cancer cells, were determined by RT-qPCR. miR-26a expression was
significantly decreased in lung cancer tissues compared with in
normal tissues (Fig. 2A; P<0.05).
Expression levels of miR-26a in eight lung cancer cell lines,
including PAa, A549, H460, 95D, H1299, H520, PLAM and H446, was
significantly decreased, compared with in 16 HBE cells (Fig. 2B).
Expression levels of DAL-1 in lung
cancer tissues and cell lines
Expression levels of DAL-1 protein in lung cancer
tissues, matched adjacent normal lung tissues and various lung
cancer cells, were evaluated using western blot analysis. DAL-1
protein expression levels demonstrated a decrease in lung cancer
tissues compared with in normal lung tissues (Fig. 2C). Additionally, expression levels of
DAL-1 in lung cancer cells significantly decreased compared with 16
HBE cells (Fig. 2D; *P<0.05).
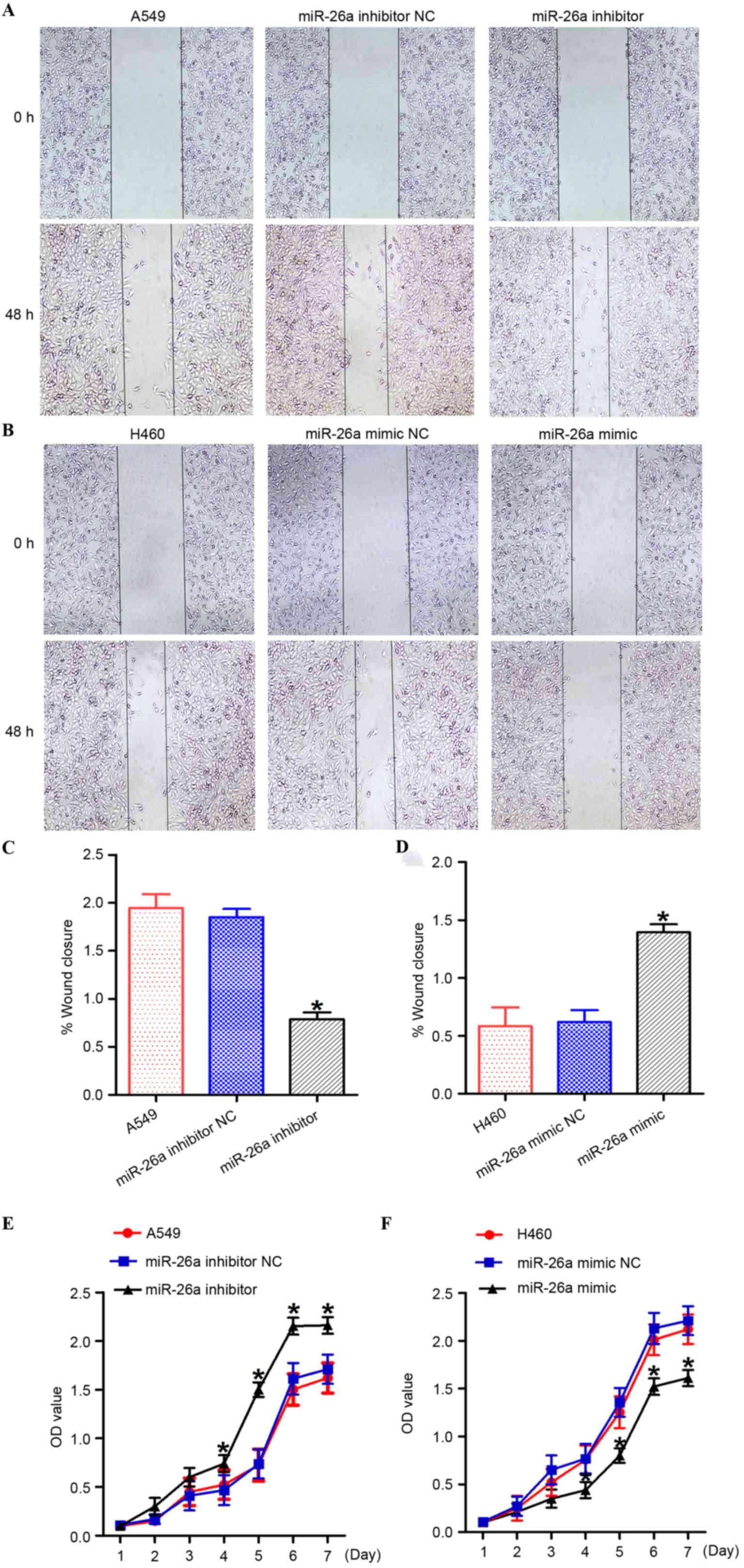
miR-26a suppress lung cancer cell
growth in vitro
In order to investigate the function of miR-26a in
the regulation of lung cancer cell growth, A549 cells were
transfected with the miR-26a inhibitor/NC, and H460 cells were
transfected with the miR-26a mimic/NC (Fig. 3). Suppression of miR-26a by the
miR-26a inhibitor significantly promoted the growth of A549 cells
(Fig. 3E; P<0.05). However,
overexpression of miR-26a induced by the miR-26a mimic
significantly inhibited the growth of H460 cells (Fig. 3F; P<0.05).
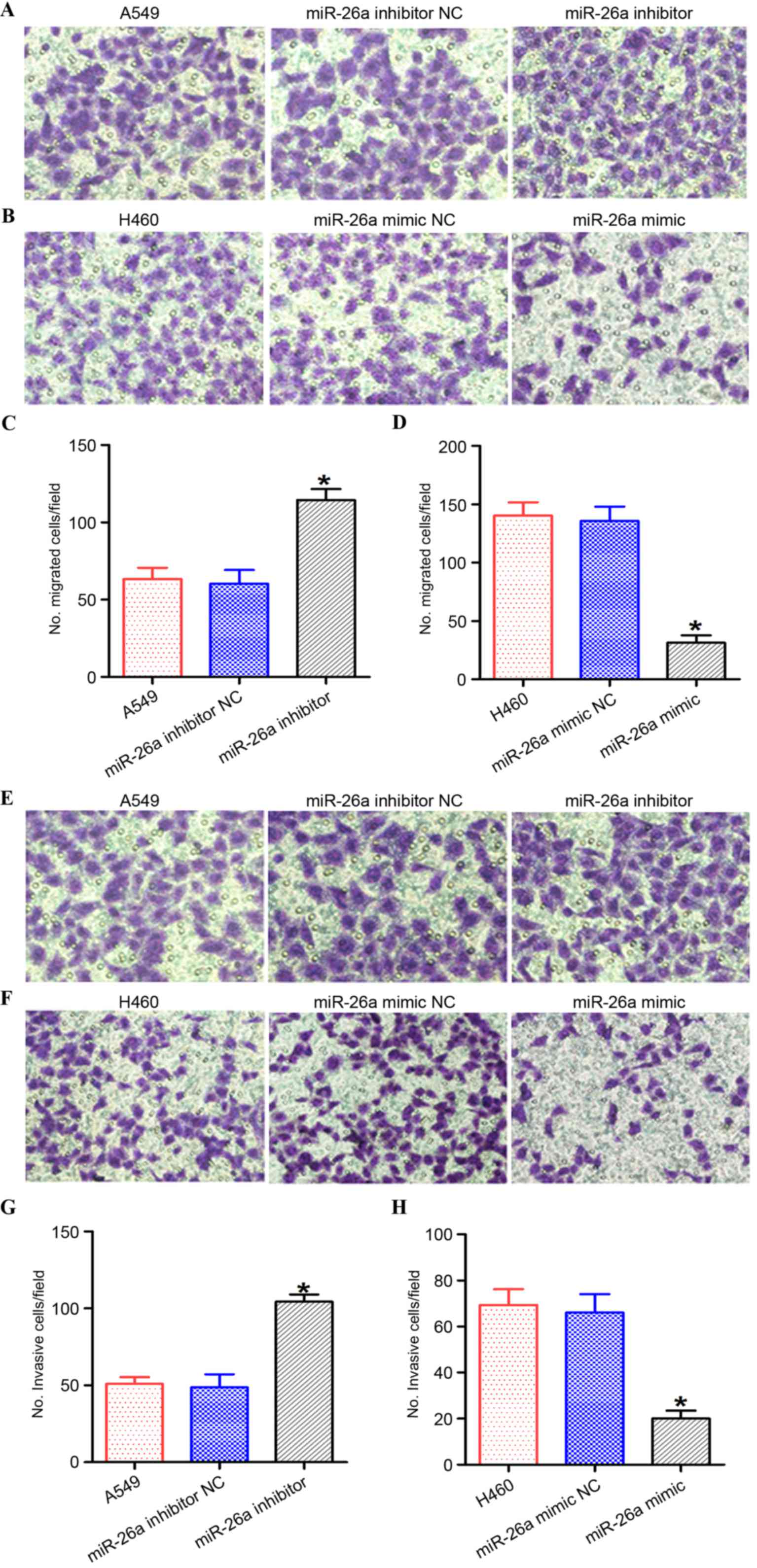
miR-26a suppress lung cancer cell
migration and invasion
In order to investigate the effect of miR-26a on the
motility of lung cancer cells, the miR-26a inhibitor/NC was
transfected into A549 cells, and the miR-26a mimic/NC was
transfected into H460 cells (Figs. 3
and 4). Inhibition of miR-26a
promoted the migration and invasion capabilities of A549 cells
compared with the negative control cells (Figs. 3A and B, and 4A, B, E and F; P<0.05). However,
overexpression of miR-26a suppressed the migration and invasion
capabilities of lung cancer cells compared with the negative
control cells (Figs. 3C and D, and
4C, D, G and H; P<0.05).
miR-26a is negatively associated with
DAL-1 in A549 and H460 cells
The expression levels of DAL-1 and miR-26a in lung
cancer cells were determined and the results revealed that the
DAL-1 protein expression level in A549 cells was low, compared with
H460 cells (Fig. 2C). The miR-26 mRNA
expression level in A549 cells was higher compared with H460 cells
(Fig. 2B).
DAL-1 is not a direct target of
miR-26a in lung cancer cells
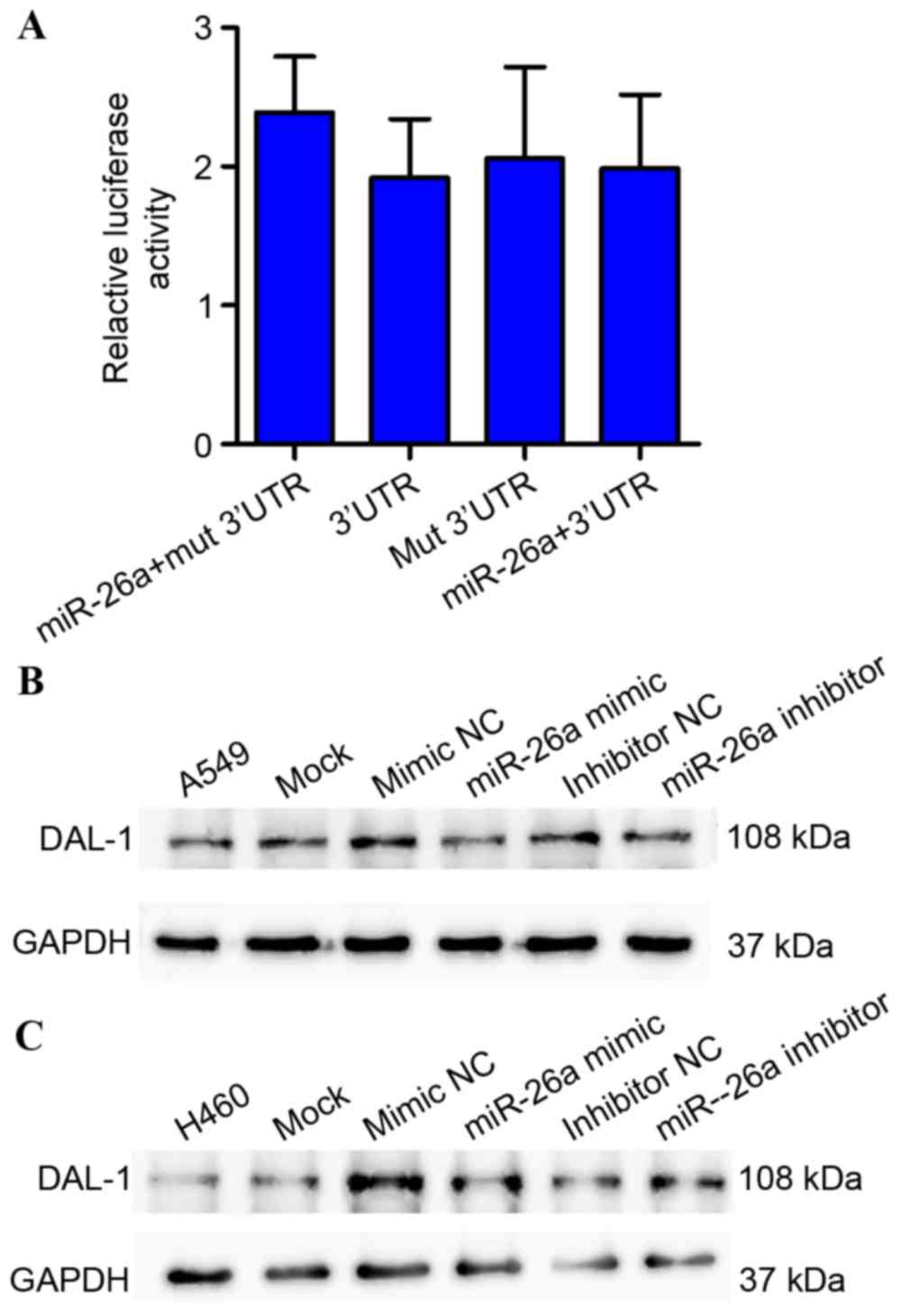
Relative luciferase activity assays demonstrated
that miR-26a may not inhibit the luciferase activity of the wild
type or mutation (Mut)-3′UTR of DAL-1 in 293T cells (Fig. 5A). In addition, inhibition or
overexpression of miR-26a does not alter DAL-1 expression levels
(P>0.05; Fig. 5B and C). This
result suggests that miR-26a is not the direct regulator of
DAL-1.
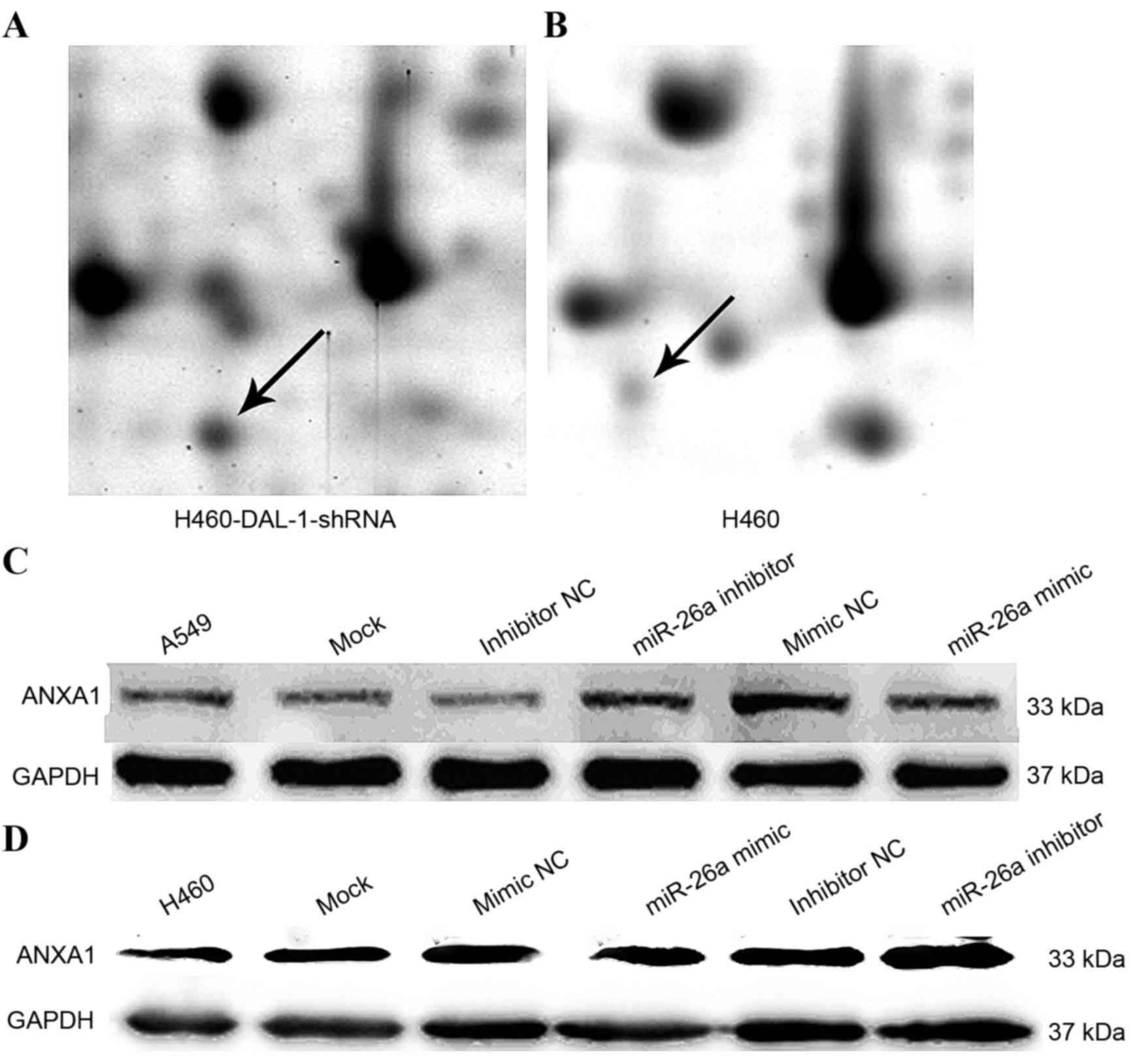
ANXA1 is a DAL-1-associated protein
regulated by miR-26a
The results of two-dimensional gel electrophoresis
assays indicated that the ANXA1 protein expression level in lung
cancer was significantly increased (P<0.05) following DAL-1
silencing by shRNA compared with the control (Fig. 6A and B). In addition, compared with
H460-miR-26a mimic NC group, the overexpression of miR-26a
decreased the expression level of ANXA1; and compared with
A549-miR-26a inhibitor NC group, the inhibition of miR-26a
increased the expression level of ANXA1 (Fig. 6C and D).
Discussion
The initiation of lung cancer is a complicated
process with numerous genes and stages of development (18). A previous study demonstrated that
DAL-1 is a fundamental tumor suppressor gene and serves an
important role in the development of lung cancer (19). A previous study revealed that the
expression level of DAL-1 was decreased in lung cancer tissues and
the silencing of the DAL-1 gene may enhance the growth, migration
and invasion capabilities of lung cancer cells (20). DAL-1 directly associates with the
E-cadherin promoter and the subsequent regulation of its expression
may induce impairment of EMT and decrease cell migration and
invasion (3). The present study
utilized shRNA to silence the DAL-1 gene by using a two-dimensional
gel electrophoresis assay to identify DAL-1 associated proteins.
There are five main upregulated proteins (stathmin, ANXA1,
chaperonin containing TCP1 subunit 2, heterogeneous nuclear
ribonucleoprotein H1 and moesin) and four main downregulated
proteins (peroxredoxin I, peroxredoxin II, peroxredoxin III and
mitochondrial antioxidant manganese superoxide dismutase). DAL-1
may regulate the expression level of ANXA1 protein and participate
in the progress of lung cancer, as demonstrated by the results of
the present study. However, the detailed role of DAL-1 in lung
cancer remains poorly understood.
Previously, tumor suppressive miR-26a has been
studied in numerous types of human cancer, and it was revealed to
serve a key role as a cancer suppressor gene in liposarcoma
(21), thyroid carcinoma (22) and breast cancer (23). miR-26a was demonstrated to be
upregulated in glioma, and may enhance cancer cell growth and
colony formation (24). The function
of miR-26a in lung cancer remains to be elucidated. miR-26a was
selected in the present study for investigation as it was suggested
that it may combine with DAL-1 mRNA 3′UTR and regulate DAL-1 gene
expression, according to the bioinformatics predictions presented
in Fig. 1.
The present study investigated the function of
miR-26a in the progression of lung cancer. The validation
experiments revealed that the mature form of miR-26a was
downregulated in lung cancer tissues and lung cancer cells. The
biological behavior-inhibitory effects of miR-26a on two NSCLC cell
lines, A549 and H460, were also investigated. In vitro
experiment results demonstrated that miR-26a significantly
inhibited the proliferation, migration and invasion capabilities of
NSCLC cells. These results suggest that miR-26a serves an important
role in NSCLC and may be exploited for adjuvant therapeutic
application.
Furthermore, to confirm the association between the
expression levels of miR-26a and DAL-1 in lung cancer tissues and
cells, the expression of DAL-1 was decreased in lung cancer tissues
and various lung cancer cell lines. This result indicated that a
decreased expression level of DAL-1 may promote the development of
lung cancer. Furthermore, the present study utilized shRNA to
silence the DAL-1 gene using a two-dimensional gel electrophoresis
assay to identify DAL-1 associated proteins. Congruent with a
previous study (25), it was revealed
that DAL-1 may regulate the expression levels of ANXA1 protein, and
ANXA1 may also participate in the lung cancer process.
It has previously been revealed that the expression
level of ANXA1 protein is decreased or not present in head and neck
cancer (26), esophageal squamous
cell carcinoma (27) and prostate
cancer (28), whereas it was revealed
to be upregulated in breast cancer (25), glioma (29) and oropharyngeal cancer (30). The molecular mechanisms underlying
ANXA1 modulation of cellular responses have not been fully
determined. The previous study revealed that ANXA1 upregulation in
lung cancer, and suppressing ANXA1 gene expression by the
lentiviral vector LV-ANXA1-RNAi, may inhibit the proliferation,
migration and invasion capabilities of lung cancer cells (31).
The present study demonstrated an inverse
correlation between the expression levels of miR-26a and DAL-1 in
A549 and H460 cells. However, the relative luciferase activity in
293T cells co-transfected with miR-26a mimics did not reveal a
significant decrease following 48 h or subsequent to transfection
with the miR-26a mimics or inhibitor, significant changes were also
not observed in DAL-1 protein levels. These results suggest that
DAL-1 may not be the target gene of miR-26a. However, the present
study demonstrated that ANXA1 protein levels were decreased
following transfection of the miR-26a mimic into A549 and H460
cells, and increased subsequent to transfection with the miR-26a
inhibitor.
miR-26a is decreased in numerous types of human
cancer and has anti-oncogenic roles. A previous study demonstrated
that miR-26a targets the bone morphogenetic protein (BMP)/mothers
against decapentaplegic homolog 4 pathway and regulated
neovascularization (32). miR-26a was
revealed to suppress enhancer of zest homolog 2 (33), myeloid cell leukemia sequence 1
(23) and fibroblast growth factor 9
(34), which inhibited cancer cell
migration and invasion. miR-26a regulated the proliferation and
growth of cancer cells by targeting chromodomain helicase DNA
binding protein 1 (35), growth
regulation by estrogen in breast cancer 1 (35) and β-catenin (36). A number of studies demonstrated that
the expression levels of miR-26a decreased in various types of
tumor (23,37,38),
whereas ANXA1 protein was overexpressed (39–41).
Combined with the results of the present study suggesting that the
miR-26a mimic may reduce the expression levels of ANXA1, a negative
correlation was identified between miR-26a and ANXA1 in numerous
tumors. Furthermore, the present study revealed that overexpression
of miR-26a or suppression of ANXA1 may inhibit the ability of
growth, invasion and migration of lung cancer cells. The
aforementioned results suggest that miR-26a may regulate ANXA1 in
lung cancer.
In conclusion, miR-26a expression was decreased in
lung cancer and the altered expression level of miR-26a may affect
various biological processes in lung cancer cells, including
proliferation, migration and invasion. miR-26a regulated the
expression levels of ANXA1 and not DAL-1. miR-26a may function as a
biomarker for lung cancer, providing a novel strategy for the
treatment of lung cancer.
Acknowledgements
The authors would like to thank Dr. Ruobing Xu for
the two-dimensional gel electrophoresis assay experiment.
References
|
1
|
Boyero L, Sánchez-Palencia A, Miranda-Leon
MT, Hernandez-Escobar F, Gómez-Capilla JA and Fárez-Vidal ME:
Survival, classifications, and desmosomal plaque genes in non-small
cell lung cancer. Int J Med Sci. 10:1166–1173. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen X, Guan X, Zhang H, Xie X, Wang H,
Long J, Cai T, Li S, Liu Z and Zhang Y: DAL-1 attenuates
epithelial-to mesenchymal transition in lung cancer. J Exp Clin
Cancer Res. 34:32015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: At the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Floor S, van Staveren WC, Larsimont D,
Dumont JE and Maenhaut C: Cancer cells in epithelial-to-mesenchymal
transition and tumor-propagating-cancer stem cell: Distinct,
overlapping or same populations. Oncogene. 30:4609–4621. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shih JY and Yang PC: The EMT regulator
slug and lung carcinogenesis. Carcinogenesis. 32:1299–1304. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang J, Liu Y, Liu Z, Wang XM, Yin DT,
Zheng LL, Zhang DY and Lu XB: Differential expression profiling and
functional analysis of microRNAs through stage I–III papillary
thyroid carcinoma. Int J Med Sci. 10:585–592. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li X, Zhang Y, Zhang H, Liu X, Gong T, Li
M, Sun L, Ji G, Shi Y, Han Z, et al: miRNA-223 promotes gastric
cancer invasion and metastasis by targeting tumor suppressor
EPB41L3. Mol Cancer Res. 9:824–833. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lim LH and Pervaiz S: Annexin 1: The new
face of an old molecule. FASEB J. 21:968–975. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hongsrichan N, Rucksaken R, Chamgramol Y,
Pinlaor P, Techasen A, Yongvanit P, Khuntikeo N, Pairojkul C and
Pinlaor S: Annexin A1: A new immunohistological marker of
cholangiocarcinoma. World J Gastroenterol. 19:2456–2465. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: Targets and exression. Nucleic
Acids Res. 36:D149–D153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective miroRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar
|
|
13
|
Anders G, Mackowiak SD, Jens M, Maaskola
J, Kuntzagk A, Rajewsky N, Landthaler M and Dieterich C: doRiNA: A
database of RNA interactions in post-transcriptional regulation.
Nucleic Acids Res. 40:D180–D186. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Benson DA, Karsch-Mizrachi I, Lipman DJ,
Ostell J and Wheeler DL: GenBank. Nucleic Acids Res. 33:D34–D38.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Altschul SF, Gish W, Miller W, Myers EW
and Lipman DJ: Basic local alignment search tool. J Mol Biol.
215:1–410. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu Y, Zhou J, Zhang X, Zheng X, Jiang X,
Shi L, Yin W and Wang J: Optimized sample preparation for
two-dimensional gel electrophoresis of soluble proteins from
chicken bursa of fabricius. Proteome Sci. 7:382009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao X, Wang N, Zhang M, Xue S, Shi K and
Chen Z: Detaction of methylation of the RAR-β gene in patients with
non-small cell lung cancer. Oncol Lett. 3:654–658. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tran YK, Bögler O, Gorse KM, Wieland I,
Green MR and Newsham IF: A novel member of the NF2/ERM/4.1
superfamily with growth suppressing properties in lung cancer.
Cancer Res. 59:35–43. 1999.PubMed/NCBI
|
|
20
|
Xu RB, Zhang YJ and Guo AL: Primary survey
of effects of DAL-1 in procession of proliferation, migration and
invasion of NSCLC cell line. Chin J Cancer Prev Treat.
16:1139–1143. 2009.(In Chinese).
|
|
21
|
Lee DH, Amanat S, Goff C, Weiss LM, Said
JW, Doan NB, Sato-Otsubo A, Ogawa S, Forscher C and Koeffler HP:
Overexpression of miR-26a-2 in human liposarcoma is correlated with
poor patient survival. Oncogenesis. 2:e472013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lv M, Zhang X, Li M, Chen Q, Ye M, Liang
W, Ding L, Cai H, Fu D and Lv Z: miR-26a and its target CKS2
modulate cell growth and tumorigenesis of papillary thyroid
carcinoma. PLoS One. 8:e675912013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gao J, Li L, Wu M, Liu M and Xie X, Guo J,
Tang H and Xie X: miR-26a inhibits proliferation and migration of
breast cancer through repression of MCL-1. PLoS One. 8:e651382013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qian X, Zhao P, Li W, Shi ZM, Wang L, Xu
Q, Wang M, Liu N, Liu LZ and Jiang BH: MicroRNA-26a promotes tumor
growth and angiogenesis in glioma by directly targeting prohibitin.
CNS Neurosci Ther. 19:804–812. 2013.PubMed/NCBI
|
|
25
|
de Graauw M, van Miltenburg MH, Schmidt
MK, Pont C, Lalai R, Kartopawiro J, Pardali E, Le Dévédec SE, Smit
VT, van der Wal A, et al: Annexin A1 regulates TGF-beta signaling
and promotes metastasis formation of basal-like breast cancer
cells. Proc Natl Acad Sci USA. 107:pp. 6340–6345. 2010; View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Suh YE, Raulf N, Gäken J, Lawler K, Urbano
TG, Bullenkamp J, Gobeil S, Huot J, Odell E and Tavassoli M:
MicroRNA-196a promotes an oncogenic effect in head and neck cancer
cells by suppressing annexin A1 and enhancing radioresistance. Int
J Cancer. 137:1021–1034. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Han G, Tian Y, Duan B, Sheng H, Gao H and
Huang J: Association of nuclear annexin A1 with prognosis of
patients with esophageal squamous cell carcinoma. Int J Clin Exp
Pathol. 7:751–759. 2014.PubMed/NCBI
|
|
28
|
Geary LA, Nash KA, Adisetiyo H, Liang M,
Liao CP, Jeong JH, Zandi E and Roy-Burman P: CAF-secreted annexin
A1 induces prostate cancer cells to gain stem cell-like features.
Mol Cancer Res. 12:607–621. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cheng SX, Tu Y and Zhang S: FoxM1 promotes
glioma cells progression by up-regulating anxa1 expression. PLoS
One. 8:e723762013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Queiroz CJ, Nakata CM, Solito E and Damazo
A: Relationship between HPV and the biomarkers annexin A1 and p53
in oropharyngeal cancer. Infect Agent Cancer. 9:132014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fang Y, Guan X, Cai T, Long J, Wang H, Xie
X and Zhang Y: Knockdown of ANXA1 supresses the biological behavior
of human NSCLC cells in vitro. Mol Med Rep. 13:3858–3866. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Icli B, Wara AK, Moslehi J, Sun X, Plovie
E, Cahill M, Marchini JF, Schissler A, Padera RF, Shi J, et al:
MicroRNA-26a regulates pathological and physiological angiogenesis
by targeting BMP/SMAD1 signaling. Circ Res. 113:1231–1241. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu L, Lu J, Zhang B, Liu X, Wang L, Li SY,
Peng XH, Xu X, Tian WD and Li XP: miR-26a inhibits invasion and
metastasis of nasopharyngeal cancer by targeting EZH2. Oncol Lett.
5:1223–1228. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Deng M, Tang HL, Lu XH, Liu MY, Lu XM, Gu
YX, Liu JF and He ZM: miR-26a suppresses tumor growth and
metastasis by targeting FGF9 in gastric cancer. PLoS One.
8:e726622013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tan S, Ding K, Li R, Zhang W, Li G, Kong
X, Qian P, Lobie PE and Zhu T: Identification of miR-26 as a key
mediator of estrogen stimulated cell proliferation by targeting
CHD1, GREB1 and KPNA2. Breast Cancer Res. 16:R402014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang J, Han C and Wu T: MicroRNA-26a
promotes cholangiocarcinoma growth by activating β-catenin.
Gastroenterology. 143:246–256 e248. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ghanbari R, Mosakhani N, Asadi J, Nouraee
N, Mowla SJ, Yazdani Y, Mohamadkhani A, Poustchi H, Knuutila S and
Malekzadeh R: Downregulation of plasma MiR-142-3p and MiR-26a-5p in
patients with colorectal carcinoma. Iran J Cancer Prev.
8:e23292015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhuang C, Jiang W, Huang D, Xu L, Yang Q,
Zheng L, Wang X and Hu L: Serum miR-21, miR-26a and miR-101 as
potential biomarkers of hepatocellular carcinoma. Clin Res Hepatol
Gastroenterol. 40:386–396. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ydy LR, do Espirito Santo GF, de Menezes
I, Martins MS, Ignotti E and Damazo AS: Study of the annexin A1 and
its associations with carcinoembryonic antigen and mismatch repair
proteins in colorectal cancer. J Gastrointest Cancer. 47:61–68.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sobral-Leite M, Wesseling J, Smit VT,
Nevanlinna H, van Miltenburg MH, Sanders J, Hofland I, Blows FM,
Coulson P, Patrycja G, et al: Annexin A1 expression in a pooled
breast cancer series: Association with tumor subtypes and
prognosis. BMC Med. 13:1562015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lin Y, Lin G, Fang W, Zhu H and Chu K:
Increased expression of annexin A1 predicts poor prognosis in human
hepatocellular carcinoma and enhances cell malignant phenotype. Med
Oncol. 31:3272014. View Article : Google Scholar : PubMed/NCBI
|